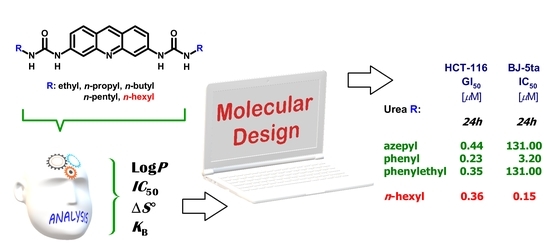
Synthesis of Novel Biologically Active Proflavine Ureas Designed on the Basis of Predicted Entropy Changes
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Remarks
3.2. Synthesis
3.2.1. Synthesis of 3,6-Diisothiocyanatoacridine (9)
3.2.2. General Protocol for the Synthesis of Thioureas 10a–10k
3.2.3. General Protocol for the Synthesis of the Ureas 11a–11l
3.3. Biology
3.3.1. Cell Line
3.3.2. Agilent xCELLigence Real-Time Cell Analysis
Statistical Analysis
3.3.3. Screening of Anticancer Activity-NCI-60 Panels
3.4. Molecular Modeling
3.4.1. Normal Mode Analysis
3.4.2. Calculation of ΔS° Value
3.4.3. Calculation of LogP Value
3.4.4. Calculation of Score Value
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Gensicka-Kowalewska, M.; Cholewiński, G.; Dzierzbicka, K. Recent developments in the synthesis and biological activity of acridine/acridone analogues. RSC Adv. 2017, 7, 15776–15804. [Google Scholar] [CrossRef] [Green Version]
- Demeunynck, M. Antitumour acridines. Expert Opin. Ther. Patents 2004, 14, 55–70. [Google Scholar] [CrossRef]
- Lerman, L.S. The structure of the DNA-acridine complex. Proc. Natl. Acad. Sci. USA 1963, 49, 94–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, A. Crystal structures of acridines complexed with nucleic acids. Curr. Med. Chem. 2002, 9, 1667–1675. [Google Scholar] [CrossRef] [PubMed]
- Belmont, P.; Dorange, I. Acridine/acridone: A simple scaffold with a wide range of application in oncology. Expert Opin. Ther. Patents 2008, 18, 1211–1224. [Google Scholar] [CrossRef]
- Blackburn, G.M.; Gait, M.J.; Loakes, D.; Williams, D.M. Nucleic Acids in Chemistry and Biology, 3rd ed.; The Royal Society of Chemistry: London, UK, 2006. [Google Scholar]
- Belmont, P.; Bosson, J.; Godet, T.; Tiano, M. Acridine and acridone derivatives, anticancer properties and synthetic methods: Where are we now? Anti-Cancer Agent. Med. Chem. 2007, 7, 139–169. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.J.; Gowan, S.M.; Kelland, L.R.; Neidle, S. Human telomerase inhibition by substituted acridine derivatives. Bioorg. Med. Chem. Lett. 1999, 9, 2463–2468. [Google Scholar] [CrossRef]
- Harrison, R.J.; Cuesta, J.; Chessari, G.; Read, M.A.; Basra, S.K.; Reszka, A.P.; Morrell, J.; Gowan, S.M.; Incles, C.M.; Tanious, F.A.; et al. Trisubstituted acridine derivatives as potent and selective telomerase inhibitors. J. Med. Chem. 2003, 46, 4463–4476. [Google Scholar] [CrossRef] [PubMed]
- Hamissa, M.F.; Niederhafner, P.; Šafařík, M.; Telus, M.; Kolářová, L.; Koutná, L.; Šestáková, H.; Souček, R.; Šebestík, J. Total synthesis of inubosin B. Tetrahedron Lett. 2020, 61, 152641. [Google Scholar] [CrossRef]
- Gatasheh, M.K.; Kannan, S.; Hemalatha, K.; Imrana, N. Proflavine an acridine DNA intercalating agent and strong antimicrobial possessing potential properties of carcinogen. Karbala Int. J. Mod. Sci. 2017, 3, 272–278. [Google Scholar] [CrossRef]
- Wainwright, M. Acridine—A neglected antibacterial chromophore. J. Antimicrob. Chemoth. 2001, 47, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Valdés, A.F.-C. Acridine and acridinones: Old and new structures with antimalarial activity. Open Med. Chem. J. 2011, 5, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Kožurková, M.; Sabolová, D.; Janovec, L.; Mikeš, J.; Koval’, J.; Ungvarský, J.; Štefanišinová, M.; Fedoročko, P.; Kristian, P.; Imrich, J. Cytotoxic activity of proflavine diureas: Synthesis, antitumor, evaluation and DNA binding properties of 1′,1″-(acridin-3,6-diyl)-3′,3″-dialkyldiureas. Bioorg. Med. Chem. 2008, 16, 3976–3984. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- ACD. ChemSketch Package 2020.2.0. Available online: www.acdlabs.com (accessed on 27 July 2021).
