Novel Perbutyrylated Glucose Derivatives of (–)-Epigallocatechin-3-Gallate Inhibit Cancer Cells Proliferation by Decreasing Phosphorylation of the EGFR: Synthesis, Cytotoxicity, and Molecular Docking
Abstract
:1. Introduction
2. Results and Discussion
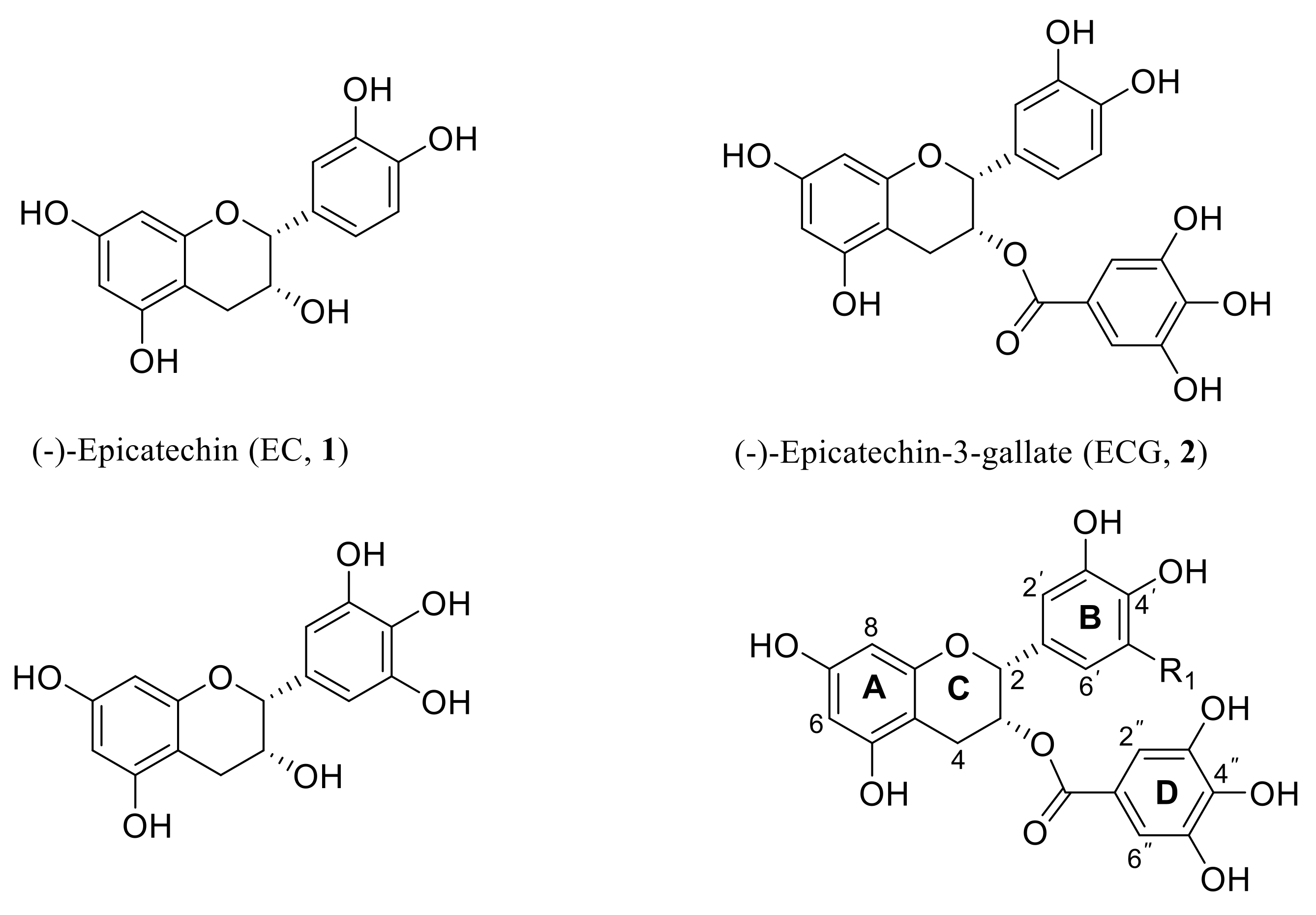
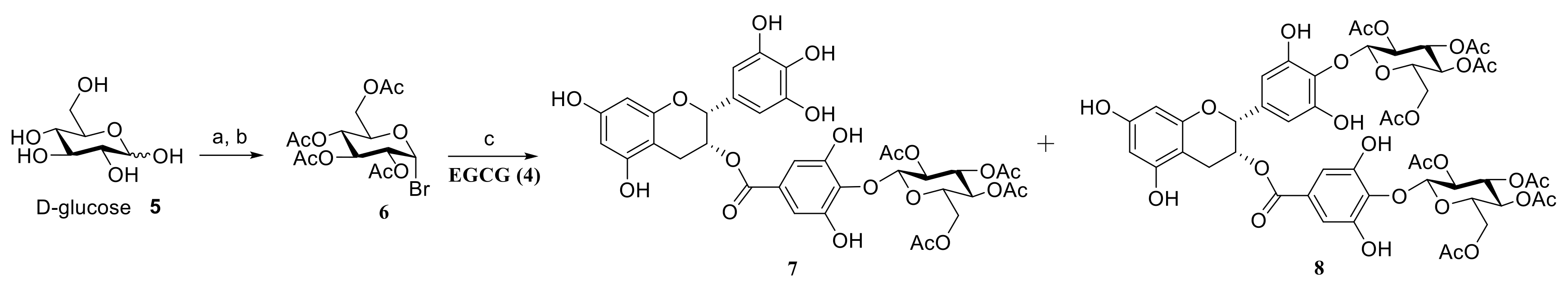
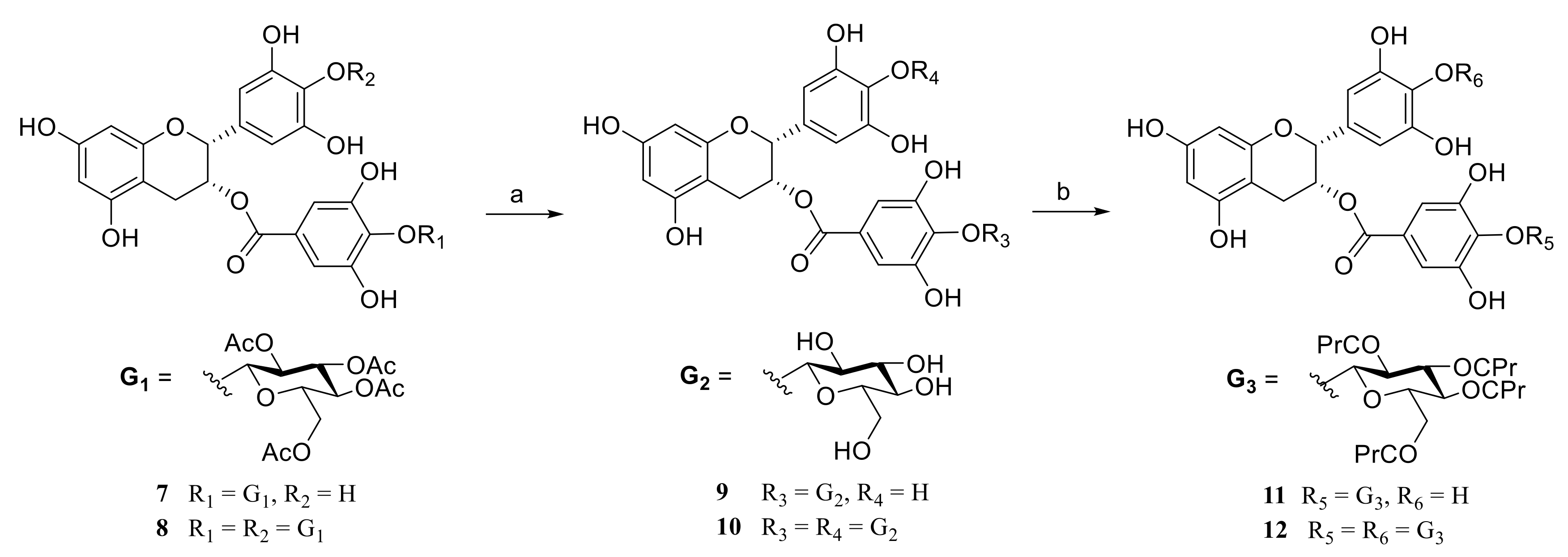
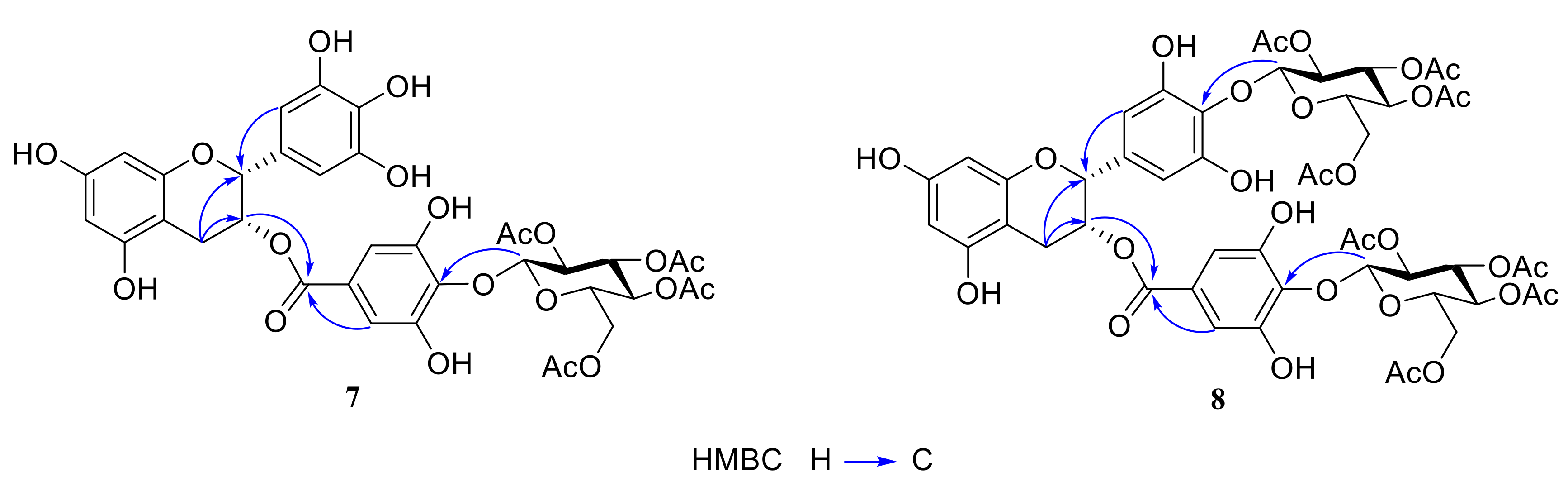
2.1. Chemistry
2.2. Evaluation of Anticancer Activity
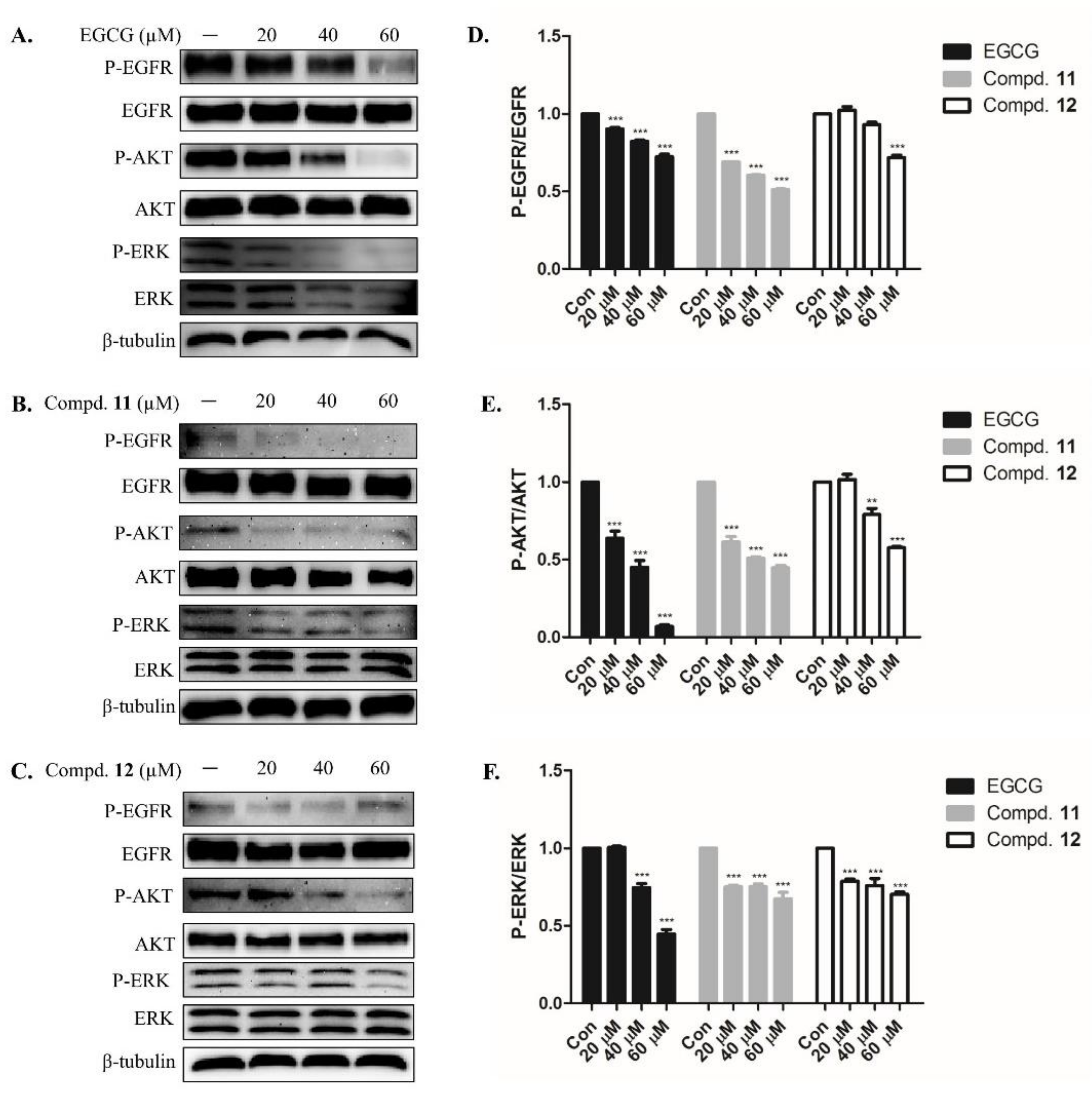
2.3. EGCG Derivatives Inhibited Phosphorylation of EGFR and Downstream Signaling Protein in A-549 Cells
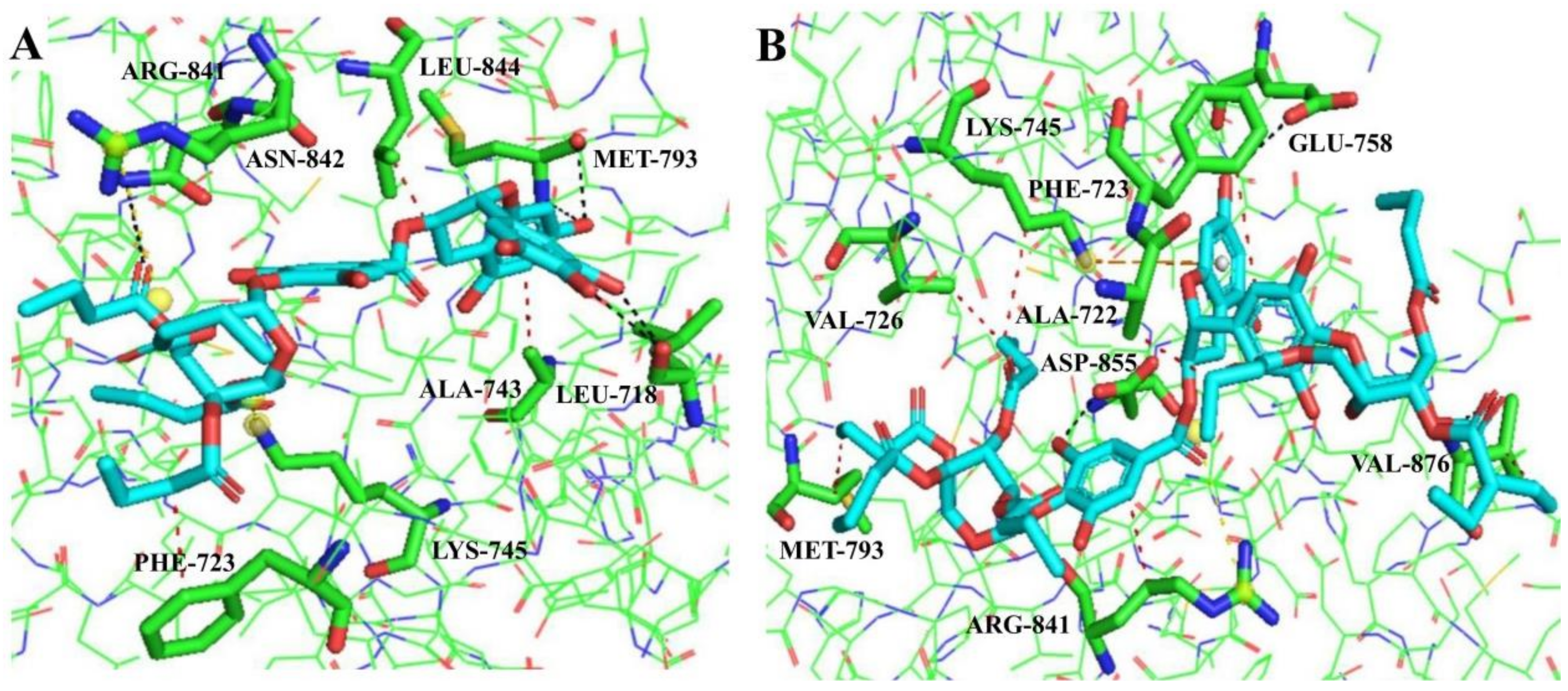
2.4. Molecular Docking Analysis
3. Experimental
3.1. Materials and Methods
3.2. Chemistry
3.3. Cytotoxicity Assay
3.4. Western Blotting
3.5. Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Torr, L.A.; Siegel, R.L.; Jemal, A. Lung cancer statistics. Adv. Exp. Med. Biol. 2016, 893, 1–19. [Google Scholar]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–536. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortettieulent, J.; Jemal, A. 2012 Global cancer statistics. CA Cancer J. Clin. 2012, 65, 87–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, I.; Morise, M.; Miyazawa, A.; Kodama, Y.; Tamiya, Y.; Gen, S.; Matsui, A.; Hase, T.; Hashimoto, N.; Sato, M.; et al. Potential benefits of bevacizumab combined with platinum-based chemotherapy in advanced non-small-cell lung cancer patients with EGFR mutation. Clin. Lung Cancer 2020, 21, 273–280. [Google Scholar] [CrossRef]
- Wang, J.; Sun, P.Y.; Wang, Q.; Zhang, P.; Wang, Y.N.; Zi, C.T.; Wang, X.J.; Sheng, J. (-)-Epigallocatechin-3-gallate derivatives combined with cisplatin exhibit synergistic inhibitory effects on non-small-cell lung cancer cells. Cancer Cell Int. 2019, 19, 1–16. [Google Scholar] [CrossRef]
