Abstract
The extraordinary cellular diversity and the complex connections established within different cells types render the nervous system of vertebrates one of the most sophisticated tissues found in living organisms. Such complexity is ensured by numerous regulatory mechanisms that provide tight spatiotemporal control, robustness and reliability. While the unusual abundance of long noncoding RNAs (lncRNAs) in nervous tissues was traditionally puzzling, it is becoming clear that these molecules have genuine regulatory functions in the brain and they are essential for neuronal physiology. The canonical view of RNA as predominantly a ‘coding molecule’ has been largely surpassed, together with the conception that lncRNAs only represent ‘waste material’ produced by cells as a side effect of pervasive transcription. Here we review a growing body of evidence showing that lncRNAs play key roles in several regulatory mechanisms of neurons and other brain cells. In particular, neuronal lncRNAs are crucial for orchestrating neurogenesis, for tuning neuronal differentiation and for the exact calibration of neuronal excitability. Moreover, their diversity and the association to neurodegenerative diseases render them particularly interesting as putative biomarkers for brain disease. Overall, we foresee that in the future a more systematic scrutiny of lncRNA functions will be instrumental for an exhaustive understanding of neuronal pathophysiology.
1. Introduction
Despite being initially considered as ‘transcriptional noise’, lncRNAs are currently recognized as modulators of cellular functions in every branch of life and have been implicated in a large number of biological processes including X-chromosome silencing, genetic imprinting, regulation of cell state, coordination of differentiation and possibly disease modulation (reviewed in [1,2,3,4,5,6]). This concept is supported by the carefully-regulated subcellular localization of lncRNAs, with several examples of lncRNAs that are restricted to the nucleus [7], while others are transported into the cytoplasm [8,9]. Likewise, the number of lncRNAs in different organisms seems to correlate with genome complexity [1], and in complex tissues such as the nervous system, lncRNAs show extreme spatiotemporal regulation and cell specificity [8,10,11,12]. In this review we focus on the incipient roles of lncRNAs as flexible regulators of neuronal activity and brain pathophysiology.
2. General Characteristics of LncRNAs
While only a small portion (~3%) of the vertebrate genome encodes for proteins, it has been proposed that ~80% can lead to the production of a variety of noncoding RNAs (ncRNAs) [13,14,15,16,17]. It is still debated whether the pervasive transcription leading to the production of these molecules is noise related to the regulation of the expression of neighboring genes, a way to provide ‘raw-material’ for the evolution of de novo genes or rather leads to the production of functional molecular species [18,19,20,21,22,23,24,25]. The large family of ncRNA comprises several RNA species including (i) the classical ncRNAs involved in translation and pre-mRNA processing (such as tRNAs, rRNAs and small nuclear RNAs); (ii) molecules of 21–28 nucleotides that regulate transcription and mRNA silencing (small interfering RNAs and miRNAs) and (iii) a more heterogenous family of longer transcripts (>200 nucleotides) collectively defined as long noncoding RNAs (lncRNAs) [26,27,28]. Several lncRNAs share similarities with conventional mRNAs, such as being generally synthetized by RNA polymerase II, undergoing capping, splicing and polyadenylation. However, lncRNAs are usually less abundant than most of conventional mRNAs and by definition display virtually non-existent coding potential [6,29]. The absence of coding potential needs to be considered carefully when studying newly-discovered lncRNAs, as it is becoming clear that appeared initially devoid of functional open reading frames are actually involved in the production of small peptides, revealing a hidden ‘short proteome‘ which has important cellular functions, such as transcription regulation or cell signaling [30,31,32,33,34]. In general, a real challenge in this field is to distinguish transcriptional noise from molecules that exert a ‘true molecular function’. In this review, whenever possible, we have tried to follow experimental evidences that would help us to distinguish these two scenarios (noise vs. function). While for some few extensively studied lncRNAs this is possible, the task is complicated by the fact that often there are few follow up works on the same lncRNA and the literature is growing exponentially with newly found lncRNAs. Here we have summarized the more relevant lncRNAs in brain function.
3. LncRNAs in the Nervous System
The brain of vertebrates, and in particular of birds and mammals, is one of the most complex systems in biology where an elaborate network established between different cell types allows advanced cognitive tasks [35]. During brain development, an intricate series of processes is required for cell differentiation and construction of refined neuronal circuits. Even in the adult brain, processes such as memory recall, learning and the underlying orchestration of cell activities require an exceptionally precise spatiotemporal tuning of gene expression and protein production. Thanks to their structural plasticity and intrinsic ability to facilitate protein-genome interactions, it is becoming evident that lncRNAs might be optimally suited for the regulation of brain gene expression, as we outline in this review.
The importance of lncRNAs in the central nervous system is exemplified by the high number of brain-specific transcripts compared to other tissues [36,37], i.e., up to 40% of the differentially expressed lncRNAs are brain-specific [38]. Several transcriptome studies identified thousands of lncRNAs expressed during brain development [39,40,41,42,43]. Measures of RNA abundance also revealed changes of lncRNA expression during neurogenesis [40,42,44], and specific expression patterns in distinct neuronal sub-types suggest that they could be involved in cell fate choices [37,45]. Numerous lncRNAs that vary during development are embedded in the genome in the vicinity of neurodevelopmental regulators [46]. While this observation could also be explained by leaky transcription in the vicinity of active genomic regions, several examples suggest that lncRNAs tend to be co-regulated with other genes, as often happens with ncRNAs whose expression is coordinated in gene clusters [47,48].
Results from in situ hybridization and lacZ reporter mouse lines have revealed that lncRNAs are excellent markers for brain developmental stages, brain sub-region identity and even reliable markers for neuronal subtypes, further indicating that there is a tight regulation in their expression profile and that they are intentional transcription products [8,11,12,49]. Moreover, besides being modulated during differentiation, hundreds of lncRNAs are dynamically controlled by neuronal activity and are associated with the dynamic regulation of synapses [50,51].
Despite the overall low level of conservation of lncRNAs, brain-specific lncRNAs display high sequence conservation when compared to lncRNAs from other tissues [46,52]. Even their spatiotemporal expression is maintained among some species as confirmed by a set of human lncRNAs showing transient expression during primate organoid differentiation, as well as cell-specific expression shown by single-cell sequencing [53]. Even more astoundingly, some neurospecific lncRNAs are conserved across species with larger brains, suggesting a specific role in the evolution of the primate neocortex [54]. Consistently, there are significant structural similarities between the ~3000 multi-exonic lncRNAs found in humans with those of great apes and also in their expression profile in cortical development [53].
Altogether, these observations remain correlative, raising the question of whether lncRNA expression is just an ‘epiphenomenon’, a casual correlate of other processes such as gene expression regulation, or if it plays a direct causative role in the regulation of brain morphogenesis and functioning. To answer this question, in the next sections we will summarize the molecular roles of the most relevant lncRNAs and their functions in neurons and other cells for the central nervous system.
4. Functional Classification of Neuronal LncRNAs
Among the numerous possible classifications for lncRNAs proposed [55,56,57,58,59,60,61], here we decided to divide brain lncRNAs in six main functional classes. These include: (1) Enhancer lncRNAs (eRNAs), which are the product of bidirectional DNA transcription taking place in the region of active enhancers [62]; (2) Antisense lncRNAs (AS-lncRNAs; also known as natural antisense transcripts) produced by antisense transcription of a protein-coding sequence which can be degraded following the formation of double strand RNAs [63]; (3) Cis-acting nuclear lncRNAs, thus acting in the same genome locus from which they originate and that interfere with and/or regulate the transcription of neighboring genes [64]; (4) Trans-acting nuclear lncRNAs involved in epigenetic regulation; (5) LncRNAs that modulate gene expression by post transcriptional mechanisms and affect mRNA stability, translation, or splicing [65,66,67] and finally (6) LncRNAs directly modulating protein functions that are generally functioning in association with RNA binding proteins (RBPs), in some cases also outside the nucleus. This classification is summarized in Figure 1.
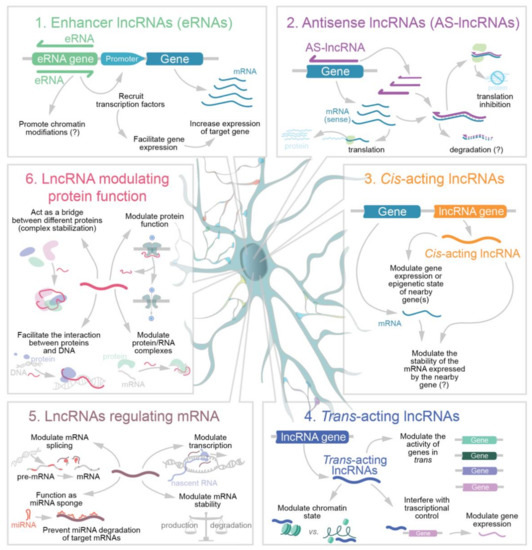
Figure 1.
Schematic representation of the functional classes of neuronal lncRNAs.
4.1. Enhancer LncRNAs: More Than Markers of Actively Transcribed Regions
Even though eRNAs are sometimes excluded from the class of lncRNAs because they can be shorter than 200 nucleotides, the vast majority are longer than 200 nucleotides. Moreover, these ncRNAs do not behave as most other shorter ncRNAs. For these reasons, and for their predominant abundance in the brain, we believe they should be discussed here.
