Design, Synthesis and Biological Evaluation of Novel Pyrazolo[1,2,4]triazolopyrimidine Derivatives as Potential Anticancer Agents
Abstract
1. Introduction
2. Results
2.1. Chemistry
2.2. Biological Activity Evaluation
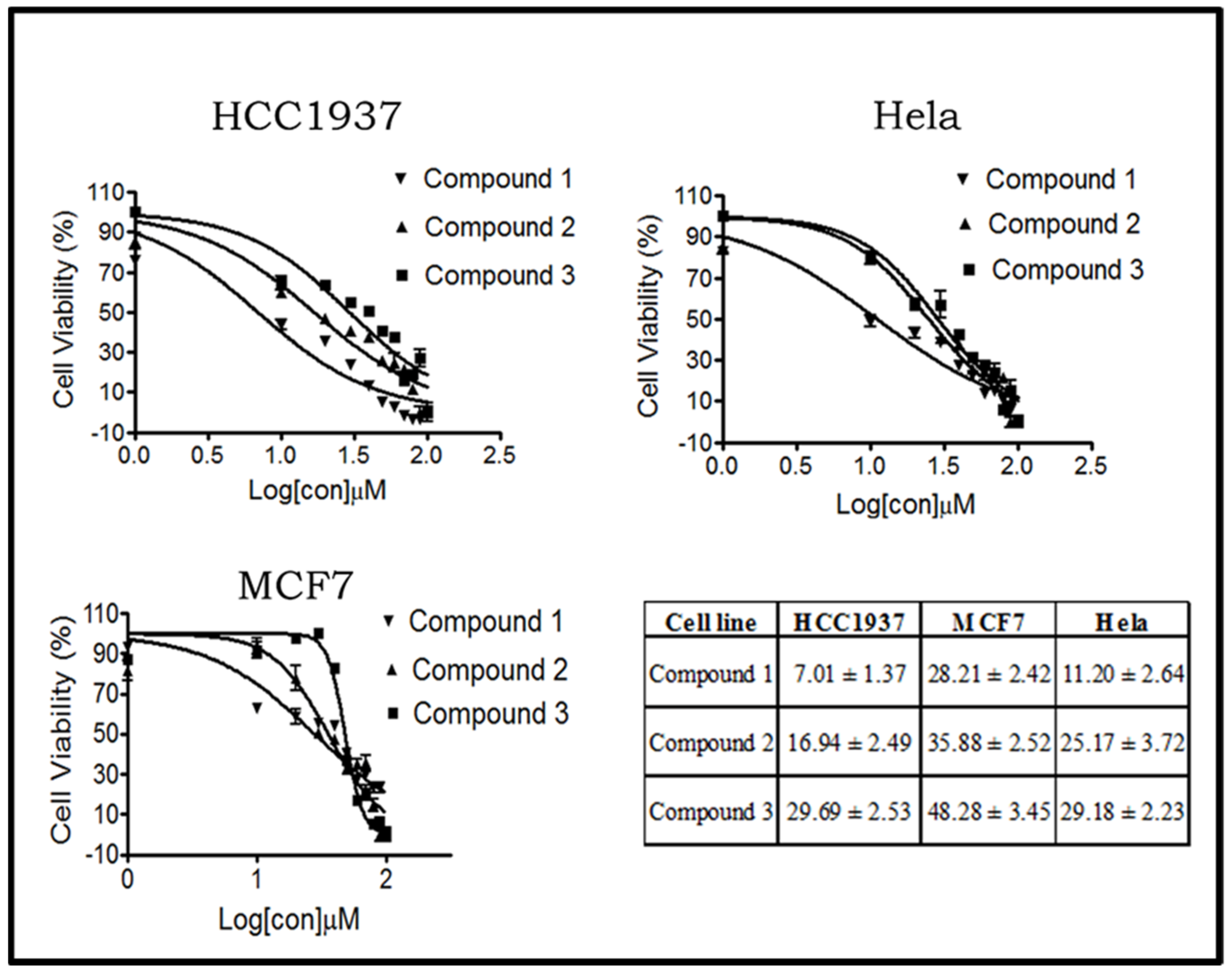
2.2.1. Compounds 1–3 Show Different Cytotoxic Activities on Human Breast and Cervical Cancer Cells
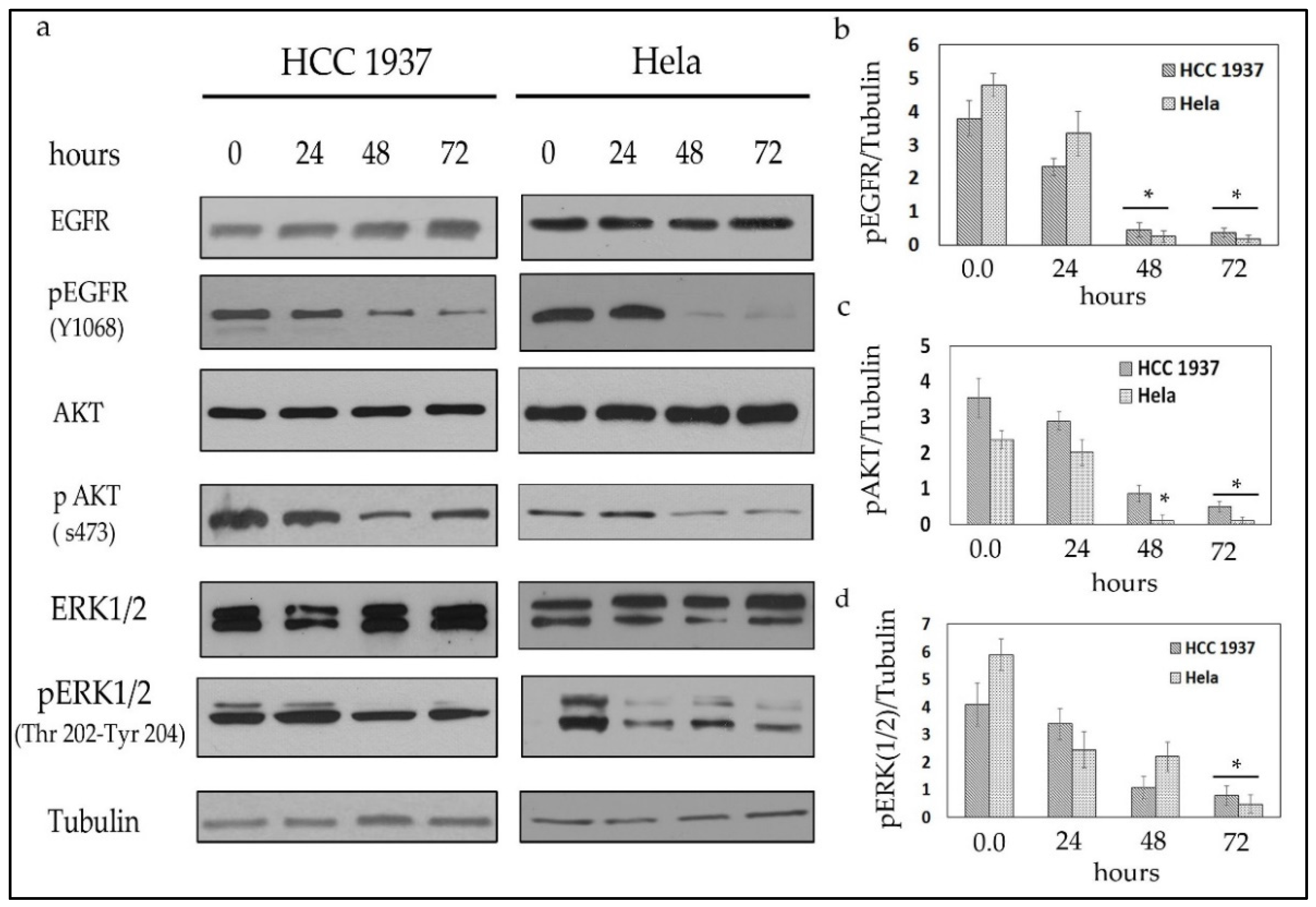
2.2.2. Compound 1 Inhibits the EGFR/AKT Pathway in Both Breast and Cervical Cancer Cells
2.2.3. Compound 1 Induces Cell Cycle Arrest and Apoptosis in Both Breast and Cervical Cancer Cells
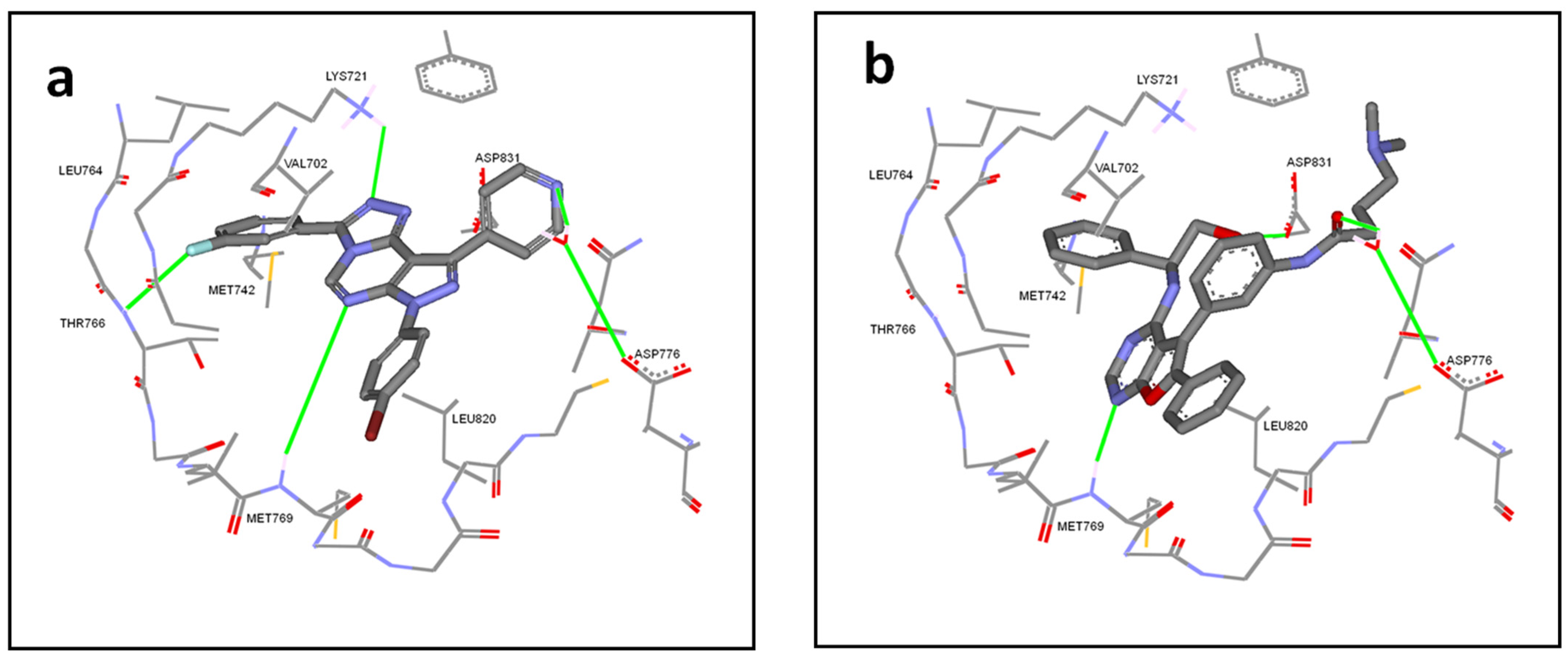
2.3. Molecular Docking Simulations
2.4. Calculation of Molecular Properties and Drug-Likeliness
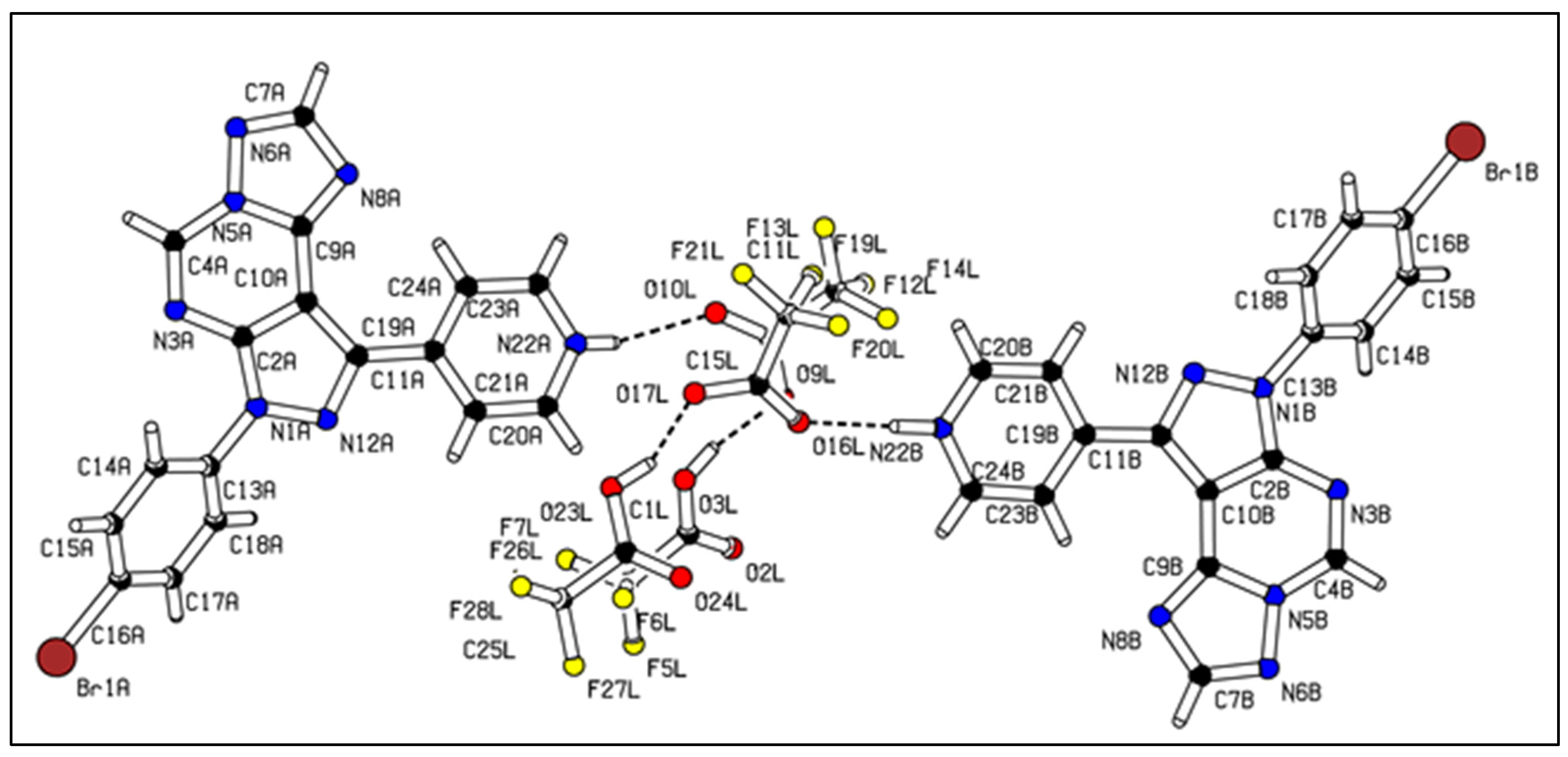
2.5. X-ray Crystallography
3. Discussion
4. Materials and Methods
4.1. Computational Methodology (Molecular Modeling)
4.1.1. Docking Experiment
4.1.2. Calculation of Physicochemical Properties
4.2. Chemistry
General Procedure of the Synthesis of Pyrazolo[1,2,4]triazolopyrimidine Derivatives 1, 2, and 3
4.3. X-ray Determination of Compound 3
4.4. Cell Culture and Treatments
4.5. MTT Assay for the Anti-Proliferative Activity In Vitro
4.6. Western Blotting
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Roskoski, R. Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol. Res. 2019, 139, 395–411. [Google Scholar] [CrossRef]