- Sharma, S.V.; Bell, D.W.; Settleman, J. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 2007, 7, 169–181. [Google Scholar] [CrossRef]
- Wu, C.H.; Coumar, M.S.; Chu, C.Y.; Lin, W.H.; Chen, Y.R.; Chen, C.T.; Shiao, H.Y.; Rafi, S.; Wang, S.Y.; Hsu, H.; et al. Design and synthesis of tetrahydropyridothieno[2,3-d]pyrimidine scaffold based pidermal growth factor receptor (EGFR) kinase inhibitors: The role of side chain chirality and Michael acceptor group for maximal potency. J. Med. Chem. 2010, 53, 7316–7326. [Google Scholar] [CrossRef]
- Janku, F.; Stewart, D.J.; Kurzrock, R. Targeted therapy in nonsmall-cell lung cancer-is it becoming a reality. Nat. Rev. Clin. Oncol. 2011, 8, 384. [Google Scholar] [CrossRef] [Green Version]
- Cohen, M.H.; Williams, G.A.; Sridhara, R.; Chen, G.; Pazdur, R. FDA drug approval summary: Gefitinib (ZD1839) (Iressa) tablets. Oncologist 2003, 8, 303–306. [Google Scholar] [CrossRef]
- Gridelli, C.; Rossi, A.; Carbone, D.P.; Guarize, J.; Karachaliou, N.; Mok, T.; Petrella, F.; Spaggiari, L.; Rosell, R. Non-small-cell lung cancer. Nat. Rev. 2015, 1, 1–16. [Google Scholar] [CrossRef]
- Cohen, M.H.; Johnson, J.R.; Chen, Y.F.; Sridhara, R.; Pazdur, R. FDA drug approval summary: Erlotinib (Tarceva) tablets. Oncologist 2005, 10, 461–466. [Google Scholar] [CrossRef]
- Shi, B.Y.; Wang, Z.H.; Wang, X.J.; Sheng, J.; Zi, C.T. Recent Advances on Small-Molecule Epidermal Growth Factor Receptor Inhibitors. Med. Res. 2020, 4, 200011. [Google Scholar] [CrossRef]
- Wang, S.H.; Cang, S.D.; Liu, D.L. Third-generation inhibitors targeting EGFR T790M mutation in advanced non-small cell lung cancer. J. Hematol. Oncol. 2016, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Ambrogio, L.; Shimamura, T.; Kubo, S.; Takahashi, M.; Chirieac, L.R.; Padera, R.F.; Shapiro, G.I.; Baum, A.; Himmelsbach, F.; et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene 2008, 27, 4702–4711. [Google Scholar] [CrossRef] [Green Version]
