The Recognition of and Reactions to Nucleic Acid Nanoparticles by Human Immune Cells
Abstract
:1. Introduction
Nucleic Acid Nanoparticles
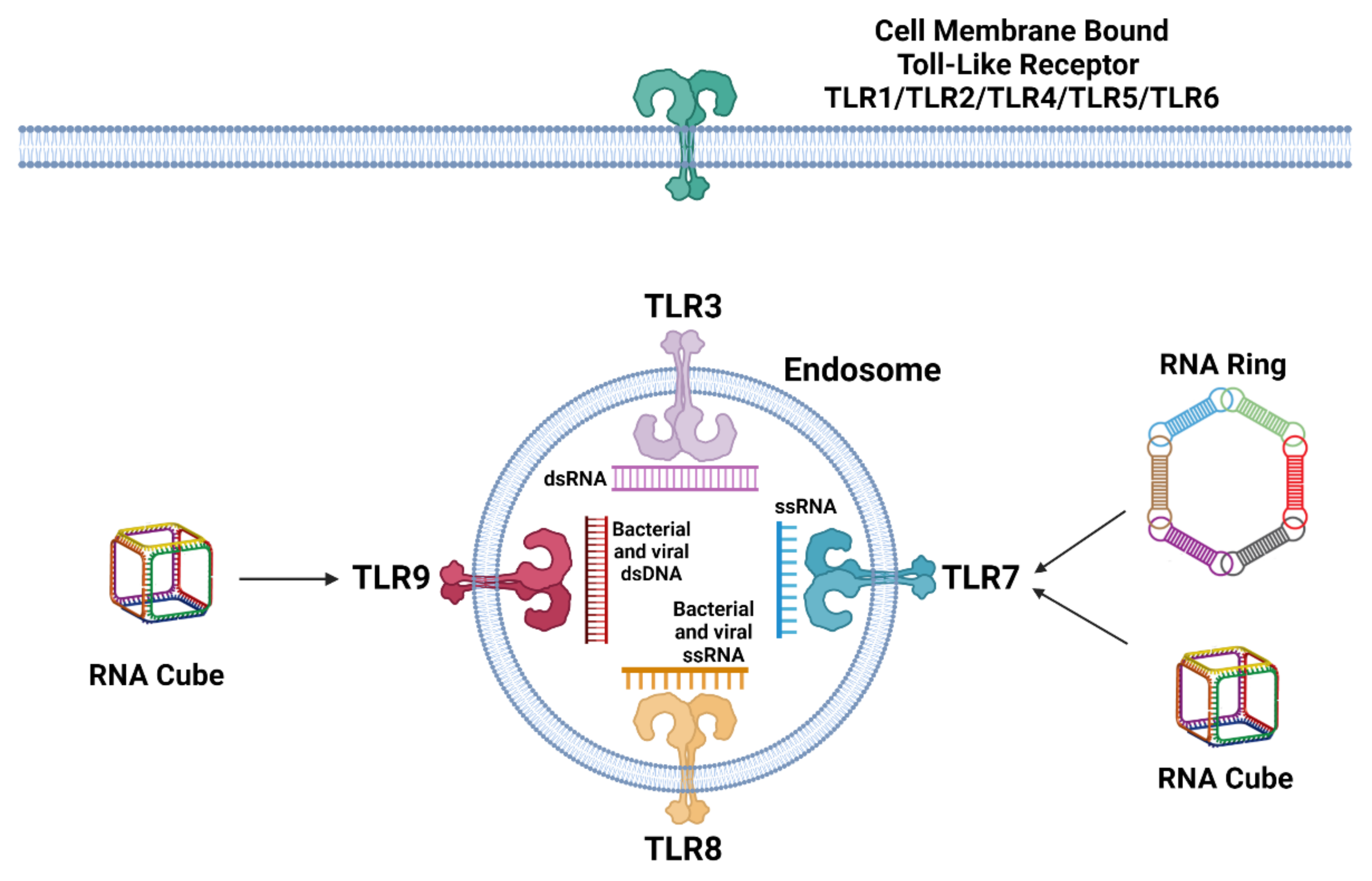
2. Recognition and Reaction of Immune Cells to Nucleic Acids
3. Recognition and Reaction of Immune Cells to NANPs
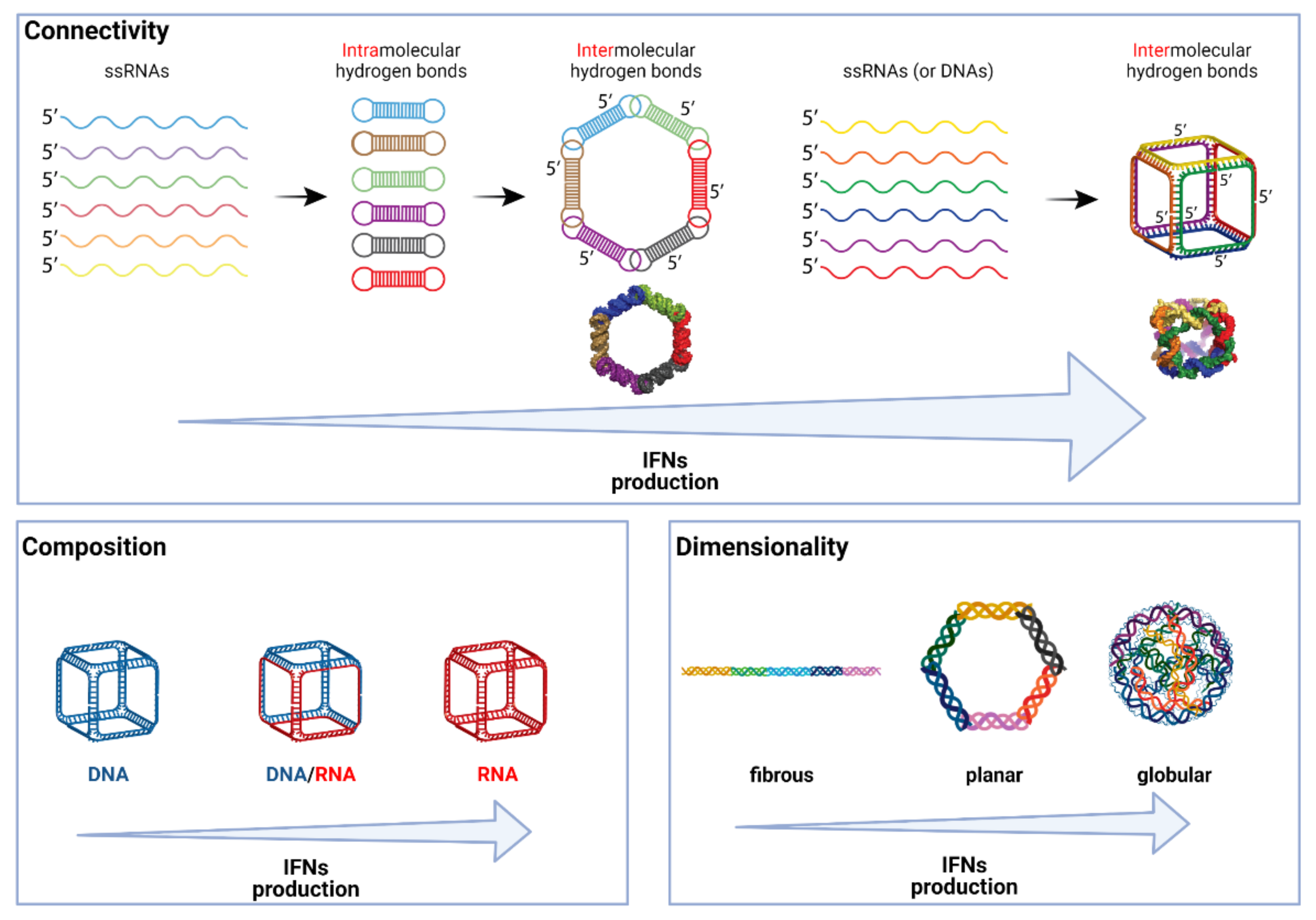
4. What Makes NANPs Immunostimulatory?
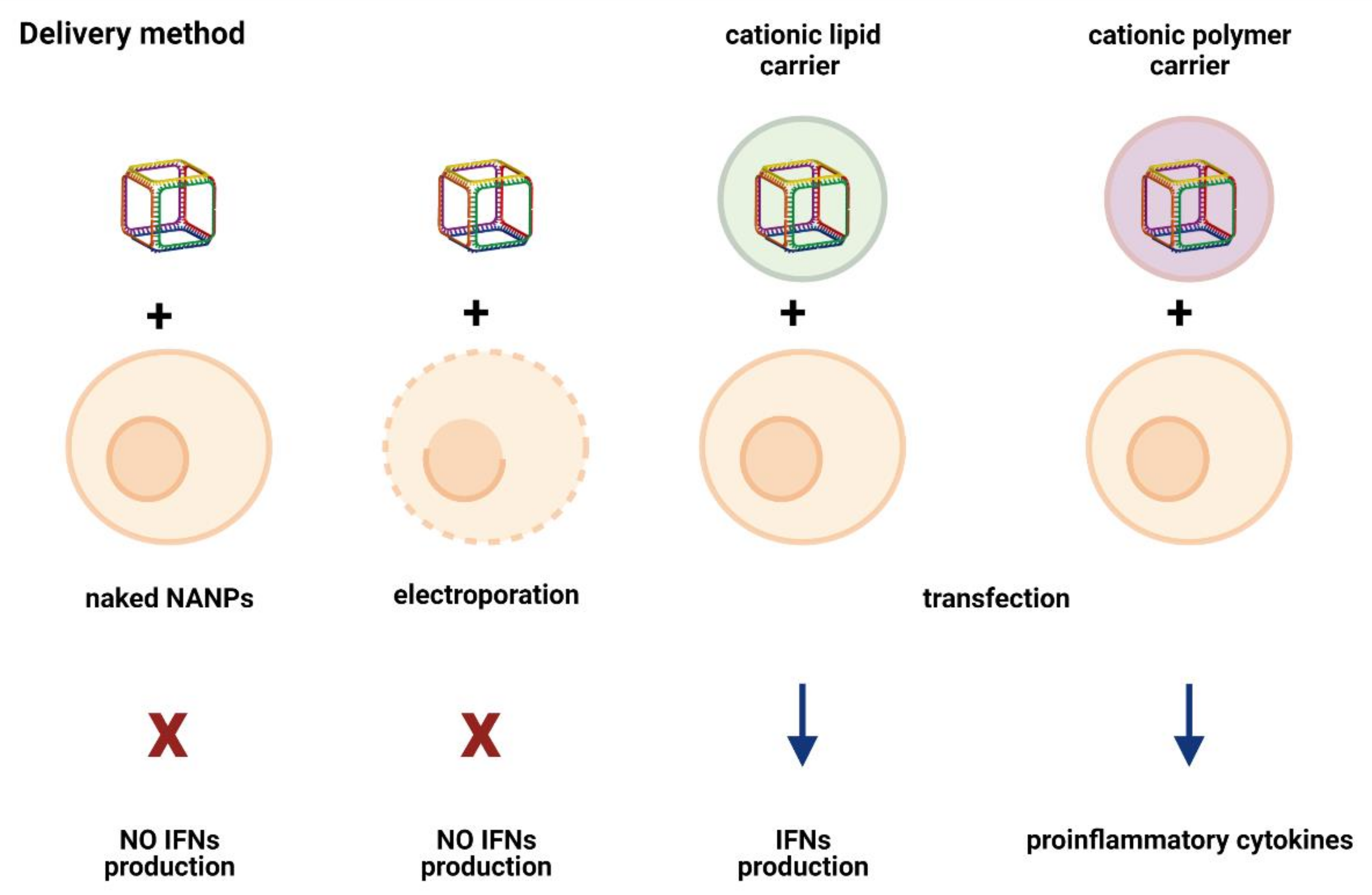
5. Delivery Method/Carrier: An Unexpected Immunomodulator
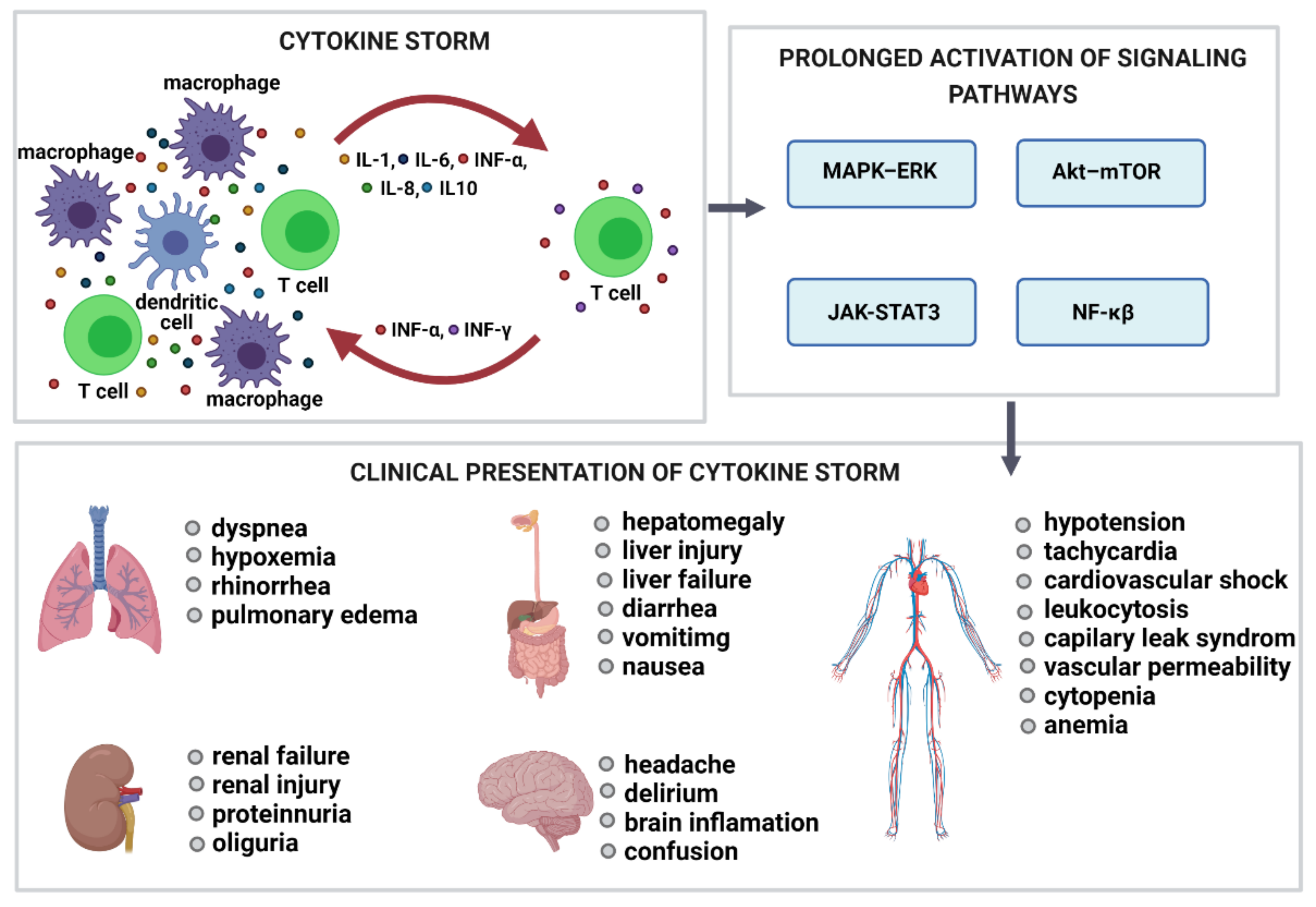
6. Complement Activation-Related Pseudoallergy (CARPA) and Cytokine Release Syndrome (CRS)
7. Conclusions
Funding
Conflicts of Interest
References
- Afonin, K.A.; Viard, M.; Koyfman, A.Y.; Martins, A.N.; Kasprzak, W.K.; Panigaj, M.; Desai, R.; Santhanam, A.; Grabow, W.W.; Jaeger, L.; et al. Multifunctional RNA nanoparticles. Nano Lett. 2014, 14, 5662–5671. [Google Scholar] [CrossRef] [Green Version]
- Khisamutdinov, E.F.; Li, H.; Jasinski, D.L.; Chen, J.; Fu, J.; Guo, P. Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic Acids Res. 2014, 42, 9996–10004. [Google Scholar] [CrossRef] [Green Version]
- Rackley, L.; Stewart, J.M.; Salotti, J.; Krokhotin, A.; Shah, A.; Halman, J.; Juneja, R.; Smollett, J.; Roark, B.; Viard, M.; et al. RNA Fibers as Optimized Nanoscaffolds for siRNA Coordination and Reduced Immunological Recognition. Adv. Funct. Mater. 2018, 28. [Google Scholar] [CrossRef]