- Kennedy, S.P.; Hastings, J.F.; Han, J.Z.R.; Croucher, D.R. The Under-Appreciated Promiscuity of the Epidermal Growth Factor Receptor Family. Front. Cell Dev. Biol. 2016, 4, 88. [Google Scholar] [CrossRef] [PubMed]
- Fichter, C.D.; Gudernatsch, V.; Przypadlo, C.M.; Follo, M.; Schmidt, G.; Werner, M.; Lassmann, S. ErbB targeting inhibitors repress cell migration of esophageal squamous cell carcinoma and adenocarcinoma cells by distinct signaling pathways. J. Mol. Med. 2014, 92, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Moroni, M.; Veronese, S.; Benvenuti, S.; Marrapese, G.; Sartore-Bianchi, A.; Di Nicolantonio, F.; Gambacorta, M.; Siena, S.; Bardelli, A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: A cohort study. Lancet Oncol. 2005, 6, 279–286. [Google Scholar] [CrossRef]
- Rehmani, H.S.; Issaeva, N. EGFR in head and neck squamous cell carcinoma: Exploring possibilities of novel drug combinations. Ann. Transl. Med. 2020, 8, 813. [Google Scholar] [CrossRef]
- Chen, X.; Liang, R.; Zhu, X. Anti-EGFR therapies in nasopharyngeal carcinoma. Biomed. Pharmacother. 2020, 131, 110649. [Google Scholar] [CrossRef] [PubMed]
- Von Deimling, A.; Louis, D.N.; Von Ammon, K.; Petersen, I.; Hoell, T.; Chung, R.Y.; Martuza, R.L.; Schoenfeld, D.A.; Yasargil, M.G.; Wiestler, O.D.; et al. Association of epidermal growth factor receptor gene amplification with loss of chromosome 10 in human glioblastoma multiforme. J. Neurosurg. 