- Odogwu, L.; Mathieu, L.; Goldberg, K.B.; Blumenthal, M.B.; Larkins, E.; Fiero, M.H.; Rodriguez, L.; Bijwaard, K.; Lee, E.Y.; Philip, R.; et al. FDA benefit-risk assessment of osimertinib for the treatment of metastatic non-small cell lung cancer harboring epidermal growth factor receptor T790M mutation. Oncologist 2018, 23, 353–359. [Google Scholar] [CrossRef] [Green Version]
- Hinterding, K.; Knebel, A.; Herrlich, P.; Waldmann, H. Synthesis and biological evaluation of aeroplysinin analogues: A new class of receptor tyrosine kinase inhibitors. Bioorg. Med. Chem. 1998, 6, 1153–1162. [Google Scholar] [CrossRef]
- Singh, F.; Gao, D.; Lebwohl, M.G.; Wei, H. Shikonin modulates cell proliferation by inhibiting epidermal growth factor receptor signaling in human epidermoid carcinoma cells. Cancer Lett. 2003, 200, 115–121. [Google Scholar] [CrossRef]
- Siddiqui, I.A.; Asim, M.; Hafeez, B.B.; Adhami, V.M.; Tarapore, R.S.; Mukhtar, H. Green tea polyphenol EGCG blunts androgen receptor function in prostate cancer. FASEB J. 2010, 25, 1198–1207. [Google Scholar] [CrossRef] [Green Version]
- Bettuzzi, S.; Brausi, M.; Rizzi, F.; Castagnetti, G.; Peracchia, G.; Corti, A. Chemoprevention of human prostate cancer by oral administration of green tea catechins in volunteers with high-grade prostate intraepithelial neoplasia: A preliminary report from a one-year proof-of-principle study. Cancer Res. 2006, 66, 1234–1240. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.S.; Wang, X.; Lu, G.; Picinich, S.C. Cancer prevention by tea: Animal studies, molecular mechanisms and human relevance. Nat. Rev. Cancer 2009, 9, 429–439. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Afaq, F.; Saleem, M.; Ahmad, N.; Mukhtar, H. Targeting multiple signaling pathways by green tea polyphenol (-)-epigallocatechin-3-gallate. Cancer Res. 2006, 66, 2500–2505. [Google Scholar] [CrossRef] [Green Version]