Several eRNAs are produced in the vicinity of genes as bidirectional transcripts, whose levels correlate with the local gene expression [68,69]. For this reason eRNAs were originally considered markers of actively transcribed loci [68]. In 2010, Greenberg and collaborators showed that in neurons ~12,000 enhancers are actively transcribed and give rise to eRNAs upon neuronal activation in a process requiring polymerase II, the p300-CBP coactivator, and triggering the mono-methylation of Histone 3 at lysine 4 [68]. In neurons, the synthesis of eRNAs facilitates the recruitment of activity-regulated transcription factors including CREB, SRF, NPAS4 and leads to the formation of enhancer-promoter loops, driving the transcription of activity-regulated genes such as c-Fos and Nr4a2 [68,70,71].
For the activity-regulated cytoskeleton-associated gene (Arc), it was described that eRNAs can directly modulate in cis the elongation of the corresponding mRNA by acting as decoys for the negative elongation factor NELF, thus facilitating the progression of RNA polymerase II at the promoter of the target gene [72]. Similar mechanisms could underlie the function of utNgn1. eRNA utNgn1 is expressed in concomitance with Neurogenin-1 (NGN1), an essential transcription factor for neuronal differentiation [73], and loss of function experiments indicate that utNgn1 might control the levels of NGN1 [73].
Altogether these findings point to a specific biological function of eRNAs, suggesting that their transcription is not just a side effect of the change in the local DNA conformation and epigenetic state, but that they might directly recruit protein complexes. They might even function as bridges between far DNA regions. This is exemplified by Evf2, a highly-conserved eRNA with a role in brain development. Loss of Evf2 in mice is accompanied by a reduction in GABAergic interneurons in the newborn hippocampus followed by impaired GABA circuitry in adult animals. In the developing forebrain, Evf2 has a complex regulatory mechanism since it can simultaneously localize and activate a gene distant ~1.5 Mb on chr6 (Umad1), while repressing another gene that is ~25 Mb distant (Akr1b8). Moreover, its overexpression activates Lsm8, also on the same chromosome but ~12 Mb apart [74]. These effects are probably mediated through complex changes of three-dimensional chromosome topology, although other effects in trans cannot be excluded, since it has been observed that Evf2 can also recruit transcriptional activators (such as DLX1/2) and repressors (such as MECP2) that can interfere with the regulatory regions of developmental genes such as Dlx5, Dlx6 and Gad1, explaining its effect on GABAergic differentiation [75].
Overall, a comprehensive study of the molecular functions and structural effects of eRNAs is largely missing. Remarkably, several single nucleotide polymorphisms on eRNAs near synaptic activity-regulated genes are associated with autism spectrum disorders, indicating that their sequences contain structural motifs required for a specific scaffolding function [76].
4.2. Antisense LncRNAs Regulate Key Genes in the Brain
Around 70% of genes are transcribed in the antisense direction, giving rise to AS-lncRNAs implicated in post-transcriptional regulation of the corresponding ‘sense’ transcript by degradation of the complementary mRNA molecules or by modulating their expression or stability [77,78,79,80,81].
In the brain, several mRNAs are under the control of a corresponding AS-lncRNA, including for example the genes encoding the CAMK2N1 subunit of the calmodulin-dependent protein kinase and neurogranin (NRGN), which are both implicated in the regulation of postsynaptic signal transduction pathways and synaptic long-term potentiation [82].
Another notable example is Kcna2-AS that targets the mRNA of the ‘shaker type’ voltage-dependent potassium channel (Kcna2), which is relevant for human pathology since its mutations are linked to ataxia and focal epilepsies [83]. Indeed, the overexpression of Kcna2-AS in experimental models inversely correlates with the levels of KCNA2 [84], confirming that this AS-lncRNA can regulate its respective protein. In rats, peripheral nerve injury increases the expression of Kcna2-AS as a result of transcription activation mediated by the myeloid zinc finger protein 1 (MZF1) that leads to a ~50% decrease of active channels [84]. As a consequence, the induction of Kcna2-AS during nerve damage reduces the voltage-gated potassium current, and increases the excitability of dorsal root ganglia neurons producing neuropathic pain symptoms. While these observations hold great potential for therapeutic strategies aimed at decreasing neuropathic pain following injury, this does not entirely explain the physiological roles of Kcna2-AS in the regulation of neuronal excitability which will require further studies.
During the development of the brain cortex, several AS-lncRNA are expressed to subtly regulate the amounts of transcription factors. Examples include Sox (SRY-related HMG-box) genes that encode high mobility groups that can bind and bend the DNA during the regulation of embryonic development and influence cell fate. In particular two members of the Sox family, Sox4 and Sox11, have corresponding AS-lncRNAs that are highly expressed during the development of the mouse cortex, suggesting they might have a specific function in cerebral corticogenesis and cell differentiation [85].
An eminent AS-lncRNAs in the brain is BDNF-AS, which controls the levels of the brain-derived neurotrophic factor (BDNF), a neurotrophin implicated in the regulation of several key processes including neurogenesis, precursor cell proliferation, neuronal differentiation, maturation, plasticity and with a role in pathophysiology of psychiatric disorders [86,87,88,89]. BDNF-AS overexpression decreases the levels of BDNF, both in vitro and in vivo [63]. The exact mechanisms of BDNF-AS function are not entirely known, although it seems that its expression affects in cis the Bdnf gene by recruiting EZH2, an abundant histone methyltransferase that causes the inactivation of the BDNF promoter. As expected, BDNF-AS knockdown results in increased neuronal cell number, differentiation and more prominent neurite outgrowth [63,90]. The postmortem amygdala of alcoholics showed that the expression of BDNF-AS is increased only in individuals who started to drink before 21 years of age, suggesting that this BDNF-AS might affect epigenetic reprogramming in an age-dependent manner [91]. Deficits of BDNF signaling in the amygdala decreased the levels of the activity-regulated cytoskeleton-associated protein (ARC) and reduced synaptic plasticity while increasing the risk of alcohol dependence in adulthood [91].
Among AS-lncRNAs associated with human pathologies, it is worth mentioning BACE1-AS which, in contrast to most AS-lncRNAs, stabilizes its related transcript Bace1 which encodes the β-secretase enzyme 1 implicated in Alzheimer’s disease (BACE1) [80]. This enzyme cuts and processes the amyloid precursor protein (APP), and the BACE1-AS lncRNA is thus involved in a feed-forward loop that is induced by high levels of the amyloid-beta 1–42 fragment itself. The molecular details of this unusual process are not known and, although it has been suggested that RNA duplex formation could alter the structure of Bace1 RNA and thus increase its stability, this possibility remains speculative. To reinforce its connection to the pathology, the concentration of BACE1-AS is higher in patients with Alzheimer’s disease than in matched controls [80] and correlates with Aβ production and plaque deposition in animal models [92].
Angelman syndrome is a pathology where an AS-lncRNA, named Ube3a-ATS, might become a possible target for therapeutic intervention. This transcript is nuclear, neurospecific and regulates the gene Ube3a [93]. Angelman syndrome is a neurodevelopment disorder where patients develop intellectual disability, speaking and movement problems often associated with what looks like a happy demeanor [94]. The gene Ube3a codes for a ubiquitin ligase, that ubiquitinates and regulates the stability of several important proteins in neuronal function such as MAPK1 and β-catenin. The paternal Ube3a is usually repressed by Ube3a-ATS and the pathology manifests itself upon deletion of the maternal gene. As a consequence of paternal imprinting, cells that in principle contain a copy of the functioning gene, do not actually express the protein UBE3A. For this reason, downregulation of the paternal Ube3a-ATS could be a rescue strategy for this pathology [95,96].
A number of AS-lncRNAs appear to have acquired additional functions besides regulating the expression of their sense gene. One example is Six3OS, which is transcribed from the opposite strand of the Six3 gene, a homeodomain transcription factor involved in forebrain development and retinal cell specification [97]. Besides regulating the expression of Six3, Six3OS also acts as a molecular scaffold for the transcriptional co-regulator of SIX3 (EYA) and the histone-lysine N-methyltransferase enzyme EZH2, both implicated in the regulation of SIX3 target genes [98]. Interaction of Six3OS with EZH2 possibly changes the chromatin structure by triggering the H3K27me3 modification, which is usually associated with decreased expression of nearby genes [98]. Furthermore, loss of function studies revealed that Six3OS is implicated in the glial-neuronal lineage specification of adult neural stem cells in vivo [41]. Due to technical challenges related to the study of antisense RNA, it is conceivable that there is some skepticism for results that could also be simply due to an effect on the target gene (i.e., Six3). An additional confounding factor could be the several isoforms of Six3OS, some of which probably code for short ~100 amino acid proteins [99]. Nevertheless, it can be hypothesized that lncRNAs that were initially expressed for the direct regulation of their respective mRNA with an antisense mechanism acquired during evolution additional specific functions in cis in the vicinity of the gene, similar to cis-acting lncRNAs which will be discussed in the next section.