1992, 77, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Papageorgio, C.; Perry, M.C. Epidermal Growth Factor Receptor-Targeted Therapy for Pancreatic Cancer. Cancer Investig. 2007, 25, 647–657. [Google Scholar] [CrossRef]
- Roskoski, R. ErbB/HER protein-tyrosine kinases: Structures and small molecule inhibitors. Pharmacol. Res. 2014, 87, 42–59. [Google Scholar] [CrossRef]
- Yu, H.A.; Sima, C.S.; Huang, J.; Solomon, S.B.; Rimner, A.; Pietanza, M.C.; Azzoli, C.G.; Rizvi, N.A.; Krug, L.M.; Vincent, A.; et al. Local therapy with continued EGFR tyrosine kinase inhibitor therapy as a treatment strategy in EGFR mutant advanced lung cancers that have developed acquired resistance to EGFR tyrosine kinase inhibitors. J. Thorac. Oncol. 2013, 8, 346–351. [Google Scholar] [CrossRef]
- Topics, C.; TAKEUCHI, K.; ITO, F.; Topics, C. Target Therapy for Cancer: Anti-cancer Drugs Targeting Growth-Factor Signaling Molecules Receptor Tyrosine Kinases and Targeted Cancer Therapeutics. Biol. Pharm. Bull. 2011, 34, 1774–1780. [Google Scholar]
- Wu, J.; Chen, W.; Xia, G.; Zhang, J.; Shao, J.; Tan, B.; Zhang, C.; Yu, W.; Weng, Q.; Liu, H.; et al. Design, Synthesis, and Biological Evaluation of Novel Conformationally Constrained Inhibitors Targeting EGFR. ACS Med. Chem. Lett. 2013, 4, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Xiang, M.; Lei, K.; Fan, W.; Lin, Y.; He, G.; Yang, M.; Chen, L.; Mo, Y. In