- Avila, Y.I.; Chandler, M.; Cedrone, E.; Newton, H.S.; Richardson, M.; Xu, J.; Clogston, J.D.; Liptrott, N.J.; Afonin, K.A.; Dobrovolskaia, M.A. Induction of cytokines by nucleic acid nanoparticles (NANPs) depends on the type of delivery carrier. Molecules 2021, 26, 652. [Google Scholar] [CrossRef]
- Leontis, N.B.; Stombaugh, J.; Westhof, E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002, 30, 3497–3531. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskaia, M.A.; Afonin, K.A. Use of human peripheral blood mononuclear cells to define immunological properties of nucleic acid nanoparticles. Nat. Protoc. 2020, 15, 3678–3698. [Google Scholar] [CrossRef] [PubMed]
- Delebecque, C.J.; Lindner, A.B.; Silver, P.A.; Aldaye, F.A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 2011, 333, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Panigaj, M.; Johnson, M.B.; Ke, W.; McMillan, J.; Goncharova, E.A.; Chandler, M.; Afonin, K.A. Aptamers as modular components of therapeutic nucleic acid nanotechnology. ACS Nano 2019, 13, 12301–12321. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Zhao, Z.; Lv, Y.-F.; Huan, S.-Y.; Fu, T.; Zhang, X.-B.; Shen, G.-L.; Yu, R.-Q. DNAzyme-based biosensors and nanodevices. Chem. Commun. 2015, 51, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Franco, E. RNA nanotechnology in synthetic biology. Curr. Opin. Biotechnol. 2020, 63, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Guo, P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010, 5, 833–842. [Google Scholar] [CrossRef]