- Cabrera, C.; Gimenez, R.; Lopez, M.C. Determination of tea components with antioxidant activity. J. Agric. Food Chem. 2003, 51, 4427–4435. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.D.; Kim, M.S.; Shin, B.A.; Chay, K.O.; Ahn, B.W.; Liu, W.; Ducans, C.D.; Gallick, G.E.; Ellis, L.M. EGCG, a major component of green tea, inhibits tumour growth by inhibiting VEGF induction in human colon carcinoma cells. Br. J. Cancer 2001, 84, 844–850. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Tang, H.; Hou, B.; Zhang, P.; Wang, Q.; Zhang, B.L.; Huang, Y.W.; Wang, Y.; Xiang, Z.M.; Zi, C.T.; et al. Synthesis, antioxidant activity, and density functional theory study of catechin derivatives. RSC Adv. 2017, 7, 54136–54141. [Google Scholar] [CrossRef] [Green Version]
- Mukhtar, H.; Ahmad, N. Tea polyphenols: Prevention of cancer and optimizing health. Am. J. Clin. Nutr. 2000, 71, 1698–1702. [Google Scholar] [CrossRef] [Green Version]
- Rady, I.; Mohamed, H.; Rady, M.; Siddiqui, I.A.; Mukhtara, H. Cancer preventive and therapeutic effects of EGCG, the major polyphenol in green tea. Egypt. J. Basic Appl. Sci. 2018, 5, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.N.; Shankar, S.; Srivastava, R.K. Green tea catechin, epigallocatechin-3-gallate (EGCG): Mechanisms, perspectives and clinical applications. Biochem. Pharmacol. 2011, 82, 1807–1821. [Google Scholar] [CrossRef] [Green Version]
- Tyagia, N.; Dec, R.; Begun, J.; Popat, A. Cancer therapeutics with epigallocatechin-3-gallate encapsulated in biopolymeric nanoparticles. Int. J. Pharm. 2017, 518, 220–227. [Google Scholar] [CrossRef] [Green Version]