- Stewart, J.J.P. Stewart Computational Chemistry. Available online: http://openmopac.net (accessed on 27 July 2021).
- Molinspiration Cheminformatics Free Web Services. Slovensky Grob, Slovakia. Available online: https://www.molinspiration.com (accessed on 27 July 2021).
- Allouche, A.-R. Gabedit—A graphical user interface for computational chemistry softwares. J. Comput. Chem. 2011, 32, 174–182. [Google Scholar] [CrossRef] [PubMed]
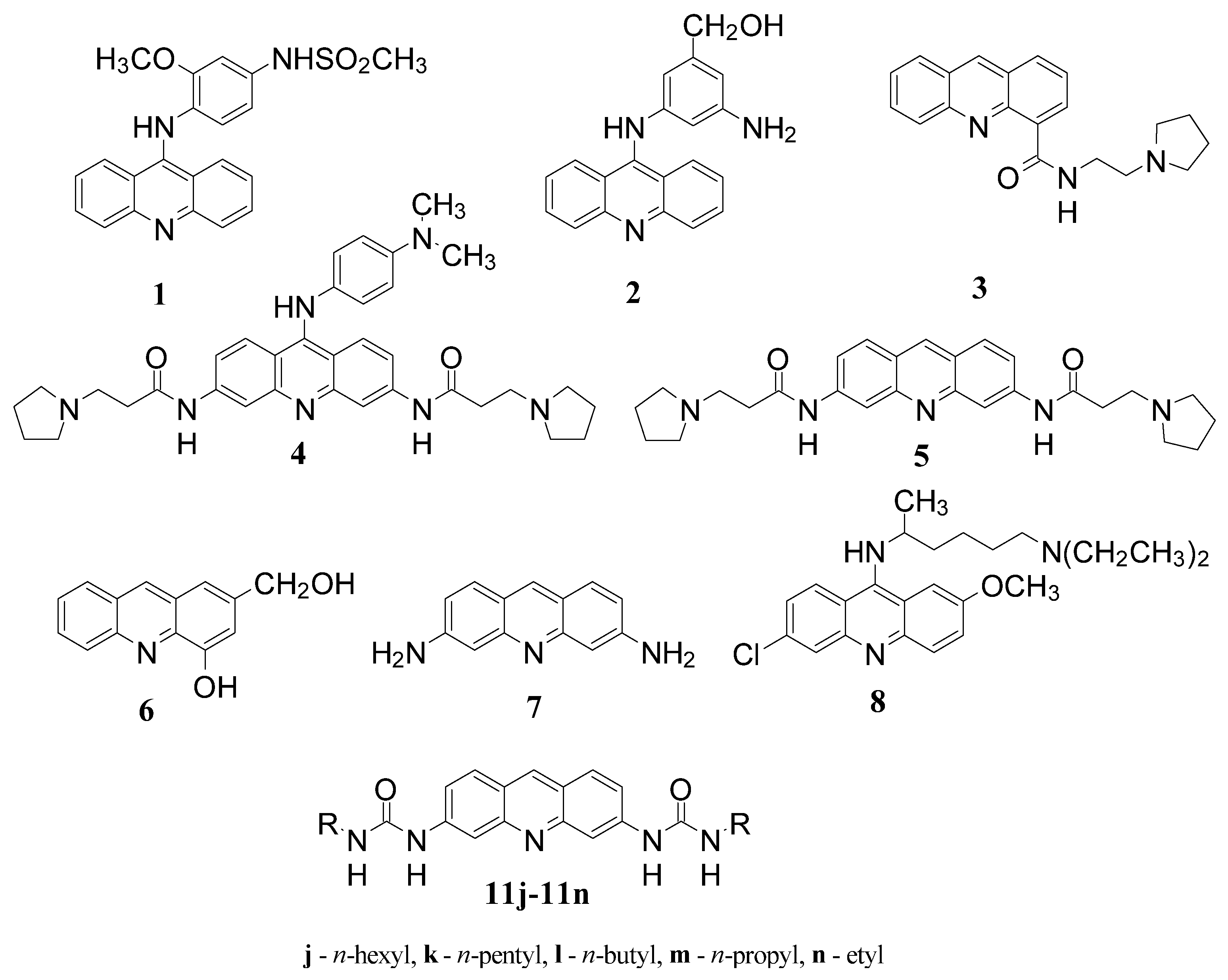
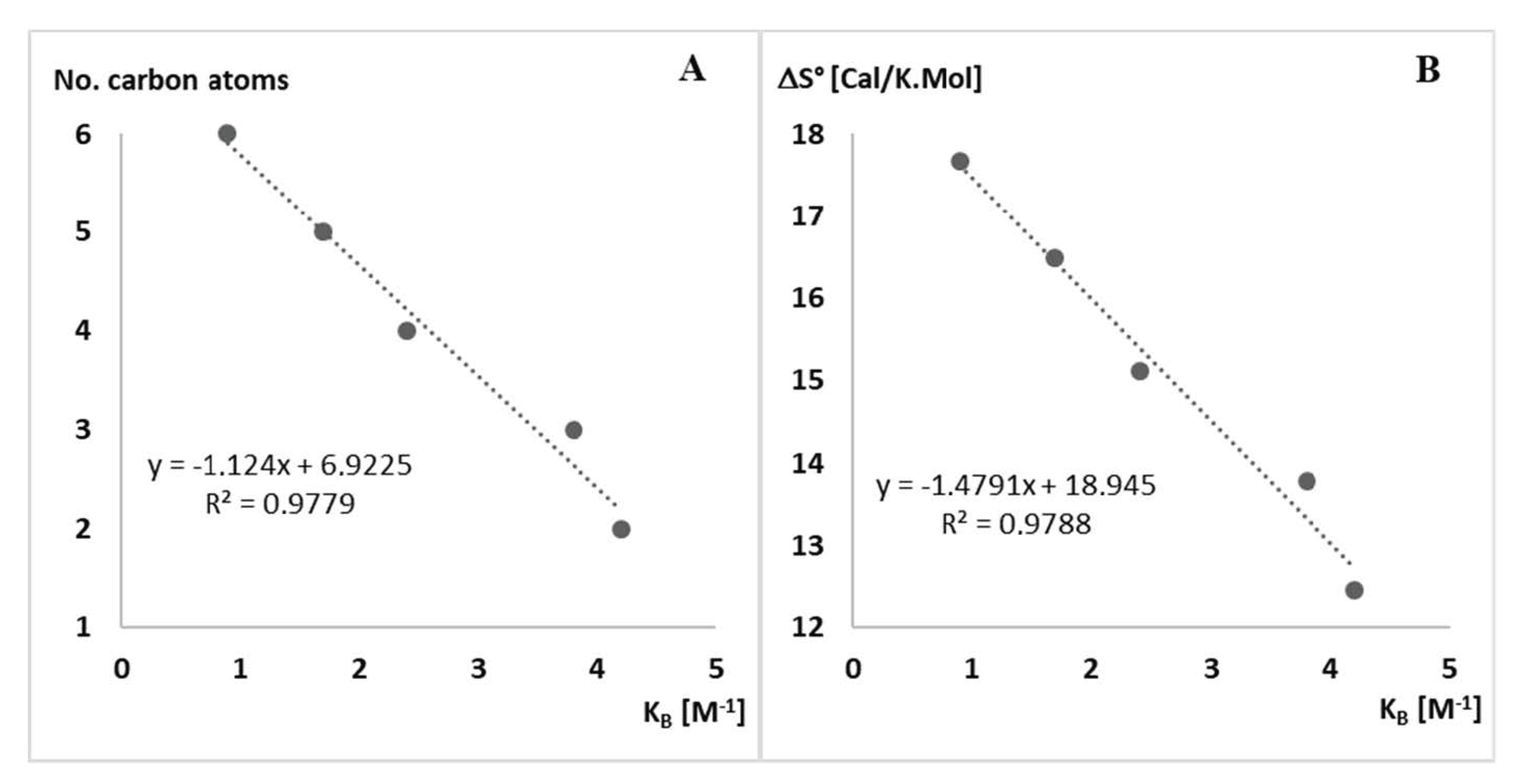
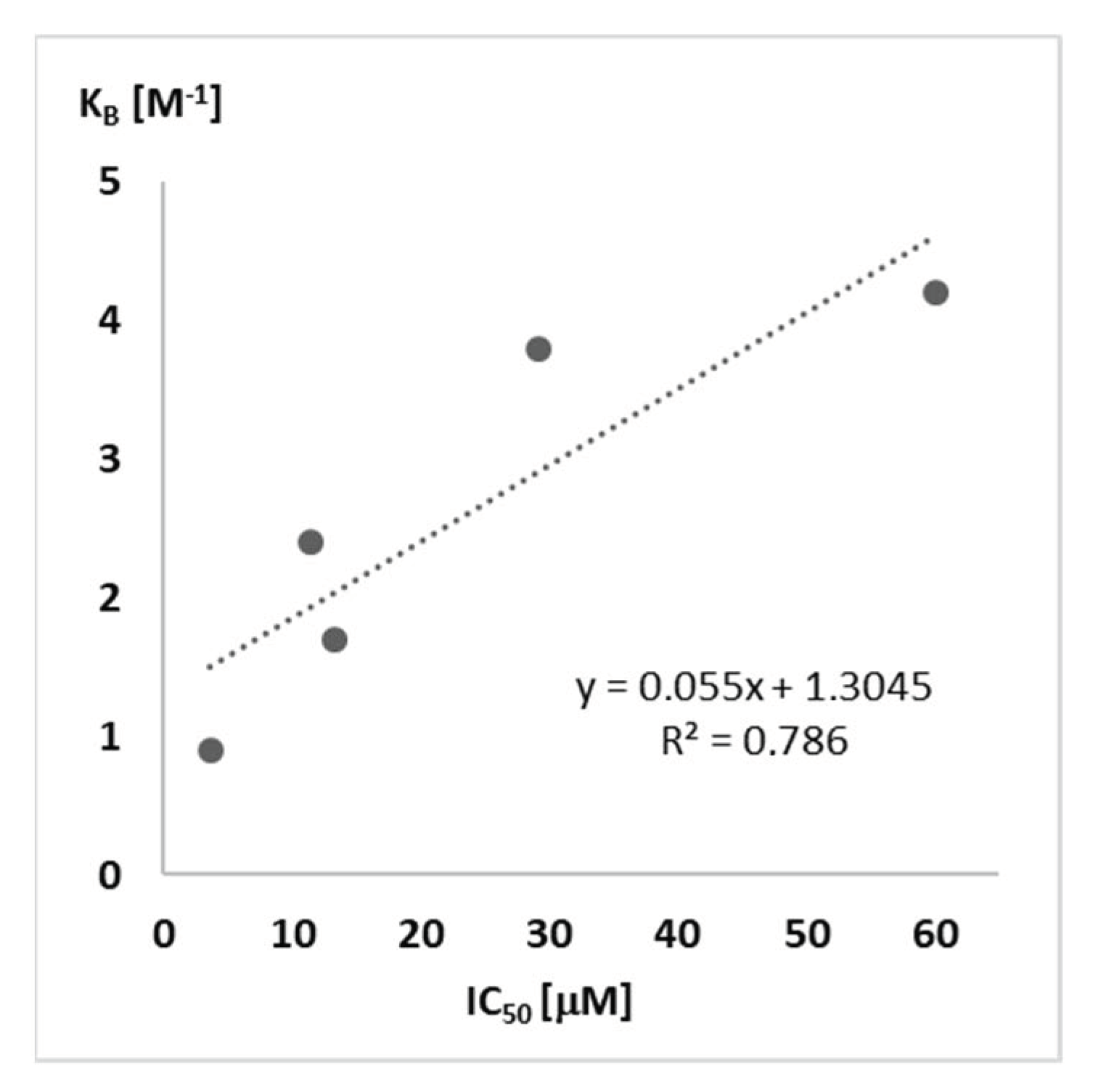
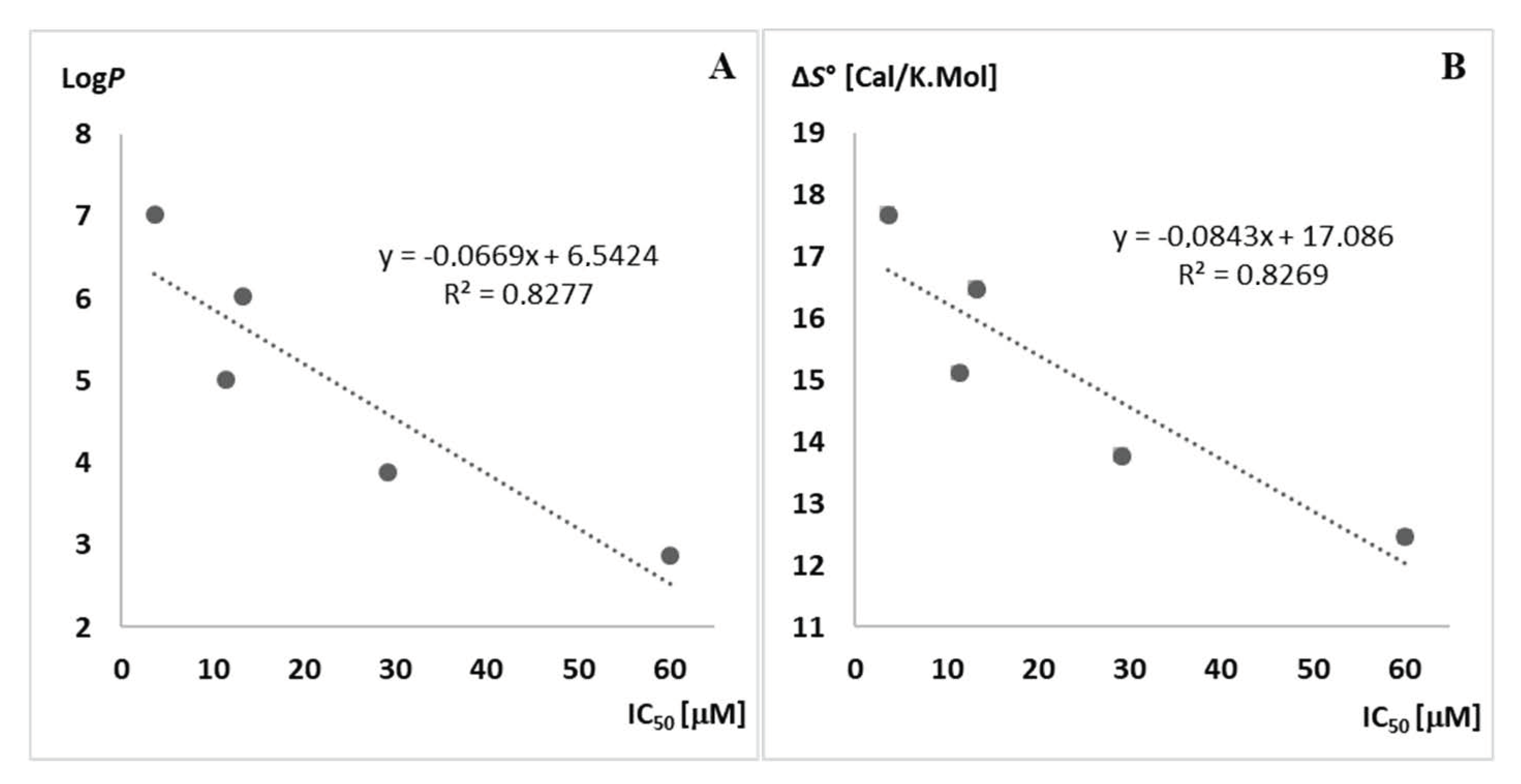

| R– a | Urea a | S°300Kb | S°340Kc | ΔS° d | LogP e | IC50 f | KB g |
|---|---|---|---|---|---|---|---|
| (Cal/K.Mol) | (Cal/K.Mol) | (Cal/K.Mol) | (μM) | (105) | |||
| n-Hexyl | 11j | 235.004 | 252.682 | 17.67 | 7.03 | 3.7 | 0.9 |