4.3. Cis-Acting LncRNAs Regulating Nearby Genes Might Evolve Additional Molecular Functions
The local regulation of gene expression exerted by lncRNAs can encompass different mechanisms, such as regulation of promoter activity but also local modulation of transcription and splicing [100,101]. The reason why lncRNAs might restrict their function at the site of their production is not entirely clear, although it might be related to the fact that once they are produced, some lncRNAs are physically engaged in local complexes, thus limiting their range of action. This might be the case for several lncRNAs that are operating locally at the chromatin interface [102,103]. Since it is known that several lncRNAs interfere with transcription and translation in the vicinity of their genes, this is the first mechanism that is investigated for lncRNAs and their regulatory role in cis has been observed and described in many cases. At the same time, in several cases as also summarized here, a more careful examination of their role has revealed that many lncRNAs also have additional roles, besides their activity in cis.
A relevant lncRNA with a cis-acting mechanism is Paupar (Pax6 upstream antisense RNA), which is transcribed upstream the Pax6 gene in the opposite direction [104]. Paupar acts in cis regulating PAX6-dependent functions since its knockdown leads to an increase in PAX6 expression and interferes with eye development, progenitor cell potency and proliferation in the brain [104,105]. The exact molecular mechanism of regulation on the Pax6 is not entirely known, although it could be speculated that its transcription affects the nearby promoter region of Pax6, either stabilizing transcription factor binding or inducing promoter-enhancer looping. In parallel, Paupar has probably evolved a more elaborate mechanism of action. In addition to regulating the expression of Pax6, it also regulates the binding of PAX6 to its neural target genes [105] and acts in trans by forming a ribonucleoprotein (RNP) complex together with PAX6 and KAP1 (coded by Trim28), which is involved in transcriptional control [104,106]. It seems that Paupar promotes the association of KAP1 with the DNA resulting in H3K9me3 modifications on the promoter of genes involved in neural stem cell renewal and differentiation, inducing heterochromatin and inactivation [106]. Loss of function studies confirm that the lack of Paupar disrupts olfactory bulb neurogenesis [106] and induces neuroblastoma cell differentiation [104].
A key transcription factor in cerebral cortex development is POU3F3 (also known as BRN1). It has been reported that POU3F3 is under the control of several lncRNAs. In its close proximity, there are two lncRNAs that have been studied in detail by the laboratory of Rinn and collaborators [11]. Deletion of one of these two lncRNAs, linc–Brn1b (also known as Pantr2), has a cis-regulatory effect on Pou3f3. The level of linc–Brn1b usually is higher in neural stem cells and its expression declines during development. Mice where both copies of linc–Brn1b are ablated exhibit a ~50% reduction in POU3F3 levels and a subsequent decrease in the number of cortex intermediate progenitor cells and abnormal lamination in the cerebral cortex [11]. While the mechanism of action of this lncRNA is not entirely known, its close proximity to the Pou3f3 gene and the effect on its expression suggest that the linc–Brn1b acts as an eRNA, although the differential expression observed in other surrounding genes might suggest that linc–Brn1b also has additional functions that surpass its local effects on DNA conformation. Dali (DNMT1-Associated Long Intergenic RNA) is yet another lncRNA that interferes with the expression of Pou3f3, which is located ~50 Kb upstream. Additionally, similarly to Paupar, Dali has a set of supplementary functions and forms a complex with POU3F3 which, in trans, regulates a subset of genes involved in neural differentiation. Another mechanism by which Dali carries on its functions is achieved by its direct interaction with chromatin remodeling factors such as BRG1, SIN3A and DNMT1 [64]. Through these interactions Dali affects the epigenetic reprogramming of gene promoters, as also shown by its knockdown which increases the methylation of CpG islands on the promoters of genes involved in neuronal differentiation [64]. There are also several other lncRNAs which have a role in the epigenetic modulation of the brain that will be discussed more in detail in the next section.
4.4. Trans-Acting LncRNAs Involved in Epigenetic and Transcriptional Regulation
In several tissues, nuclear lncRNAs often act as molecular scaffolds that recruit chromatin modifiers such as the polycomb repressive complex (PRC2) and the DNA methyltransferase 3 (DNMT3) to regulatory genomic elements and modulate the expression of target genes (Reviewed in Refs. [107,108]). In the brain, global dynamic modification of chromatin is often observed during cell lineage commitment, development, terminal differentiation and in more complex cognitive functions such as learning [109,110,111]. While lncRNAs do not have overt ribozyme-like ‘enzymatic’ functions, their interaction with the epigenetic machinery and the target DNA sequences coupled to their ability to act as a scaffold for the formation of large multimeric complexes, can lead to precise covalent modification of histones and/or DNA and result in the control of gene expression. As an example, lncOL1 is a lncRNA with epigenetic functions, which is exclusively expressed in oligodendrocytes, a specific type of glial cells deputed to myelination in the central nervous system. lncOL1 promotes oligodendrocyte differentiation and speeds the onset of myelination forming a complex with SUZ12, a zinc finger that is part of the polycomb repressive complex 2. The lncOL1-SUZ12 complex silences genes that in undifferentiated cells repress oligodendrocyte genes promoting the formation of H3K27me3 modifications on their regulatory elements. As a result, lncOL1 overexpression promotes myelination and even more strikingly favors re-myelination following injury, suggesting a possible therapeutic use of this lncRNA in the treatment of demyelinating conditions [112].
Regulating neuronal differentiation and neurogenesis, examples of lncRNAs include the Hox-encoded HOXA Transcript Antisense RNA, Myeloid-Specific 1 (HOTAIRM1) and Tcl1 upstream neuron-associated lincRNA (Tuna). HOTAIRM1 modulates Neurogenin 2, a basic helix-loop-helix transcription factor that acts as a master regulator of neurogenesis [113]. Downregulation of Tuna, a highly expressed and evolutionary conserved lncRNA that has been studied in the developing mouse brain, is sufficient to block neurogenesis in both mouse and human cell models [114].
Tuna affects neuronal development by recruiting two heterogenous nuclear ribonucleoproteins (HNRNPK and PTBP1) and Nucleolin, which can regulate chromatin condensation by bringing histone H1 in the proximity of genes to be silenced [114]. The overexpression of Tuna results in the deposition of the active histone modification H3K4me3 to the promoters of key genes in neuronal specification such as Nanog, Fgf4 and Sox2, indicating that the lncRNA might also modulate the activity of some histone methyltransferases [114].
Transcriptional regulation can take place independently from epigenetic remodeling as possibly observed in the case of the lncRNA GM12371. This lncRNA is nuclear and more abundantly expressed in the hippocampus. Its depletion in primary hippocampal neurons determines the reduction of spontaneous excitatory postsynaptic currents. Furthermore, decreasing GM12371 interferes with spine density and dendritic arborization. Mechanistically, GM12371 is regulated by cAMP-PKA signaling and, once produced, it binds to active chromatin in trans to regulate genes which are involved in neuronal growth and development. The expression of GM12371 is strictly required in neurons to induce changes in excitatory synaptic transmission mediated by cAMP-PKA signaling [115].
Another example of a lncRNA involved in transcriptional regulation is RMST, which was originally discovered as a rhabdomyosarcoma associated transcript. During neuronal differentiation, RMST is significantly upregulated in human embryonic stem cells and it is able to modulate the expression of neurogenic transcription factors [116,117]. RMST is under the control of the RE1-silencing transcription factor (REST), which is a master repressor of thousands of neurospecific genes [118]. During neuronal differentiation, the repression of REST on RMST expression is released and RMST interacts with SOX2, the transcription factor that also interacts with Tuna and that has an important role in neural fate determination [114]. The complex RMST-SOX2 regulates a large number of downstream genes which are involved in neurogenesis, and RMST is particularly required for the binding activity of SOX2 to the promoter of target genes [117].
4.5. LncRNAs Regulating mRNA Stability, Translation, or Splicing
Another mechanism employed by lncRNAs to interfere with cellular functions is to regulate RNA splicing. The lncRNA Malat1 is expressed in various tissues and in the brain its levels are particularly high in the latest stages of development, when Malat1 is found within nuclear speckles [119]. Malat1 is able to recruit pre-mRNA-splicing factors to active transcription sites [120]. Gene expression profiling of Malat1-depleted HeLa cells shows that this lncRNA can interfere with the splicing of pre-mRNAs including the RNA of the calcium/calmodulin dependent protein kinase II beta (Camk2b), which is crucial for postsynaptic signaling, plasticity and synaptogenesis [67]. DNA microarray analysis in Malat1-depleted neuronal cells also showed that this lncRNA affects the expression of presynaptic proteins (such as SNAP25 and VAMP2) and other proteins relevant for neuronal activity and development, including the activity-regulated cytoskeleton-associated protein (ARC) and the axon guidance co-receptor Neuropilin-1. Additionally, Malat1 regulates synapse formation by altering transcript levels of the synaptic cell adhesion molecule-1 (SynCAM1) and the postsynaptic cell surface protein Neuroligin-1 [120] and neuritogenesis interfering with the mir-30/Spastin axis [121]. The observation that Malat1 is important for the regulation of mRNAs encoding synaptic proteins is further confirmed by the fact that synaptic density is reduced by Malat1 knock-down, while, when Malat1 is overexpressed, the density of presynaptic boutons increases [120]. Puzzlingly, Malat1 knockout mice show no distinct phenotype or histological abnormalities [122,123,124] and its depletion does not affect pre-mRNA splicing or global gene expression pattern. To further complicate this picture, a cis-regulatory effect of Malat1 was observed on the close locus of the lncRNA NEAT1 [124]. The exact molecular mechanism of action of Malat1 remain to be established.