- Afonin, K.A.; Dobrovolskaia, M.A.; Church, G.; Bathe, M. Opportunities, barriers, and a strategy for overcoming translational challenges to therapeutic nucleic acid nanotechnology. ACS Nano 2020, 14, 9221–9227. [Google Scholar] [CrossRef]
- Panigaj, M.; Dobrovolskaia, M.A.; Afonin, K.A. 2021: An immunotherapy odyssey and the rise of nucleic acid nanotechnology. Nanomedicine 2021. [Google Scholar] [CrossRef]
- Johnson, M.B.; Halman, J.R.; Miller, D.K.; Cooper, J.S.; Khisamutdinov, E.F.; Marriott, I.; Afonin, K.A. The immunorecognition, subcellular compartmentalization, and physicochemical properties of nucleic acid nanoparticles can be controlled by composition modification. Nucleic Acids Res. 2020, 48, 11785–11798. [Google Scholar] [CrossRef]
- Juneja, R.; Vadarevu, H.; Halman, J.; Tarannum, M.; Rackley, L.; Dobbs, J.; Marquez, J.; Chandler, M.; Afonin, K.; Vivero-Escoto, J.L. Combination of Nucleic Acid and Mesoporous Silica Nanoparticles: Optimization and Therapeutic Performance In Vitro. ACS Appl. Mater. Interfaces 2020, 12, 38873–38886. [Google Scholar] [CrossRef]
- Jasinski, D.; Haque, F.; Binzel, D.W.; Guo, P. Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano 2017, 11, 1142–1164. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.B.; Chandler, M.; Afonin, K.A. Nucleic acid nanoparticles (NANPs) as molecular tools to direct desirable and avoid undesirable immunological effects. Adv. Drug. Deliv. Rev. 2021, 173, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.B.; Halman, J.R.; Satterwhite, E.; Zakharov, A.V.; Bui, M.N.; Benkato, K.; Goldsworthy, V.; Kim, T.; Hong, E.; Dobrovolskaia, M.A.; et al. Programmable Nucleic Acid Based Polygons with Controlled Neuroimmunomodulatory Properties for Predictive QSAR Modeling. Small 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Bui, M.; Johnson, M.; Viard, M.; Satterwhite, E.; Martins, A.; Li, Z.; Marriott, I.; Afonin, K.; Khisamutdinov, E. Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1137–1146. [Google Scholar] [CrossRef]
- Hong, E.; Halman, J.R.; Shah, A.; Cedrone, E.; Truong, N.; Afonin, K.A.; Dobrovolskaia, M.A. Toll-like receptor-mediated recognition of nucleic acid nanoparticles (NANPs) in human primary blood cells. Molecules 2019, 24, 1094. [Google Scholar] [CrossRef] [Green Version]
- Hong, E.; Halman, J.R.; Shah, A.B.; Khisamutdinov, E.F.; Dobrovolskaia, M.A.; Afonin, K.A. Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles. Nano Lett. 2018, 18, 4309–4321. [Google Scholar] [CrossRef]
- Ke, W.; Afonin, K.A. Exosomes as natural delivery carriers for programmable therapeutic nucleic acid nanoparticles (NANPs). Adv. Drug Deliv. Rev. 2021. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.; Panigaj, M.; Rolband, L.; Afonin, K. Challenges in optimizing RNA nanostructures for large-scale production and controlled therapeutic properties. Nanomedicine 2020, 15, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.; Johnson, M.; Panigaj, M.; Afonin, K. Innate immune responses triggered by nucleic acids inspire the design of immunomodulatory nucleic acid nanoparticles (NANPs). Curr. Opin. Biotechnol. 