| n-Pentyl | 11k | 215.063 | 231.541 | 16.47 | 6.02 | 13.3 | 1.7 |
| n-Butyl | 11l | 197.441 | 212.554 | 15.11 | 5.01 | 11.5 | 2.4 |
| n-Propyl | 11m | 188.083 | 201.859 | 13.77 | 3.89 | 29.2 | 3.8 |
| Ethyl | 11n | 172.036 | 184.490 | 12.45 | 2.88 | 60.1 | 4.2 |
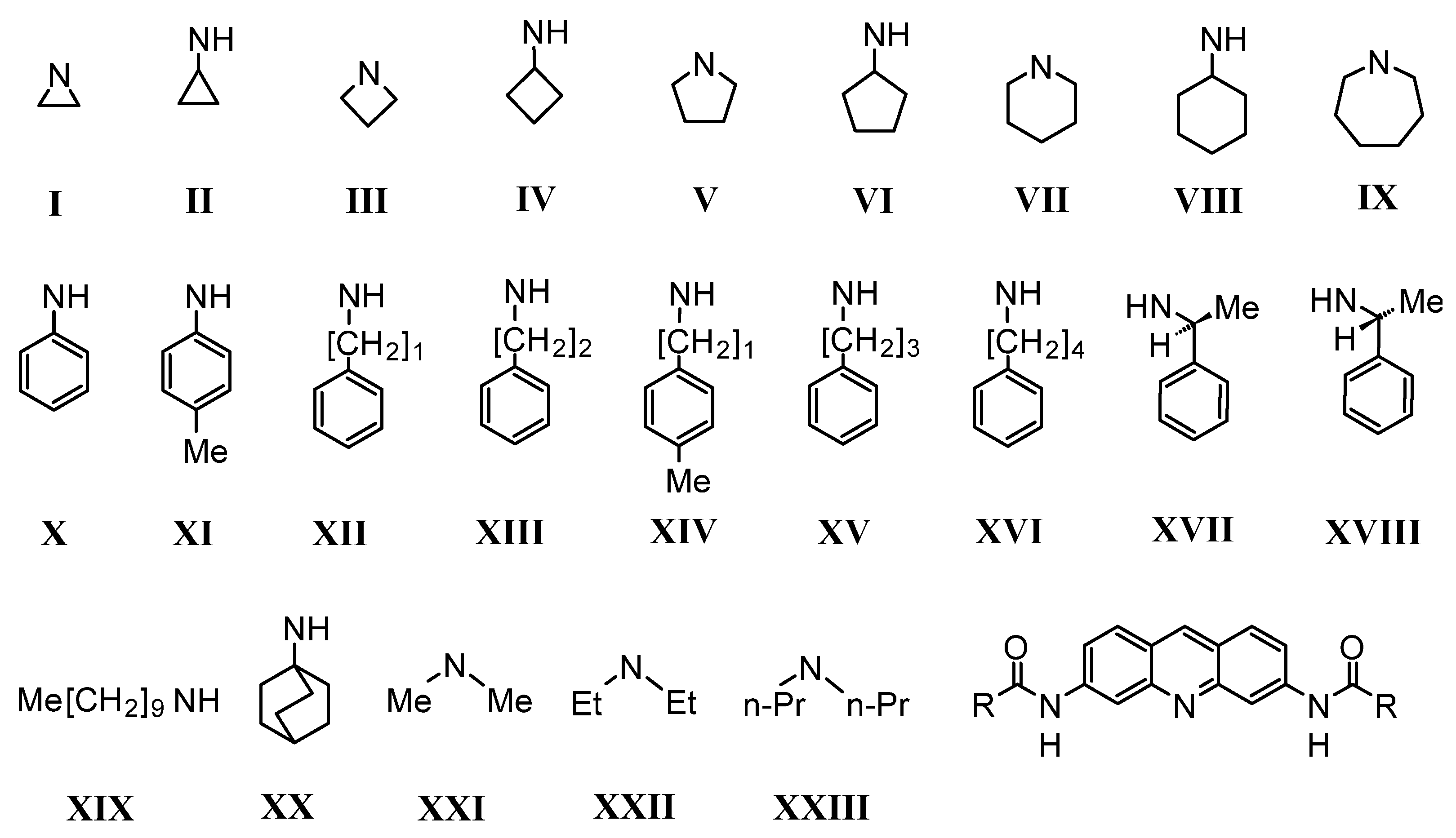
| R– a | Model a | S°300K b | S°340K c | ΔS° d | LogP e | Score f |
|---|---|---|---|---|---|---|
| (Cal/K.Mol) | (Cal/K.Mol) | (Cal/K.Mol) | ||||
| Aziridine | I | 153.9 | 165.7 | 11.4 | 2.35 | 13.75 |
| N,N’-Dimethylamine | XXI | 171.9 | 184.5 | 12.5 | 1.84 | 14.34 |
| Azetidine | III | 163.9 | 176.4 | 12.6 | 2.89 | 15.49 |
| Cyklopropylamine | II | 176.9 | 189.8 | 12.9 | 2.86 | 15.76 |
| Pyrolidine | V | 184.1 | 197.8 | 13.7 | 3.43 | 17.13 |
| Cyklobutylamine | IV | 186.1 | 200.1 | 14 | 3.45 | 17.45 |