Similar to Malat1, the lncRNA Gomafu localizes in the nucleus and is expressed in pyramidal neurons in the cerebral cortex and CA1 pyramidal neurons in the hippocampus [125]. Early studies described the role of Gomafu in mammalian retinal development and oligodendrocyte lineage specification [125,126]. A significant activity-dependent downregulation of Gomafu and subsequent release of splicing factors was observed in transcriptome analyses upon neuronal depolarization in both primary mouse neurons and induced pluripotent stem cell (iPSC)-derived neurons systems [50]. Studies in vivo revealed that Gomafu acts as a scaffold for the splicing proteins QKI and SRSF1, and interferes with the splicing of a set of genes involved in embryonic brain development and neural progenitor proliferation including Wnt7b, Disc1 and Erbb4 [40,50]. In this context Gomafu functions to balance proliferation and differentiation of neural progenitor cells through regulating splice variations of genes involved in neuronal cell fate determination [40].
The lncRNA Pinky (Pnky) is also involved in the early stages of neuronal differentiation. This lncRNA is highly expressed in the ventricular and the subventricular zone of adult mice and it has high levels in the developing brain. Pnky regulates neural stem cell self-renewal and its expression is decreased during the differentiation of neural stem cells. Loss of function experiments showed that Pnky knockdown increased neurogenesis in both the developing embryonic and postnatal cortex [127]. Functional characterization revealed that Pnky interacts with a splicing factor, PTBP1, known as a repressor of neuronal differentiation and its deletion in vivo has no effect on the expression of the nearby Pou3f2 transcript, advocating for a bona fide trans mechanism of activity [128].
LncRNAs can also indirectly increase the expression levels of target mRNAs by acting as molecular sponges or decoys for microRNAs (miRNAs) that, if not blocked, would lead to mRNA degradation. For instance, the neurodevelopment lncRNA ‘LncND’ is expressed in neuron progenitor cells and its levels decrease during the differentiation of neurons. LncND works as a sponge for miR-143-3p and is particularly efficient since it contains 16 complementary elements for this miRNA. As an example, by blocking miR-143-3p, LncND regulates the levels of the Notch signaling receptors NOTCH-1 and NOTCH-2. Although LncND is absent in mice, its overexpression in mouse radial glia cells (RGC) resulted in the expansion of PAX6-positive RGC by interacting with the highly conserved miRNA-143-3p in the developing mouse cortex. These results imply the role of LncND in cerebral cortex expansion through miR-mediated regulation of Notch signaling [129]. Another miRNA decoy, implicated in brain development, is lncRNA-1604. This lncRNA is necessary for ectoderm differentiation of mouse ESCs. Functional analyses reveal that lncRNA-1604 is a decoy for miR-200c, and it reduces the degradative potential of miR-200c on the mRNAs of the transcription factor ZEB1/2, which promotes neural differentiation [130]. Another recent example of a lncRNA that functions as a miRNA sponge is Synage, which regulates synapse development in the cerebellum by titrating miR-325-3p [131]. Through this mechanism, Synage increases the expression of the Cerebellin 1 Precursor transcript (Cbln1), which is essential for maintaining climbing fiber-Purkinje cell synapses. The KO of Synage in mice leads to cerebellar atrophy and neuron loss. Synage is one of the few examples of predominantly cytosolic lncRNAs, and it has been suggested to have an additional function as organizer of the synaptic scaffold [131]. Among other examples of lncRNAs acting in a similar manner, it is worth mentioning Arrl1, which acts as a sponge of miR-761 that would decrease the levels of Cdkn2b and thus inhibit neurite outgrowth after neuronal injury [132].
4.6. LncRNAs Directly Modulating Protein Function
It is more and more recognized that some lncRNAs have evolved the ability to sequester proteins and interfere with their function [133,134]. In the brain, one of the few examples of lncRNAs able to directly regulate extranuclear protein function is the lncRNA NEAT1, which is also found in paraspeckles with Malat1 [123]. NEAT1 expression is activity-dependent and its levels are high at rest while they decrease upon neuronal depolarization. Outside the nucleus, NEAT1 binds two potassium channel-interacting proteins, KCNAB2 and KCNIP1. Downregulation of NEAT1 combined with neuronal depolarization results in the increase of the cytoplasmic concentration of these channel-interacting proteins, thus modulating neuronal excitability. As observed in other cases, the function of lncRNAs seems to be pleiotropic, as exemplified in this case by the observation that NEAT1 controls the expression of ion channel related genes [134] and that it can interfere with gene transcription and histone 3 lysine 9 dimethylation (H3K9me2), a histone modification that represses gene function impairing memory formation in young adult mice [135].
During the characterization of lncRNAs implicated in neurotransmitter and synaptic vesicle release, our group has identified neuroLNC, a nuclear lncRNA that is highly neuron-specific and that is conserved from rodents to humans [136]. NeuroLNC influences several aspects of neuronal physiology such as calcium influx, neurite elongation and neuronal migration in the developing brain [136]. Of note, we found that, in neurons, neuroLNC functions by interacting with the RNA-binding protein TAR DNA binding protein-43 (TDP-43), leading to the specific stabilization of mRNAs encoding for presynaptic proteins. Mutating the UG-repeats which are essential for the binding of neuroLNC to TDP-43 ablates the ability of neuroLNC to promote synaptic vesicle release when overexpressed. Likewise, the down-regulation of TDP-43 abolishes the effects of neuroLNC, reveling a clear dependency between the function of neuroLNC and TDP-43. Another recent example of lncRNAs associated to neuronal activity modulation is ADEPTR, which can be quickly produced and transported to spines in a KIF2A-dependent mechanism requiring cAMP/PKA signaling [137]. ADEPTR interacts with spectrin 1 and ankyrin B to regulate their localization and tune spine dynamics [137].
In neurons, the tuning of local translation is associated with activity-dependent synaptic plasticity to regulate the expression of mRNAs encoding synaptic proteins [138]. Deregulation of neuronal protein synthesis increases neuronal hyperexcitability and seizure susceptibility [139,140]. Cytoplasmic lncRNA-like molecules might be crucial players as they display roles in the regulation of local mRNA translation. As an example, BC1 or its primate analog, BC200, two neuron-specific non-coding RNAs, have been proposed to locally regulate protein translation in dendrites. BC1 measures 152 nucleotides and BC200 200 nucleotides, and although often referred to as lncRNAs, since lncRNAs are defined as molecules > 200 nucleotides, we refer to these as ‘non-protein coding RNAs’. These molecules were initially described as taking part in a dendritic ribonucleoprotein (RNP) complex in, regulating translation initiation in response to synaptic stimuli by interacting with SRP9/14, eIF4A, eIF4B, PABP, FMRP and SYNCRIP that interfere with 48S initiation complex formation [141,142,143,144,145,146,147,148]. Moreover, it was shown that BC1/BC200 RNA can inhibit translation of dendritic protein synthesis affecting ARC and CaMKII levels [145]. It is important to underline that the relevance of these results needs to be very carefully weighted, since the interaction of FMRP with BC1/BC200 has been clearly confuted by several subsequent works, indicating that FMRP and BC1/BC200 act independently and do not in fact bind each other specifically, neither in vitro nor in vivo [149,150,151]. Similarly, the interaction between BC200 and SYNCRIP has also been argued [152]. In any case, BC1/BC200 seems to be implicated in neuronal function since the knockout mice show hyperexcitability and neuronal plasticity changes which can probably be reconducted to altered translation of group I metabotropic glutamate receptors (mGluRs) [153].
In all these cases showcasing examples of direct interaction between lncRNAs and proteins, only few direct examples of extranuclear interactions of lncRNAs and proteins are found. It is tantalizing to speculate that some lncRNAs might be implicated in the direct regulation of phase separation and condensate formation at the synapse [154], in a similar manner to what has been observed in the nucleus [155], although direct proof is still missing. The most relevant lncRNAs acting in the brain and in neurons are summarized in Table 1.

Table 1.
Summary of relevant neuronal lncRNAs including interactors, molecular targets and function.
5. LncRNAs in Brain Disease and Neurodegeneration: Biomarkers or Active Players?
Considering the importance of lncRNAs in neuronal physiology it is not surprising that dysregulation of lncRNAs in the brain might be linked to neurodevelopmental disorders [160,161], autism spectrum disorders [162,163,164], substance use [165], intellectual disability [166], neurodegenerative diseases and neurological disorders [167,168]. A growing number of studies report that lncRNAs are associated with the pathology and development of neurodegenerative diseases [80,114,169,170]. Since the therapeutic windows for neurodegenerative diseases are narrow and current treatments only delay their progression, understanding the etiology of these diseases would provide new therapeutic approaches. Importantly, lncRNAs can be detected in easily-accessible biological fluids including blood and plasma by sequencing technologies that are sensitive and compatible with the clinical environment, hence they seem to be reliable biomarkers as also observed in the field of cancer biology [171].
Biomarker signatures based on lncRNAs could be used for detecting neurodegenerative processes very early, when clinical symptoms are not yet apparent, and degenerative processes are not yet irreversible. Moreover, due to the difficult accessibility of brain tissue in patients, monitoring the expression of biomarker lncRNAs from peripheral fluids would allow to more reliably follow the progression of the pathology and possibly even understand which cell types are more actively contributing to the neurodegenerative process. Overall, for these reasons, lncRNAs hold great potential as biomarkers for neurodegenerative diseases. At the same time, one aspect that still requires thorough examination, is whether the expression of lncRNAs during neurodegenerative processes just correlates with brain alterations or if in some cases it might also actively contribute to the pathogenesis. In other words, if lncRNAs have a functional role in the pathology in some instances, they could be exploited as possible therapeutic targets. We will summarize here observations suggesting that only a few lncRNAs show a direct involvement in neurodegenerative alterations.