silico identification of EGFR-T790M inhibitors with novel scaffolds: Start with extraction of common features. Drug Des. Devel. Ther. 2013, 7, 789–839. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Doebele, R.C.; Oton, A.B.; Peled, N.; Camidge, D.R.; Bunn, P.A. New strategies to overcome limitations of reversible EGFR tyrosine kinase inhibitor therapy in non-small cell lung cancer. Lung Cancer 2010, 69, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Giordano, S.H.; Lin, Y.-L.; Kuo, Y.F.; Hortobagyi, G.N.; Goodwin, J.S. Decline in the use of anthracyclines for breast cancer. J. Clin. Oncol. 2012, 30, 2232–2239. [Google Scholar] [CrossRef]
- Pastan, I.; Hassan, R.; FitzGerald, D.J.; Kreitman, R.J. Immunotoxin therapy of cancer. Nat. Rev. Cancer 2006, 6, 559–565. [Google Scholar] [CrossRef]
- Zahaf, N.-I.I.; Lang, A.E.; Kaiser, L.; Fichter, C.D.; Lassmann, S.; McCluskey, A.J.; Augspach, A.; Aktories, K.; Schmidt, G. Targeted delivery of an ADP-ribosylating bacterial toxin into cancer cells. Sci. Rep. 2017, 7, 41252. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Yuan, Y.; Qiu, N.; Peng, P.; Sheng, R.; Hu, Y. Identification of novel 4-anilinoquinazoline derivatives as potent EGFR inhibitors both under normoxia and hypoxia. Bioorg. Med. Chem. 2014, 22, 6796–6805. [Google Scholar] [CrossRef]