| N,N’-Diethylamine | XXII | 196.6 | 211.9 | 15.1 | 4.13 | 19.23 |
| Piperidine | VII | 184.6 | 199.4 | 14.8 | 4.44 | 19.24 |
| Cyklopentylamine | VI | 195.6 | 210.7 | 15.1 | 4.93 | 20.03 |
| Aniline | X | 177.7 | 192.2 | 14.5 | 5.53 | 20.03 |
| Benzylamine | XII | 178.1 | 193.3 | 15.3 | 4.93 | 20.23 |
| Azepane | IX | 194.6 | 210.6 | 16 | 5.45 | 21.45 |
| Cyklohexylamine | VIII | 210.2 | 226.4 | 16.3 | 5.94 | 22.24 |
| 4-Methylbenzylamine | XIV | 240.4 | 223.0 | 17.4 | 5.38 | 22.78 |
| 4-Methylaniline | XI | 205.5 | 221.8 | 16.36 | 6.42 | 22.78 |
| Phenylethylamine | XIII | 211.6 | 228.8 | 17.2 | 5.74 | 22.94 |
| (R/S)-Methylethylamine | XVII/XVIII | 227.6 | 245.2 | 17.6 | 6.05 | 23.65 |
| N,N’-Dipropylamine | XXIII | 239.3 | 257.2 | 17.9 | 6.14 | 24.04 |