The most common neurodegenerative disease, accounting for around 80% of dementia cases in the elderly population, is Alzheimer’s Disease (AD) [172]. In the APP/PS1 mice, which are used as AD model, several lncRNAS are differentially expressed, providing a number of possible biomarkers to screen in the human pathology [173]. Inhibition of the lncRNA BACE1-AS in vivo drives the reduction of β-amyloid synthesis and aggregation in the brain by decreasing the level of Bace1. As previously mentioned, BACE1-AS stabilizes BACE1 which is the enzyme cleaving amyloid precursor protein (APP) and producing the amyloid β precursors. It seems that BACE1-AS is involved in a positive feedback loop with β-amyloid peptides, so that high levels of BACE1-AS in AD results in amyloid beta-peptide synthesis and, subsequently, amyloid beta-peptide induces elevated BACE1-AS expression [80]. BC200, another non-protein coding RNA, negatively regulated over aging, is significantly upregulated in AD [174]. The increased level of BC200 and its delocalization in perikaryal structures was also correlated with AD severity [174]. The dysregulation of lncRNAs, such as lncRNA-17A, 51A, and Sox2OT in AD are also reported in other studies [175,176,177]. Besides BACE1-AS, in all other examples, the correlations between lncRNA levels and the presence of pathological alterations cannot be considered sufficient indication for a direct involvement in AD and targeted studies are required to clarify if the modulation of these lncRNAs has a direct role in the modulation of the pathology.
The lncRNA AS-Uchl1 is an antisense RNA that seems to increase the translation of UCHL1, a deubiquitinating enzyme whose reduction is associated with neurodegenerative diseases and mutations in the Uchl1 gene are found in familial Parkinson’s disease (PD) [178,179,180,181]. In a rotenone-induced PD mouse model, the lncRNA AS-Uchl1 was significantly downregulated in dopaminergic neurons indicating a possible direct role in the pathology [180]. Several other lncRNAs show significant changes in PD. As an example, the expression of lincRNA-p21, Malat1, SNHG1, HOTAIR and TncRNA are upregulated in PD, while the two lncRNAs upstream of the lncRNA H19 (Huc 1 and 2) are downregulated [182,183]. Malat1 in this context has been proposed to have a detrimental role and promote inflammasome activation and reactive oxygen species (ROS) production by interfering with the expression of NRF2, a transcription factor that controls the expression of antioxidant proteins during inflammation [184]. A recent work indicates that HOTAIR might act as a regulator of miR-126-5p which in turn targets the RAB3A interacting protein RAB3IP in PD cells and in a PD mouse model where the pathology is induced with the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) [185]. RAB3IP in PD cells seems to worsen the pathology via a mechanism that inhibits neuron autophagy and increases apoptosis [185]. Among the lncRNAs increased in PD, LINC-PINT was recently found to correlate with neuronal maturation and increase in levels in several brain regions of neurodegenerative diseases [158]. Its depletion in SH-SY5Y cells leads to increased cell death, suggesting that it might have a protective role and thus is being induced due to increased cellular stress during neurodegeneration [158].
Huntington disease (HD) is an inherited neurodegenerative disorder with progressive chorea and cognitive impairment. Patient postmortem HD brains exhibited dysregulated lncRNA expression such as NEAT1 upregulation which is potentially induced by ubiquitin-proteasome system impairment in HD [186]. NEAT1 upregulation under stress has implications in cell survival pathways and might represent a neuroprotective mechanism against neuronal damage in HD, as also observed in relation to its ability to meliorate the proteostasis stress caused by TDP-43 [187]. In contrast, Tuna expression is potentially negatively associated with the severity of HD as zebrafish treated with Tuna antisense showed severe locomotor defects [114]. Of note, the transcription factor REST, a pivotal repressor of key target genes in HD, negatively regulates some of the differentially expressed lncRNAs in HD such as HAR1F and HAR1R, which are decreased in the striatum of HD patients [188]. Furthermore, the genes for the lncRNA DGCR5 and MEG3 have REST binding sites and are downregulated in HD [189].
LncRNAs have been implicated in neuronal degeneration during amyotrophic lateral sclerosis (ALS) and in motoneuron pathology [190,191]. Studying a cell line expressing a mutant FUS devoid of the nuclear localization signal and ALS-patient derived fibroblasts revealed that mutant FUS results in accumulation of NEAT1 isoforms and excessive paraspeckle formation which may contribute to ALS severity [192,193]. Additional studies have also demonstrated the potential role of the lncRNA ATXN2-AS in spinocerebellar ataxia type 2 which is associated with an increased risk for ALS [194]. Screening of peripheral blood mononuclear cells from sporadic ALS patients revealed that there are 293 differentially expressed lncRNAs in patients in comparison with healthy individuals reflecting the involvement of lncRNAs in ALS [195]. In all these examples there is only a correlation between lncRNA expression and pathology.
Dysregulation of lncRNAs has been associated with synapse function and neurological disorders such as epilepsy. Evf2, as described above, is a crucial lncRNA in GABAergic synaptic transmission of medial prefrontal cortex and hippocampal neurons. In vivo analyses of Evf2 showed that its downregulation increases seizure susceptibility and severity in the adult mouse brain [74]. In contrast, NEAT1, which regulates potassium channel function, is upregulated in human epilepsy samples and increases seizure susceptibility [134]. Ion channel dysregulation is also implicated in neuropathic pain. Reduced potassium current after peripheral nerve injury contributes to the neuropathic pain via KCNA2-AS induction, as also discussed earlier. Notably, hypersensitivity and neuropathic pain could be decreased by blocking KCNA2-AS expression, opening a new therapeutic window in relieving neuropathic pain [84]. The imprinted lncRNA H19 has also been implicated in epilepsy, since its expression is strongly upregulated in a rat model of temporal lobe epilepsy [196]. Its function is probably mediated by a sponging effect on the microRNA let-7b which is a known regulator of the cell cycle in the developing cerebral cortex [196,197].
The lncRNA lnc-NR2F1 is a conserved lncRNA that was recently found to be associated with autism spectrum disorder and intellectual disability in children and it has been observed that the lnc-NR2F1 locus is frequently mutated in these patients [159]. Importantly, functional characterization of lnc-NR2F1 revealed that this lncRNA binds to a genomic region enriched with basic helix-loop-helix motifs to regulate the transcription of genes involved in neuron maturation [159].
Emerging evidence suggests that lncRNAs are involved in various other psychiatric disorders including schizophrenia. Schizophrenia is a complex psychiatric disease with aberrant splicing of genes associated with this pathology, including Disc1 and Errb4 [198,199]. It has been shown that transcripts of these genes and their splicing variants are upregulated by Gomafu knockdown through an interaction with the splicing proteins SF1, SRSF1 and QKI [50,157]. Gomafu is also downregulated in post-mortem cortex of patients with schizophrenia [50]. In addition, the gene encoding Gomafu is embedded in a schizophrenia linked chromosomal region [200,201] and its genetic variant is associated with the risk of paranoid schizophrenia in a Chinese Han population [202], which altogether suggest a potential link of Gomafu dysregulation with schizophrenia pathogenesis.
6. Conclusions and Perspectives
The study of lncRNAs in neurological disorders using animal models and in vitro strategies is promising for defining the etiology of neuropathologies, and in the future will eventually contribute to the diagnosis and the treatment of brain diseases [203]. A recent study also indicates that the lncRNA XIST can be used to trigger silencing of chromosome 21 in Down syndrome patient-derived cells, and correct a differentiation delay observed in this model [204], opening new avenues for leveraging lncRNA biology in alleviating dosage problems in the brain. Modeling some aspects of lncRNA biology in human iPSC-derived neurons and brain organoids might be a way to overcome implementation limitations, although in vivo solutions, such as chimeric mice studies [205], will also be necessary to correctly model lncRNAs. The comparative analysis of lncRNAs across different species [53] is also a powerful approach that can be used to understand lncRNA function [18], providing information about lncRNA and genome interaction [206], and broadening the computational toolbox for the study of this molecular species. At the same time, lncRNAs are often not conserved between rodents and humans [18], rendering this task particularly complex.
One important aspect is to characterize lncRNAs in a broad and unbiased manner. Along these lines, omics studies combining lncRNA knockdown or knockout with molecular phenotyping (such as deep-sequencing libraries) are becoming available for human cell lines [207], and, although technically challenging, the application of similar approaches to iPSC-derived human neurons seems a likely future development. What renders lncRNAs particularly difficult to model and study in neurons is the fact that they often act as fine modulators of sophisticated regulatory networks and the phenotypes are subtle. For this reason, whenever possible, the best approach to study lncRNAs would be to use an inducible gene knockout model, allowing to trigger lncRNA inactivation in the right time frame thus avoiding developmental adjustment or ‘vicariation’ effects that would mask the exact role of these molecules.