- Kékesi, L.; Sipos, A.; Németh, G.; Dancsó, A.; Illyés, E.; Boros, S.; Breza, N.; Nemes, Z.; Hegymegi-Barakonyi, B.; Pató, J.; et al. Pyrido[2,3-b]pyrazines inhibiting both erlotinib-sensitive and erlotinib-resistant cell lines, and their preparation via regioselective condensation reaction. Acta Pharm. Hung. 2014, 84, 91–104. [Google Scholar]
- Xia, G.; Chen, W.; Zhang, J.; Shao, J.; Zhang, Y.; Huang, W.; Zhang, L.; Qi, W.; Sun, X.; Li, B.; et al. A Chemical Tuned Strategy to Develop Novel Irreversible EGFR-TK Inhibitors with Improved Safety and Pharmacokinetic Profiles. J. Med. Chem. 2014, 57, 9889–9900. [Google Scholar] [CrossRef]
- Schenone, S.; Bruno, O.; Radi, M.; Botta, M. 4-Amino-Substituted Pyrazolo [3,4-d] Pyrimidines: Synthesis and Biological Properties. Mini. Rev. Org. Chem. 2009, 6, 220–233. [Google Scholar] [CrossRef]
- Greco, C.; Taresco, V.; Pearce, A.K.; Vasey, C.E.; Smith, S.; Rahman, R.; Alexander, C.; Cavanagh, R.J.; Musumeci, F.; Schenone, S. Development of Pyrazolo[3,4- d]pyrimidine Kinase Inhibitors as Potential Clinical Candidates for Glioblastoma Multiforme. ACS Med. Chem. Lett. 2020, 11, 657–667. [Google Scholar] [CrossRef]
- Li, M.; Liu, S.; Chen, H.; Zhou, X.; Zhou, J.; Zhou, S.; Yuan, H.; Xu, Q.L.; Liu, J.; Cheng, K.; et al. N-benzylpiperidinol derivatives as novel USP7 inhibitors: Structure–activity relationships and X-ray crystallographic studies. Eur. J. Med. Chem. 2020, 199, 112279. [Google Scholar] [CrossRef]