| Phenylpropylamine | XV | 230.2 | 248.6 | 18.48 | 6.78 | 25.26 |
| Phenylbutylamine | XVI | 235.5 | 255.2 | 19.73 | 7.32 | 27.05 |
| Adamantylamine | XX | 210.2 | 229.3 | 19.2 | 7.98 | 27.18 |
| n-Dekylamine | XIX | 283.5 | 306.3 | 22.8 | 11.07 | 33.87 |
| HL-60 | NCI-H522 | HCT-116 | U251 | LOXIMVI | OVCAR-8 | RXF393 | DU-145 | HS578T | |
|---|---|---|---|---|---|---|---|---|---|
| GI50 (μM) | |||||||||
| Hexyl-11j | 0.26 | 0.32 | 0.36 | 0.65 | 0.22 | 0.45 | 1.41 | 2.00 | 0.63 |
| Azepyl-11b | 0.23 | 1.58 | 0.44 | 0.54 | 1.41 | 1.70 | 0.22 | 1.74 | 0.78 |
| Phenyl-11c | 1.20 | 1.58 | 0.23 | 0.98 | 0.47 | 1.15 | 1.48 | 2.40 | 1.51 |
| Phenylethyl-11f | 0.26 | 0.32 | 0.35 | 0.74 | 0.20 | 0.81 | 1.32 | 1.23 | 0.28 |
| Fluorouracil | 4.57 | 11.48 | 0.49 | 2.24 | 0.49 | 2.40 | 2.88 | 0.63 | 14.13 |