- Sajadimajd, S.; Bahramsoltani, R.; Iranpanah, A.; Patra, J.K.; Das, G.; Gaouda, S.; Rahimi, R.; Rezaeiamiri, E.; Cao, H.; Giampieri, F.; et al. Advances on Natural Polyphenols as Anticancer Agents for Skin Cancer. Pharmacol. Res. 2020, 151, 104584. [Google Scholar] [CrossRef]
- Zhang, W.J.; Zhang, W.J.; Sun, L.L.; Xiang, L.M.; Lai, X.F.; Li, Q.H.; Sun, S.L. The effects and mechanisms of epigallocatechin-3-gallate on reversing multidrug resistance in cancer. Trends Food Sci. Technol. 2019, 93, 221–233. [Google Scholar] [CrossRef]
- Ma, Y.C.; Li, C.; Gao, F.; Xu, Y.; Jiang, Z.B.; Liu, J.X.; Jin, L.Y. Epigallocatechin gallate inhibits the growth of human lung cancer by directly targeting the EGFR signaling pathway. Oncol. Rep. 2014, 31, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.B.; Hu, C.P.; Xiong, Z.; Yang, H.P.; Li, Y.Y. Treatment with EGCG in NSCLC leads to decreasing interstitial fluid pressure and hypoxia to improve chemotherapy efficacy through rebalance of Ang-1 and Ang-2. Chin. J. Nat. Med. 2013, 11, 245–253. [Google Scholar] [CrossRef]
- Liang, Y.C.; Lin-Shiau, S.Y.; Chen, C.F.; Lin, J.K. Suppression of extracellular signals and cell proliferation through EGF receptor binding by (-)-epigallocatechin gallate in human A431 epidermoid carcinoma cells. J. Cell Biochem. 1997, 67, 55–65. [Google Scholar] [CrossRef]
- Hong, J.; Lu, H.; Meng, X.; Ryu, J.H.; Hara, Y.; Yang, C.S. Stability, cellular uptake, biotransformation, and efflux of tea polyphenol (-)-epigallocatechin-3-gallate in HT-29 human colon adenocarcinoma cells. Cancer Res. 2002, 62, 7241–7246. [Google Scholar]
- Baba, S.; Osakabe, N.; Natsume, M.; Muto, Y.; Takizawa, T.; Terao, J. In vivo comparison of the bioavailability of (+)-catechin, (-)-epicatechin and their mixture in orally administered rats. J. Nutr. 2001, 131, 2885–2891. [Google Scholar] [CrossRef] [Green Version]