- Dolzhenko, A.V.; Tan, K.; Anna, V.; Koh, L.; Pastorin, G. Experimental pyrimidin-5-amine methanol disolvate. Acta Crystallogr. Sect. E Struct. Rep. 2010, E66, o1835–o1836. [Google Scholar] [CrossRef]
- Thaher, B.A.; Arnsmann, M.; Totzke, F.; Ehlert, J.E.; Kubbutat, M.H.G.; Schächtele, C.; Zimmermann, M.O.; Koch, P.; Boeckler, F.M.; Laufer, S.A. Tri- and Tetrasubstituted Pyrazole Derivates: Regioisomerism Switches Activity from p38MAP Kinase to Important Cancer Kinases. J. Med. Chem. 2012, 55, 961–965. [Google Scholar] [CrossRef]
- Tyurin, R.V.; Vorob, E.V.; Minyaeva, L.G.; Krasnikov, V.V.; Mezheritskii, V.V. [1,5-c] pyrimidine System. Russ. J. Org. Chem. 2005, 41, 916–921. [Google Scholar] [CrossRef]
- Dolzhenko, A.V.; Pastorin, G.; Dolzhenko, A.V.; Chui, W.K. [1,5-c] pyrimidines. Tetrahedron Lett. 2009, 50, 5617–5621. [Google Scholar] [CrossRef]
- Shaban, M.A.E.; Morgaan, A.E.A. The Chemistry of 1, 2, 4-Triazolopyrimidines 1:1,2,4-Triazolo [4,3-a] Pyrimidines. Adv. Heterocycl. Chem. 1999, 73, 131–176. [Google Scholar]
- Shurrab, N.K.; Alhendawi, H.; Kerrit, M.A. pyrimidines acyclo C -nucleosides via oxidative cyclisation. J. Chem. Res. 2018 2018, 42, 601–603. [Google Scholar]