In the brain, a notable recent example of the fact that lncRNAs can be part of subtle regulation of neuronal function is Cyrano, which promotes the degradation of miR-7 and thus favors the accumulation of Cdr1as, a circular RNA that modulates neuronal activity [47]. In the absence of Cyrano, Cdr1as is depleted in neurons, in concomitance with the help of a second miRNA, miR-671 [47]. This is just one example, and more generally the study of lncRNAs will require the concomitant measure of several omics layers to truly narrow down their most relevant molecular functions [136]. Of note, lncRNAs in the brain have been linked to memory formation and behavior, indicating that their role is important at the whole organism level, and not only for the fine tuning of neuronal activity [135,208]. Moreover, due to the limited number of functional studies, compared to other molecular species, lncRNAs lack extensive resources for their interpretation [209,210,211], although efforts are being made and these tools will become more accessible in the future.
One aspect that very often remains obscure in lncRNA biology is whether these molecules only show correlations with a specific pathology or phenotype or whether they are active players. This can only be addressed if their ablation or overexpression can recapitulate some of the observed changes that do correlate with their expression levels. In order to exclude that their effect is overestimated, direct KO in mice or cells with CRISPR approaches is necessary, although in some cases it leads to puzzling results, as in the case of Malat1 KO mice which show no obvious differences [122,123,124]. In light of the complex interactions between lncRNAs with the genome and their roles in nuclear organization, it is necessary to apply and further develop methods for capturing these transient interactions in the most reliable manner, using crosslinking conditions that avoid artifacts [212], and more generally to adapt and further validate the techniques to reliably study these elusive biomolecules.
For the examination of neurodegenerative diseases in clinical settings, lncRNAs hold great potential since brain lncRNAs can be detected in peripheral fluids [213] and can thus be explored as biomarkers for both following disease progression and evaluating the outcome of therapeutic regimes. It is also possible that the study of the regenerative potential of the peripheral nervous system, where several lncRNAs are expressed during the regenerative period that follows injury [214], will provide new targets that could be leveraged for driving the repair of the damaged central nervous system or possibly to revert some of the changes observed in aging [215].
Overall, we foresee that a more systematic study of lncRNAs in the pathophysiology of the brain will provide a more comprehensive representation of the complexity of brain function.
Author Contributions
Conceptualization, S.K. and E.F.F.; writing—review and editing, S.K., V.K. and E.F.F.; visualization, E.F.F.; supervision, E.F.F.; funding acquisition, E.F.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by a Schram Stiftung (T0287/35359/2020) and a Deutsche Forschungsgemeinschaft DFG grant (FO 1342/1-3) to E.F.F.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We apologize to colleagues whose work could not be cited or discussed due to space limitations. We sincerely thank Elisa Bonnin for carefully reading and commenting the final version of the manuscript. We would also like to thank Silvio O. Rizzoli and the SFB 1286 for the continuous support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.A.; Chang, H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 2014, 14, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chang, H.Y. Physiological roles of long noncoding RNAs: Insight from knockout mice. Trends Cell Biol. 2014, 24, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Mueller, A.C.; Dutta, A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription 2014, 5, e944014. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef]
- Lv, J.; Liu, H.; Huang, Z.; Su, J.; He, H.; Xiu, Y.; Zhang, Y.; Wu, Q. Long non-coding RNA identification over mouse brain development by integrative modeling of chromatin and genomic features. Nucleic Acids Res. 2013, 41, 10044–10061. [Google Scholar] [CrossRef]
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2013, 2, e01749. [Google Scholar] [CrossRef]
- Lai, K.-M.V.; Gong, G.; Atanasio, A.; Rojas, J.; Quispe, J.; Posca, J.; White, D.; Huang, M.; Fedorova, D.; Grant, C.; et al. Diverse Phenotypes and Specific Transcription Patterns in Twenty Mouse Lines with Ablated LincRNAs. PLoS ONE 2015, 10, e0125522. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Hon, C.-C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.L.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef]
- Ulitsky, I. Evolution to the rescue: Using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016, 17, 601–614. [Google Scholar] [CrossRef]
- Villegas, V.E.; Zaphiropoulos, P.G. Neighboring gene regulation by antisense long non-coding RNAs. Int. J. Mol. Sci. 2015, 16, 3251–3266. [Google Scholar] [CrossRef]
- Cao, H.; Wahlestedt, C.; Kapranov, P. Strategies to Annotate and Characterize Long Noncoding RNAs: Advantages and Pitfalls. Trends Genet. 2018, 34, 704–721. [Google Scholar] [CrossRef]
- Guo, X.; Gao, L.; Wang, Y.; Chiu, D.K.Y.; Wang, T.; Deng, Y. Advances in long noncoding RNAs: Identification, structure prediction and function annotation. Brief. Funct. Genomics 2016, 15, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Graur, D.; Zheng, Y.; Price, N.; Azevedo, R.B.R.; Zufall, R.A.; Elhaik, E. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol. Evol. 2013, 5, 578–590. [Google Scholar] [CrossRef] [PubMed]
- Brosius, J. Waste not, want not—Transcript excess in multicellular eukaryotes. Trends Genet. 2005, 21, 287–288. [Google Scholar] [CrossRef] [PubMed]
- Brosius, J.; Raabe, C.A. What is an RNA? A top layer for RNA classification. RNA Biol. 2016, 13, 140–144. [Google Scholar] [CrossRef]
- Graur, D. Corrigendum to: “An upper limit on the functional fraction of the human genome”. Genome Biol. Evol. 2019, 11, 3158. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Hernandez, N. Small nuclear RNA genes: A model system to study fundamental mechanisms of transcription. J. Biol. Chem. 2001, 276, 26733–26736. [Google Scholar] [CrossRef]
- Ip, J.Y.; Nakagawa, S. Long non-coding RNAs in nuclear bodies. Dev. Growth Differ. 2012, 54, 44–54. [Google Scholar] [CrossRef]
- Bazzini, A.A.; Johnstone, T.G.; Christiano, R.; Mackowiak, S.D.; Obermayer, B.; Fleming, E.S.; Vejnar, C.E.; Lee, M.T.; Rajewsky, N.; Walther, T.C.; et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014, 33, 981–993. [Google Scholar] [CrossRef]
- Choi, S.W.; Kim, H.W.; Nam, J.W. The small peptide world in long noncoding RNAs. Brief. Bioinform. 2019, 20, 1853–1864. [Google Scholar] [CrossRef]
- Li, J.; Liu, C. Coding or noncoding, the converging concepts of RNAs. Front. Genet. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, J.; Lian, X.; Sun, L.; Meng, K.; Chen, Y.; Sun, Z.; Yin, X.; Li, Y.; Zhao, J.; et al. A hidden human proteome encoded by “non-coding” genes. Nucleic Acids Res. 2019, 47, 8111–8125. [Google Scholar] [CrossRef]
- Hartford, C.C.R.; Lal, A. When long non-coding becomes protein-coding. Mol. Cell. Biol. 2020, 40, e00528-19. [Google Scholar] [CrossRef]
- Roth, G. Convergent evolution of complex brains and high intelligence. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015, 370, 20150049. [Google Scholar] [CrossRef]
- Chodroff, R.A.; Goodstadt, L.; Sirey, T.M.; Oliver, P.L.; Davies, K.E.; Green, E.D.; Molnár, Z.; Ponting, C.P. Long noncoding RNA genes: Conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010, 11, R72. [Google Scholar] [CrossRef]
- Liu, S.J.; Nowakowski, T.J.; Pollen, A.A.; Lui, J.H.; Horlbeck, M.A.; Attenello, F.J.; He, D.; Weissman, J.S.; Kriegstein, A.R.; Diaz, A.A.; et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016, 17, 67. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Xie, C.; Yuan, J.; Li, H.; Li, M.; Zhao, G.; Bu, D.; Zhu, W.; Wu, W.; Chen, R.; Zhao, Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014, 42, D98–D103. [Google Scholar] [CrossRef]
- Aprea, J.; Prenninger, S.; Dori, M.; Ghosh, T.; Monasor, L.S.; Wessendorf, E.; Zocher, S.; Massalini, S.; Alexopoulou, D.; Lesche, M.; et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013, 32, 3145–3160. [Google Scholar] [CrossRef]