- Park, K.D.; Cho, S.J. Synthesis and antimicrobial activities of 3-O-alkyl analogues of (þ)-catechin: Improvement of stability and proposed action mechanism. Eur. J. Med. Chem. 2009, 45, 1028–1033. [Google Scholar] [CrossRef]
- Landis-Piwowar, K.R.; Kuhn, D.J.; Wan, S.B.; Chen, D.; Chan, T.H.; Dou, Q.P. Evaluation of proteasome-inhibitory and apoptosis-inducing potencies of novel (-)-EGCG analogs and their prodrugs. Int. J. Mol. Med. 2005, 15, 735–742. [Google Scholar] [CrossRef]
- Qsanai, K.; Landis-Piwowar, K.R.; Dou, Q.P.; Chan, T.H. A para-amino substituent on the D-ring of green tea polyphenol epigallocatechin-3-gallate as a novel proteasome inhibitor and cancer cell apoptosis inducer. Bioorg. Med. Chem. 2007, 5, 5076–5082. [Google Scholar]
- Mori, S.; Miyake, S.; Kobe, T.; Nakaya, T.; Fuller, S.D.; Kato, N.; Kaihatsu, K. Enhanced anti-influenza A virus activity of (-)-epigallocatechin-3-O-gallate fatty acid monoester derivatives: Effect of alkyl chain length. Bioorg. Med. Chem. Lett. 2008, 18, 4249–4252. [Google Scholar] [CrossRef]
- Calvaresi, E.C.; Hergenrother, P.J. Glucose conjugation for the specific targeting and treatment of cancer. Chem. Sci. 2013, 4, 2319–2333. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.S.; Tungpradit, R.; Sinchaikul, S.; An, F.M.; Liu, D.Z.; Phutrakul, S.; Chen, S.T. Targeting the delivery of glycan-based paclitaxel prodrugs to cancer cells via glucose transporters. J. Med. Chem. 2008, 51, 7428–7441. [Google Scholar] [CrossRef]
- Arafa, H.M.M. Possible contribution of beta-glycosidases and caspases in the cytotoxicity of novel glycoconjugates in colon cancer cells. Investig. New Drugs 2010, 28, 306–317. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Hu, J.M.; Huang, Y.W.; Wu, X.Y.; Zi, C.T.; Wang, X.J.; Sheng, J. Synthesis and biological testing of novel glucosylated epigallocatechin gallate (EGCG) derivatives. Molecules 2016, 21, 620. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.J. Cellular and physiological effects of short-chain fatty acids. Mini Rev. Med. Chem. 2004, 4, 839–845. [Google Scholar] [CrossRef]