- Zhang, F.; Wang, S.; Yin, L.; Yang, Y.; Guan, Y.; Wang, W.; Xu, H.; Tao, N. Quantification of Epidermal Growth Factor Receptor Expression Level and Binding Kinetics on Cell Surfaces by Surface Plasmon Resonance Imaging. Anal. Chem. 2015, 87, 9960–9965. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.; Pandey, V.; Dhananjaya Mohan, C.; Ting Chia, Y.; Rangappa, S.; Mathai, J.; Baburajeev, C.P.; Paricharak, S.; Mervin, L.H.; Bulusu, K.C.; et al. Novel Adamantanyl-Based Thiadiazolyl Pyrazoles Targeting EGFR in Triple-Negative Breast Cancer. Am. Chem. Soc. 2016, 1, 1412–1424. [Google Scholar] [CrossRef]
- Aliwaini, S.; Swarts, A.J.A.J.A.J.; Blanckenberg, A.; Mapolie, S.; Prince, S. A novel binuclear palladacycle complex inhibits melanoma growth in vitro and in vivo through apoptosis and autophagy. Biochem. Pharmacol. 2013, 86, 1650–1662. [Google Scholar] [CrossRef] [PubMed]
- El Guerrab, A.; Bamdad, M.; Kwiatkowski, F.; Bignon, Y.J.; Penault-Llorca, F.; Aubel, C. Anti-EGFR monoclonal antibodies and EGFR tyrosine kinase inhibitors as combination therapy for triple-negative breast cancer. Oncotarget 2016, 7, 73618–73637. [Google Scholar] [CrossRef] [PubMed]
- Kari, C.; Chan, T.O.; Rocha de Quadros, M.; Rodeck, U. Targeting the epidermal growth factor receptor in cancer: Apoptosis takes center stage. Cancer Res. 2003, 63, 1–5. [Google Scholar]
- OEDocking 3.0.1—Applications, v2020.2.2. Available online: https://docs.eyesopen.com/applications/oedocking/releasenotes/OEDocking_version3.0.1.html (accessed on 1 June 2021).