| Cisplatin | 6.61 | 9.12 | 17.78 | 11.22 | 6.61 | 28.18 | 20.42 | 6.76 | 23.44 |
| Amsacrine | 0.02 | 0.89 | 0.50 | 0.21 | 0.15 | 0.83 | 2.24 | nd * | nd * |
| Doxorubicin | 0.08 | 0.03 | 0.03 | 0.03 | 0.03 | 0.07 | 0.08 | 0.07 | 0.13 |
| Cancer Cell Line | Hexyl-11j | Azepyl-11b | Phenyl-11c | Phenylethyl-11f |
|---|---|---|---|---|
| Log10 GI50 (M) | ||||
| CCRF-CEM | −6.56 | −6.44 | −6.56 | −6.52 |
| HL-60(TB) | −6.59 | −6.64 | −5.92 | −6.58 |
| K-562 | −6.49 | −6.46 | −6.44 | −6.41 |
| MOLT-4 | −6.67 | −6.28 | −6.25 | −6.59 |
| RPMI-8226 | −5.79 | −6.5 | −5.27 | −5.65 |
| SR | −6.58 | −6.53 | −6.54 | −6.41 |
| COLO205 | −5.85 | −5.87 | −6.24 | −6.5 |
| HCC-2998 | −5.84 | −5.73 | −6.25 | −6.35 |
| HCT-116 | −6.44 | −6.36 | −6.63 | −6.46 |
| HCT-15 | −4.00 | −5.94 | −4.43 | −4.00 |
| HT29 | −5.65 | −6.37 | −5.72 | −5.86 |