- Tamura, M.; Okai, H. Synthesis of 2-acetamido-1-N[N-(tert-butoxycarbonyl)-l-aspart-1-oyl-(l-phenylalanyl-l-serine methyl ester)-4-oyl]-2-deoxy-β-d-glucopyranosylamine and analogs. Carbohydr. Res. 1984, 133, 207–218. [Google Scholar] [CrossRef]
- Zi, C.T.; Yang, D.; Dong, F.W.; Li, G.T.; Li, Y.; Ding, Z.T.; Zhou, J.; Jiang, Z.H.; Hu, J.M. Synthesis and antitumor activity of novel per-butyrylated glycosides of podophyllotoxin and its derivatives. Bioorg. Med. Chem. 2015, 23, 1437–1446. [Google Scholar] [CrossRef]
- Malich, G.; Markovic, B.; Winder, C. The sensitivity and specificity of the MTS tetrazolium assay for detecting the in vitro cytotoxicity of 20 chemicals using human cells. Toxicology 1997, 124, 179–192. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.; Goodsell, D.; Olson, A. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]

| Compounds | IC50 (μM) | |||||
|---|---|---|---|---|---|---|
| HL-60 | SMMC-7721 | A-549 | MCF-7 | SW480 | BEAS-2B | |
| 4 | >40.0 | >40.0 | >40.0 | >40.0 | >40.0 | NT |
| 7 | 29.5 ± 0.10 | >40.0 | 38.5 ± 0.08 | >40.0 | >40.0 | NT |
| 8 | 28.2 ± 0.06 | >40.0 | 34.4 ± 0.43 | 36.1 ± 0.11 | >40.0 | NT |
| 9 | >40.0 | >40.0 | 39.6 ± 0.02 | >40.0 | >40.0 | NT |
| 10 | >40.0 | >40.0 | >40.0 | >40.0 | >40.0 | NT |
| 11 | 28.4 ± 1.00 | 37.9 ± 0.12 | 32.1 ± 0.10 | 28.7 ± 0.38 | >40.0 | >40.0 |
| 12 | 22.9 ± 0.23 | 30.4 ± 1.00 | 24.4 ± 0.31 | 26.4 ± 0.12 | 39.7 ± 0.01 | >40.0 |
| Cisplatin | 0.8 ± 0.03 | 4.7 ± 0.35 | 1.9 ± 0.43 | 14.2 + 0.93 | 10.2 ± 1.20 | 12.9 ± 0.25 |
| Molecules | Hydrophobic Interactions | Hydrogen Bonds | π-Stacking | Salt Bridges |
|---|---|---|---|---|