- Ramos, A.D.; Diaz, A.; Nellore, A.; Delgado, R.N.; Park, K.-Y.; Gonzales-Roybal, G.; Oldham, M.C.; Song, J.S.; Lim, D.A. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 2013, 12, 616–628. [Google Scholar] [CrossRef]
- Lin, M.; Pedrosa, E.; Shah, A.; Hrabovsky, A.; Maqbool, S.; Zheng, D.; Lachman, H.M. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE 2011, 6, e23356. [Google Scholar] [CrossRef]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, H.; Zhang, Q.; Zhang, Y. The functions of long non-coding RNAs in neural stem cell proliferation and differentiation. Cell Biosci. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Molyneaux, B.J.; Goff, L.A.; Brettler, A.C.; Chen, H.-H.; Hrvatin, S.; Rinn, J.L.; Arlotta, P. DeCoN: Genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 2015, 85, 275–288. [Google Scholar] [CrossRef]
- Ponjavic, J.; Oliver, P.L.; Lunter, G.; Ponting, C.P. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009, 5, e1000617. [Google Scholar] [CrossRef]
- Kleaveland, B.; Shi, C.Y.; Stefano, J.; Bartel, D.P. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 2018, 174, 350–362. [Google Scholar] [CrossRef]
- Rasmussen, K.D.; Simmini, S.; Abreu-Goodger, C.; Bartonicek, N.; Di Giacomo, M.; Bilbao-Cortes, D.; Horos, R.; Von Lindern, M.; Enright, A.J.; O’Carroll, D. The miR-144/451 locus is required for erythroid homeostasis. J. Exp. Med. 2010, 207, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Goff, L.A.; Groff, A.F.; Sauvageau, M.; Trayes-Gibson, Z.; Sanchez-Gomez, D.B.; Morse, M.; Martin, R.D.; Elcavage, L.E.; Liapis, S.C.; Gonzalez-Celeiro, M.; et al. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2015, 112, 6855–6862. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Briggs, J.A.; Vanichkina, D.P.; Poth, E.M.; Beveridge, N.J.; Ratnu, V.S.; Nayler, S.P.; Nones, K.; Hu, J.; Bredy, T.W.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Lipovich, L.; Dachet, F.; Cai, J.; Bagla, S.; Balan, K.; Jia, H.; Loeb, J.A. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics 2012, 192, 1133–1148. [Google Scholar] [CrossRef]
- He, Z.; Bammann, H.; Han, D.; Xie, G.; Khaitovich, P. Conserved expression of lincRNA during human and macaque prefrontal cortex development and maturation. RNA 2014, 20, 1103–1111. [Google Scholar] [CrossRef]
- Field, A.R.; Jacobs, F.M.J.; Fiddes, I.T.; Phillips, A.P.R.; Reyes-Ortiz, A.M.; LaMontagne, E.; Whitehead, L.; Meng, V.; Rosenkrantz, J.L.; Olsen, M.; et al. Structurally conserved primate LncRNAs are transiently expressed during human cortical differentiation and influence cell-type-specific genes. Stem Cell Rep. 2019, 12, 245–257. [Google Scholar] [CrossRef]
- Lewitus, E.; Huttner, W.B. Neurodevelopmental lncRNA microsyteny conservation and mammalian brain size evolution. PLoS ONE 2015, 10, e0131818. [Google Scholar] [CrossRef][Green Version]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Chen, L.L. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long non-coding RNAs in the regulation of gene expression: Physiology and disease. Non-coding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Long noncoding RNAs: Molecular modalities to organismal functions. Annu. Rev. Biochem. 2020, 89, 283–308. [Google Scholar] [CrossRef]
- Ang, C.E.; Trevino, A.E.; Chang, H.Y. Diverse lncRNA mechanisms in brain development and disease. Curr. Opin. Genet. Dev. 2020, 65, 42–46. [Google Scholar] [CrossRef]
- Salvatori, B.; Biscarini, S.; Morlando, M. Non-coding RNAs in nervous system development and disease. Front. Cell Dev. Biol. 2020, 8, 273. [Google Scholar] [CrossRef]
- Kim, T.-K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef]
- Modarresi, F.; Faghihi, M.A.; Lopez-Toledano, M.A.; Fatemi, R.P.; Magistri, M.; Brothers, S.P.; van der Brug, M.P.; Wahlestedt, C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012, 30, 453–459. [Google Scholar] [CrossRef]
- Chalei, V.; Sansom, S.N.; Kong, L.; Lee, S.; Montiel, J.F.; Vance, K.W.; Ponting, C.P. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife 2014, 3, e04530. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Kim, T.-K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef]
- De Santa, F.; Barozzi, I.; Mietton, F.; Ghisletti, S.; Polletti, S.; Tusi, B.K.; Muller, H.; Ragoussis, J.; Wei, C.-L.; Natoli, G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010, 8, e1000384. [Google Scholar] [CrossRef]
- Bose, D.A.; Donahue, G.; Reinberg, D.; Shiekhattar, R.; Bonasio, R.; Berger, S.L. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell 2017, 168, 135–149.e22. [Google Scholar] [CrossRef]
- Carullo, N.V.N.; Simon, R.C.; Salisbury, A.J.; Revanna, J.S.; Bunner, K.D.; Savell, K.E.; Sultan, F.A.; Gersbach, C.A.; Day, J.J. Enhancer RNAs are necessary and sufficient for activity-dependent neuronal gene transcription. bioRxiv 2018, 270967. [Google Scholar] [CrossRef]
- Schaukowitch, K.; Joo, J.-Y.; Liu, X.; Watts, J.K.; Martinez, C.; Kim, T.-K. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 2014, 56, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Onoguchi, M.; Hirabayashi, Y.; Koseki, H.; Gotoh, Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc. Natl. Acad. Sci. USA 2012, 109, 16939–16944. [Google Scholar] [CrossRef] [PubMed]
- Cajigas, I.; Chakraborty, A.; Swyter, K.R.; Luo, H.; Bastidas, M.; Nigro, M.; Morris, E.R.; Chen, S.; VanGompel, M.J.W.; Leib, D.; et al. The evf2 ultraconserved enhancer lncRNA functionally and spatially organizes megabase distant genes in the developing forebrain. Mol. Cell 2018, 71, 956–972. [Google Scholar] [CrossRef]
- Bond, A.M.; Vangompel, M.J.W.; Sametsky, E.A.; Clark, M.F.; Savage, J.C.; Disterhoft, J.F.; Kohtz, J.D. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 2009, 12, 1020–1027. [Google Scholar] [CrossRef]
- Yao, P.; Lin, P.; Gokoolparsadh, A.; Assareh, A.; Thang, M.W.C.; Voineagu, I. Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nat. Neurosci. 2015, 18, 1168–1174. [Google Scholar] [CrossRef]
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, H.; Yap, C.C.; Suzuki, M.; Kawai, J.; et al. Antisense transcription in the mammalian transcriptome. Science 2005, 309, 1564–1566. [Google Scholar] [CrossRef]
- Galante, P.A.F.; Vidal, D.O.; de Souza, J.E.; Camargo, A.A.; de Souza, S.J. Sense-antisense pairs in mammals: Functional and evolutionary considerations. Genome Biol. 2007, 8, 1–14. [Google Scholar] [CrossRef]
- Spigoni, G.; Gedressi, C.; Mallamaci, A. Regulation of Emx2 expression by antisense transcripts in murine cortico-cerebral precursors. PLoS ONE 2010, 5, e8658. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; St Laurent, G.; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef]
- Scheele, C.; Petrovic, N.; Faghihi, M.A.; Lassmann, T.; Fredriksson, K.; Rooyackers, O.; Wahlestedt, C.; Good, L.; Timmons, J.A. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics 2007, 8, 74. [Google Scholar] [CrossRef]
- Ling, K.-H.; Hewitt, C.A.; Beissbarth, T.; Hyde, L.; Cheah, P.-S.; Smyth, G.K.; Tan, S.-S.; Hahn, C.N.; Thomas, T.; Thomas, P.Q.; et al. Spatiotemporal regulation of multiple overlapping sense and novel natural antisense transcripts at the Nrgn and Camk2n1 gene loci during mouse cerebral corticogenesis. Cereb. Cortex 2011, 21, 683–697. [Google Scholar] [CrossRef]
- Corbett, M.A.; Bellows, S.T.; Li, M.; Carroll, R.; Micallef, S.; Carvill, G.L.; Myers, C.T.; Howell, K.B.; Maljevic, S.; Lerche, H.; et al. Dominant KCNA2 mutation causes episodic ataxia and pharmacoresponsive epilepsy. Neurology 2016, 87, 1975–1984. [Google Scholar] [CrossRef]
- Zhao, X.; Tang, Z.; Zhang, H.; Atianjoh, F.E.; Zhao, J.-Y.; Liang, L.; Wang, W.; Guan, X.; Kao, S.-C.; Tiwari, V.; et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 2013, 16, 1024–1031. [Google Scholar] [CrossRef]
- Ling, K.-H.; Hewitt, C.A.; Beissbarth, T.; Hyde, L.; Banerjee, K.; Cheah, P.-S.; Cannon, P.Z.; Hahn, C.N.; Thomas, P.Q.; Smyth, G.K.; et al. Molecular networks involved in mouse cerebral corticogenesis and spatio-temporal regulation of Sox4 and Sox11 novel antisense transcripts revealed by transcriptome profiling. Genome Biol. 2009, 10, 1–31. [Google Scholar] [CrossRef]
- Cohen-Cory, S.; Kidane, A.H.; Shirkey, N.J.; Marshak, S. Brain-derived neurotrophic factor and the development of structural neuronal connectivity. Dev. Neurobiol. 2010, 70, 271–288. [Google Scholar] [CrossRef]