- Peng, Y.H.; Shiao, H.Y.; Tu, C.H.; Liu, P.M.; Hsu, J.T.A.; Amancha, P.K.; Wu, J.S.; Coumar, M.S.; Chen, C.H.; Wang, S.Y.; et al. Protein kinase inhibitor design by targeting the Asp-Phe-Gly (DFG) motif: The role of the DFG motif in the design of epidermal growth factor receptor inhibitors. J. Med. Chem. 2013, 56, 3889–3903. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Abraham, M.H.; Le, J.; Hersey, A.; Luscombe, C.N.; Beck, G.; Sherborne, B.; Cooper, I. Rate-limited steps of human oral absorption and QSAR studies. Pharm. Res. 2002, 19, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Wei, Y.; Tang, Y.; Zhou, Y.; Yang, Y.; Cui, Y.; Wang, X.; Wang, Y.; Liu, Y.; Liu, N.; Wang, Q.; et al. Discovery and Optimization of a Novel 2H-Pyrazolo[3,4-d]pyrimidine Derivative as a Potent Irreversible Pan-Fibroblast Growth Factor Receptor Inhibitor. J. Med. Chem. 2021. [Google Scholar] [CrossRef]
- Zhang, H.; Berezov, A.; Wang, Q.; Zhang, G.; Drebin, J.; Murali, R.; Greene, M.I. Review series ErbB receptors: From oncogenes to targeted cancer therapies. Molecules 2007, 117, 2051–2058. [Google Scholar] [CrossRef]
- MarvinSketch version 16.10.24, ChemAxon—Software Solutions and Services for Chemistry & Biology 2016. Available online: https://chemaxon.com (accessed on 24 June 2021).
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and the Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef] [PubMed]
- BIOVIA. Dassault Systèmes, Discovery Studio Visualizer, version 3.0; Sheldrick, G.M.: San Diego, CA, USA, 2020.
- SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [CrossRef] [PubMed]
- Spek, A.L. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003, 36, 7–13. [Google Scholar] [CrossRef]



| Compound | PSA | NRB | miLogP | HBA | HBD | Mol Vol | MW | %ABS |
|---|---|---|---|---|---|---|---|---|
| 1 | 73.81 | 3 | 4.44 | 7 | 0 | 359.02 | 486.31 | 83.53% |
| 2 | 73.81 | 2 | 1.84 | 7 | 0 | 274.66 | 349.30 | 83.53% |
| 3 | 73.81 | 2 | 2.19 | 7 | 0 | 282.68 | 392.22 | 83.53% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aliwaini, S.; Abu Thaher, B.; Al-Masri, I.; Shurrab, N.; El-Kurdi, S.; Schollmeyer, D.; Qeshta, B.; Ghunaim, M.; Csuk, R.; Laufer, S.; et al. Design, Synthesis and Biological Evaluation of Novel Pyrazolo[1,2,4]triazolopyrimidine Derivatives as Potential Anticancer Agents. Molecules 2021, 26, 4065. https://doi.org/10.3390/molecules26134065
Aliwaini S, Abu Thaher B, Al-Masri I, Shurrab N, El-Kurdi S, Schollmeyer D, Qeshta B, Ghunaim M, Csuk R, Laufer S, et al. Design, Synthesis and Biological Evaluation of Novel Pyrazolo[1,2,4]triazolopyrimidine Derivatives as Potential Anticancer Agents. Molecules. 2021; 26(13):4065. https://doi.org/10.3390/molecules26134065
Chicago/Turabian StyleAliwaini, Saeb, Bassam Abu Thaher, Ihab Al-Masri, Nabil Shurrab, Said El-Kurdi, Dieter Schollmeyer, Basem Qeshta, Mariam Ghunaim, René Csuk, Stefan Laufer, and et al. 2021. "Design, Synthesis and Biological Evaluation of Novel Pyrazolo[1,2,4]triazolopyrimidine Derivatives as Potential Anticancer Agents" Molecules 26, no. 13: 4065. https://doi.org/10.3390/molecules26134065
APA StyleAliwaini, S., Abu Thaher, B., Al-Masri, I., Shurrab, N., El-Kurdi, S., Schollmeyer, D., Qeshta, B., Ghunaim, M., Csuk, R., Laufer, S., Kaiser, L., & Deigner, H.-P. (2021). Design, Synthesis and Biological Evaluation of Novel Pyrazolo[1,2,4]triazolopyrimidine Derivatives as Potential Anticancer Agents. Molecules, 26(13), 4065. https://doi.org/10.3390/molecules26134065