2020, 63, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Li, H.; Ma, M.; Fu, J.; Dong, Y.; Guo, P. Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles. Mol. Ther. Nucleic Acids 2017, 9, 399–408. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, T.; Akira, S. TLR signaling. Cell Death Differ. 2006, 13, 816–825. [Google Scholar] [CrossRef] [Green Version]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Deguine, J.; Barton, G.M. MyD88: A central player in innate immune signaling. F1000Prime Rep. 2014, 6, 97. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target Ther. 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Dorrington, M.G.; Fraser, I.D.C. NF-kappaB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front. Immunol. 2019, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [Green Version]
- Yang, E.; Li, M.M.H. All About the RNA: Interferon-Stimulated Genes That Interfere With Viral RNA Processes. Front. Immunol. 2020, 11, 605024. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.K.; Gack, M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016, 14, 360–373. [Google Scholar] [CrossRef]
- Unterholzner, L.; Almine, J.F. Camouflage and interception: How pathogens evade detection by intracellular nucleic acid sensors. Immunology 2019, 156, 217–227. [Google Scholar] [CrossRef]
- Canton, J.; Neculai, D.; Grinstein, S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013, 13, 621–634. [Google Scholar] [CrossRef]
- Halman, J.R.; Satterwhite, E.; Roark, B.; Chandler, M.; Viard, M.; Ivanina, A.; Bindewald, E.; Kasprzak, W.K.; Panigaj, M.; Bui, M.N.; et al. Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic Acids Res. 2017, 45, 2210–2220. [Google Scholar] [CrossRef]
- Kariko, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Morrissey, D.V.; Lockridge, J.A.; Shaw, L.; Blanchard, K.; Jensen, K.; Breen, W.; Hartsough, K.; Machemer, L.; Radka, S.; Jadhav, V.; et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005, 23, 1002–1007. [Google Scholar] [CrossRef]
- Halman, J.R.; Kim, K.T.; Gwak, S.J.; Pace, R.; Johnson, M.B.; Chandler, M.R.; Rackley, L.; Viard, M.; Marriott, I.; Lee, J.S.; et al. A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (Nanps) for controlled gene silencing, immunostimulation, and biodistribution. Nanomedicine 2020, 23, 102094. [Google Scholar] [CrossRef] [PubMed]
- Lenz, H.J. Management and preparedness for infusion and hypersensitivity reactions. Oncologist 2007, 12, 601–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szebeni, J. Complement activation-related pseudoallergy: A new class of drug-induced acute immune toxicity. Toxicology 2005, 216, 106–121. [Google Scholar] [CrossRef]