| 11 | PHE723, ALA743, LEU844 | LEU718, MET793 ASN842 | — | LYS745, ARG841 |
| 12 | ALA722, PHE723, VAL726, LYS745, MET793, ARG841, VAL876 | GLU758, ASP855, VAL876 | LYS745 | ARG841 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Shen, X.-J.; Su, F.-W.; Xie, Y.-R.; Wang, L.-X.; Zhang, N.; Wu, Y.-L.; Niu, Y.; Zhang, D.-Y.; Zi, C.-T.; et al. Novel Perbutyrylated Glucose Derivatives of (–)-Epigallocatechin-3-Gallate Inhibit Cancer Cells Proliferation by Decreasing Phosphorylation of the EGFR: Synthesis, Cytotoxicity, and Molecular Docking. Molecules 2021, 26, 4361. https://doi.org/10.3390/molecules26144361
Wang Y, Shen X-J, Su F-W, Xie Y-R, Wang L-X, Zhang N, Wu Y-L, Niu Y, Zhang D-Y, Zi C-T, et al. Novel Perbutyrylated Glucose Derivatives of (–)-Epigallocatechin-3-Gallate Inhibit Cancer Cells Proliferation by Decreasing Phosphorylation of the EGFR: Synthesis, Cytotoxicity, and Molecular Docking. Molecules. 2021; 26(14):4361. https://doi.org/10.3390/molecules26144361
Chicago/Turabian StyleWang, Ya, Xiao-Jing Shen, Fa-Wu Su, Yin-Rong Xie, Li-Xia Wang, Ning Zhang, Yi-Long Wu, Yun Niu, Dong-Ying Zhang, Cheng-Ting Zi, and et al. 2021. "Novel Perbutyrylated Glucose Derivatives of (–)-Epigallocatechin-3-Gallate Inhibit Cancer Cells Proliferation by Decreasing Phosphorylation of the EGFR: Synthesis, Cytotoxicity, and Molecular Docking" Molecules 26, no. 14: 4361. https://doi.org/10.3390/molecules26144361
APA StyleWang, Y., Shen, X.-J., Su, F.-W., Xie, Y.-R., Wang, L.-X., Zhang, N., Wu, Y.-L., Niu, Y., Zhang, D.-Y., Zi, C.-T., Wang, X.-J., & Sheng, J. (2021). Novel Perbutyrylated Glucose Derivatives of (–)-Epigallocatechin-3-Gallate Inhibit Cancer Cells Proliferation by Decreasing Phosphorylation of the EGFR: Synthesis, Cytotoxicity, and Molecular Docking. Molecules, 26(14), 4361. https://doi.org/10.3390/molecules26144361