| KM12 | −5.71 | −5.72 | −5.83 | −5.79 |
| SW-620 | −6.25 | −5.97 | −6.48 | −6.46 |
| Average | −6.03 | −6.22 | −6.04 | −6.12 |
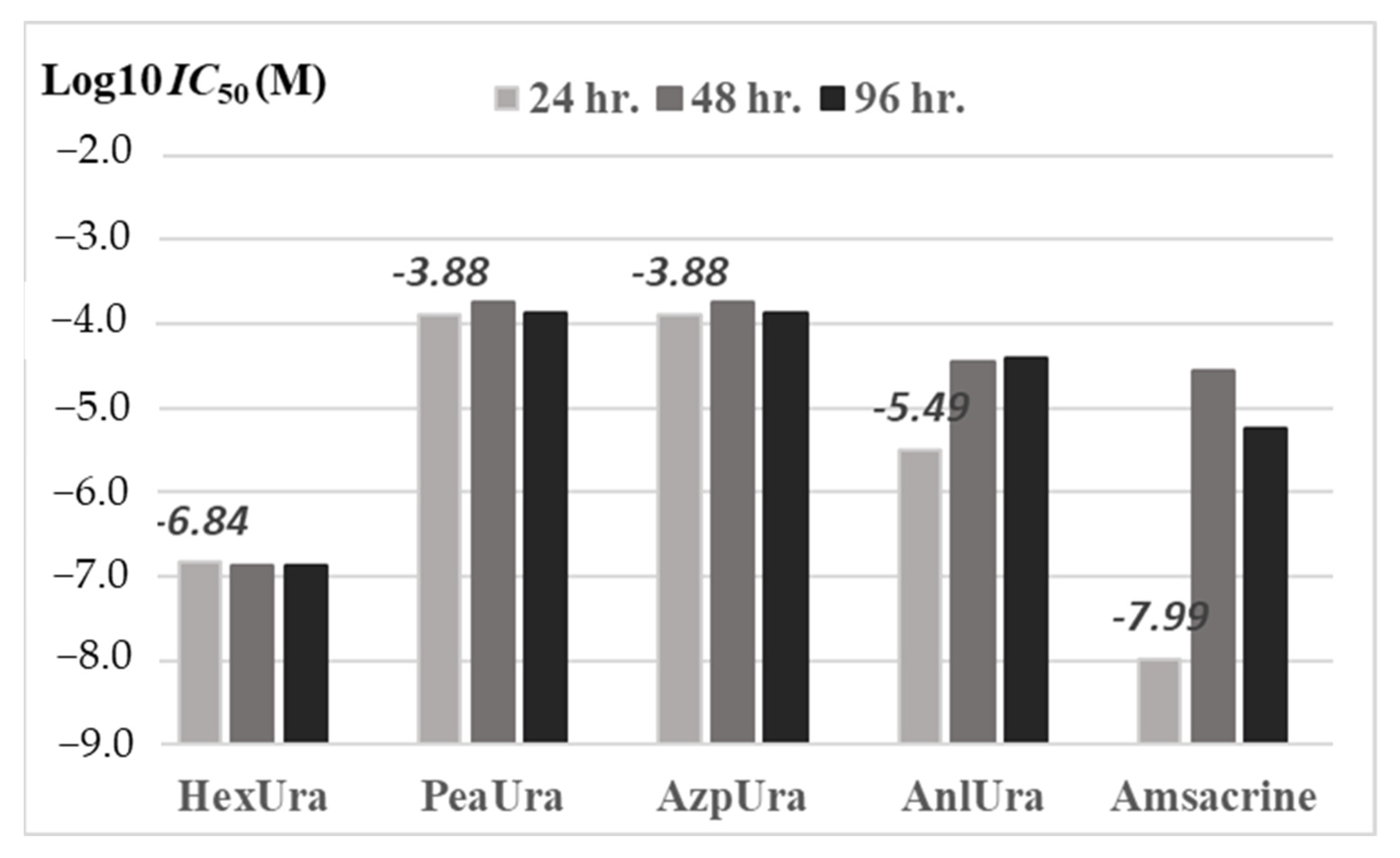
| Urea | IC50 (μM) | ||
|---|---|---|---|
| 24 h | 48 h | 96 h | |
| Hexyl-11j | 0.15 | 0.13 | 0.13 |
| Phenylethyl-11f | 131.00 | 180.00 | 137.00 |
| Azepyl-11b | 131.00 | 180.00 | 137.00 |
| Phenyl-11c | 3.20 | 34.80 | 38.50 |
| Amsacrine | 0.01 | 27.70 | 5.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janovec, L.; Kovacova, E.; Semelakova, M.; Kvakova, M.; Kupka, D.; Jager, D.; Kozurkova, M. Synthesis of Novel Biologically Active Proflavine Ureas Designed on the Basis of Predicted Entropy Changes. Molecules 2021, 26, 4860. https://doi.org/10.3390/molecules26164860
Janovec L, Kovacova E, Semelakova M, Kvakova M, Kupka D, Jager D, Kozurkova M. Synthesis of Novel Biologically Active Proflavine Ureas Designed on the Basis of Predicted Entropy Changes. Molecules. 2021; 26(16):4860. https://doi.org/10.3390/molecules26164860
Chicago/Turabian StyleJanovec, Ladislav, Eva Kovacova, Martina Semelakova, Monika Kvakova, Daniel Kupka, David Jager, and Maria Kozurkova. 2021. "Synthesis of Novel Biologically Active Proflavine Ureas Designed on the Basis of Predicted Entropy Changes" Molecules 26, no. 16: 4860. https://doi.org/10.3390/molecules26164860
APA StyleJanovec, L., Kovacova, E., Semelakova, M., Kvakova, M., Kupka, D., Jager, D., & Kozurkova, M. (2021). Synthesis of Novel Biologically Active Proflavine Ureas Designed on the Basis of Predicted Entropy Changes. Molecules, 26(16), 4860. https://doi.org/10.3390/molecules26164860