- Szebeni, J. Complement activation-related pseudoallergy caused by liposomes, micellar carriers of intravenous drugs, and radiocontrast agents. Crit. Rev. Ther. Drug Carrier Syst. 2001, 18, 567–606. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.; Abu Lila, A.S.; Shimizu, T.; Alaaeldin, E.; Hussein, A.; Sarhan, H.A.; Szebeni, J.; Ishida, T. PEGylated liposomes: Immunological responses. Sci. Technol. Adv. Mater. 2019, 20, 710–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakurai, H.; Kawabata, K.; Sakurai, F.; Nakagawa, S.; Mizuguchi, H. Innate immune response induced by gene delivery vectors. Int. J. Pharm. 2008, 354, 9–15. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bila, D.; Radwan, Y.; Dobrovolskaia, M.A.; Panigaj, M.; Afonin, K.A. The Recognition of and Reactions to Nucleic Acid Nanoparticles by Human Immune Cells. Molecules 2021, 26, 4231. https://doi.org/10.3390/molecules26144231
Bila D, Radwan Y, Dobrovolskaia MA, Panigaj M, Afonin KA. The Recognition of and Reactions to Nucleic Acid Nanoparticles by Human Immune Cells. Molecules. 2021; 26(14):4231. https://doi.org/10.3390/molecules26144231
Chicago/Turabian StyleBila, Dominika, Yasmine Radwan, Marina A. Dobrovolskaia, Martin Panigaj, and Kirill A. Afonin. 2021. "The Recognition of and Reactions to Nucleic Acid Nanoparticles by Human Immune Cells" Molecules 26, no. 14: 4231. https://doi.org/10.3390/molecules26144231
APA StyleBila, D., Radwan, Y., Dobrovolskaia, M. A., Panigaj, M., & Afonin, K. A. (2021). The Recognition of and Reactions to Nucleic Acid Nanoparticles by Human Immune Cells. Molecules, 26(14), 4231. https://doi.org/10.3390/molecules26144231