- Autry, A.E.; Monteggia, L.M. Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacol. Rev. 2012, 64, 238–258. [Google Scholar] [CrossRef]
- Kowiański, P.; Lietzau, G.; Czuba, E.; Waśkow, M.; Steliga, A.; Moryś, J. BDNF: A Key Factor with Multipotent Impact on Brain Signaling and Synaptic Plasticity. Cell. Mol. Neurobiol. 2018, 38, 579–593. [Google Scholar] [CrossRef]
- Rusconi, F.; Battaglioli, E.; Venturin, M. Psychiatric disorders and lncrnas: A synaptic match. Int. J. Mol. Sci. 2020, 21, 3030. [Google Scholar] [CrossRef]
- Pedram Fatemi, R.; Salah-Uddin, S.; Modarresi, F.; Khoury, N.; Wahlestedt, C.; Faghihi, M.A. Screening for Small-Molecule Modulators of Long Noncoding RNA-Protein Interactions Using AlphaScreen. J. Biomol. Screen. 2015, 20, 1132–1141. [Google Scholar] [CrossRef]
- Bohnsack, J.P.; Teppen, T.; Kyzar, E.J.; Dzitoyeva, S.; Pandey, S.C. The lncRNA BDNF-AS is an epigenetic regulator in the human amygdala in early onset alcohol use disorders. Transl. Psychiatry 2019, 9, 34. [Google Scholar] [CrossRef]
- Modarresi, F.; Faghihi, M.A.; Patel, N.S.; Sahagan, B.G.; Wahlestedt, C.; Lopez-Toledano, M.A. Knockdown of BACE1-AS nonprotein-coding transcript modulates beta-amyloid-related hippocampal neurogenesis. Int. J. Alzheimers. Dis. 2011, 2011, 929042. [Google Scholar] [CrossRef]
- Landers, M.; Bancescu, D.L.; Le Meur, E.; Rougeulle, C.; Glatt-Deeley, H.; Brannan, C.; Muscatelli, F.; Lalande, M. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004, 32, 3480–3492. [Google Scholar] [CrossRef]
- Maranga, C.; Fernandes, T.G.; Bekman, E.; da Rocha, S.T. Angelman syndrome: A journey through the brain. FEBS J. 2020, 287, 2154–2175. [Google Scholar] [CrossRef]
- Meng, L.; Person, R.E.; Huang, W.; Zhu, P.J.; Costa-Mattioli, M.; Beaudet, A.L. Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS Genet. 2013, 9, e1004039. [Google Scholar] [CrossRef]
- Meng, L.; Ward, A.J.; Chun, S.; Bennett, C.F.; Beaudet, A.L.; Rigo, F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 2015, 518, 409–412. [Google Scholar] [CrossRef]
- Alfano, G.; Vitiello, C.; Caccioppoli, C.; Caramico, T.; Carola, A.; Szego, M.J.; McInnes, R.R.; Auricchio, A.; Banfi, S. Natural antisense transcripts associated with genes involved in eye development. Hum. Mol. Genet. 2005, 14, 913–923. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Poth, E.M.; Zhu, H.; Blackshaw, S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011, 6, 32. [Google Scholar] [CrossRef]
- Geng, X.; Lavado, A.; Lagutin, O.V.; Liu, W.; Oliver, G. Expression of Six3 Opposite Strand (Six3os) during mouse embryonic development. Gene Expr. Patterns 2007, 7, 252–257. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Gil, N.; Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.S.; Ruthenburg, A.J. Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep. 2015, 12, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.S.; Sullivan, M.A.; Shah, R.N.; Nadadur, R.D.; Grzybowski, A.T.; Galat, V.; Moskowitz, I.P.; Ruthenburg, A.J. Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat. Struct. Mol. Biol. 2017, 24, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Sansom, S.N.; Lee, S.; Chalei, V.; Kong, L.; Cooper, S.E.; Oliver, P.L.; Ponting, C.P. The long non-coding RNA paupar regulates the expression of both local and distal genes. EMBO J. 2014, 33, 296–311. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xi, J.; Wang, G.; Guo, Z.; Sun, Q.; Lu, C.; Ma, L.; Wu, Y.; Jia, W.; Zhu, S.; et al. PAUPAR and PAX6 sequentially regulate human embryonic stem cell cortical differentiation. Nucleic Acids Res. 2021, 49, 1935–1950. [Google Scholar] [CrossRef] [PubMed]
- Pavlaki, I.; Alammari, F.; Sun, B.; Clark, N.; Sirey, T.; Lee, S.; Woodcock, D.J.; Ponting, C.P.; Szele, F.G.; Vance, K.W. The long non-coding RNA Paupar promotes KAP1-dependent chromatin changes and regulates olfactory bulb neurogenesis. EMBO J. 2018, 37, e98219. [Google Scholar] [CrossRef] [PubMed]
- Maenner, S.; Blaud, M.; Fouillen, L.; Savoye, A.; Marchand, V.; Dubois, A.; Sanglier-Cianférani, S.; Van Dorsselaer, A.; Clerc, P.; Avner, P.; et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010, 8, e1000276. [Google Scholar] [CrossRef]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef]
- Kohyama, J.; Kojima, T.; Takatsuka, E.; Yamashita, T.; Namiki, J.; Hsieh, J.; Gage, F.H.; Namihira, M.; Okano, H.; Sawamoto, K.; et al. Epigenetic regulation of neural cell differentiation plasticity in the adult mammalian brain. Proc. Natl. Acad. Sci. USA 2008, 105, 18012–18017. [Google Scholar] [CrossRef]
- Ziller, M.J.; Edri, R.; Yaffe, Y.; Donaghey, J.; Pop, R.; Mallard, W.; Issner, R.; Gifford, C.A.; Goren, A.; Xing, J.; et al. Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature 2015, 518, 355–359. [Google Scholar] [CrossRef]
- Feng, J.; Fouse, S.; Fan, G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr. Res. 2007, 61, 58–63. [Google Scholar] [CrossRef]
- He, D.; Wang, J.; Lu, Y.; Deng, Y.; Zhao, C.; Xu, L.; Chen, Y.; Hu, Y.-C.; Zhou, W.; Lu, Q.R. lncRNA Functional Networks in Oligodendrocytes Reveal Stage-Specific Myelination Control by an lncOL1/Suz12 Complex in the CNS. Neuron 2017, 93, 362–378. [Google Scholar] [CrossRef]
- Rea, J.; Menci, V.; Tollis, P.; Santini, T.; Armaos, A.; Garone, M.G.; Iberite, F.; Cipriano, A.; Tartaglia, G.G.; Rosa, A.; et al. HOTAIRM1 regulates neuronal differentiation by modulating NEUROGENIN 2 and the downstream neurogenic cascade. Cell Death Dis. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Lin, N.; Chang, K.Y.; Li, Z.; Gates, K.; Rana, Z.; Dang, J.; Zhang, D.; Han, T.; Yang, C.S.; Cunningham, T.J.; et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 2014, 53, 1005–1019. [Google Scholar] [CrossRef]
- Raveendra, B.L.; Swarnkar, S.; Avchalumov, Y.; Liu, X.-A.; Grinman, E.; Badal, K.; Reich, A.; Pascal, B.D.; Puthanveettil, S.V. Long noncoding RNA GM12371 acts as a transcriptional regulator of synapse function. Proc. Natl. Acad. Sci. USA 2018, 115, E10197–E10205. [Google Scholar] [CrossRef]
- Ng, S.-Y.; Johnson, R.; Stanton, L.W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012, 31, 522–533. [Google Scholar] [CrossRef]
- Ng, S.Y.; Bogu, G.K.; Soh, B.; Stanton, L.W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 2013, 51, 349–359. [Google Scholar] [CrossRef]
- Baldelli, P.; Meldolesi, J. The transcription repressor REST in adult neurons: Physiology, pathology, and diseases. eNeuro 2015, 2. [Google Scholar] [CrossRef]
- Mercer, T.R.; Qureshi, I.A.; Gokhan, S.; Dinger, M.E.; Li, G.; Mattick, J.S.; Mehler, M.F. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010, 11, 14. [Google Scholar] [CrossRef]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef]
- Jiang, T.; Cai, Z.; Ji, Z.; Zou, J.; Liang, Z.; Zhang, G.; Liang, Y.; Lin, H.; Tan, M. The lncRNA MALAT1/miR-30/Spastin Axis Regulates Hippocampal Neurite Outgrowth. Front. Cell. Neurosci. 2020, 14, 1–13. [Google Scholar] [CrossRef]
- Eißmann, M.; Gutschner, T.; Hämmerle, M.; Günther, S.; Caudron-Herger, M.; Groß, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Zörnig, M.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087. [Google Scholar] [CrossRef]
- Nakagawa, S.; Ip, J.Y.; Shioi, G.; Tripathi, V.; Zong, X.; Hirose, T.; Prasanth, K.V. Malat1 is not an essential component of nuclear speckles in mice. RNA 2012, 18, 1487–1499. [Google Scholar] [CrossRef]
- Zhang, B.; Arun, G.; Mao, Y.S.; Lazar, Z.; Hung, G.; Bhattacharjee, G.; Xiao, X.; Booth, C.J.; Wu, J.; Zhang, C.; et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012, 2, 111–123. [Google Scholar] [CrossRef]
- Sone, M.; Hayashi, T.; Tarui, H.; Agata, K.; Takeichi, M.; Nakagawa, S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007, 120, 2498–2506. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Poth, E.M.; Blackshaw, S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 2010, 10, 49. [Google Scholar] [CrossRef]
- Ramos, A.D.; Andersen, R.E.; Liu, S.J.; Nowakowski, T.J.; Hong, S.J.; Gertz, C.C.; Salinas, R.D.; Zarabi, H.; Kriegstein, A.R.; Lim, D.A. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 2015, 16, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.E.; Hong, S.J.; Lim, J.J.; Cui, M.; Harpur, B.A.; Hwang, E.; Delgado, R.N.; Ramos, A.D.; Liu, S.J.; Blencowe, B.J.; et al. The Long Noncoding RNA Pnky Is a Trans-acting Regulator of Cortical Development In Vivo. Dev. Cell 2019, 49, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Rani, N.; Nowakowski, T.J.; Zhou, H.; Godshalk, S.E.; Lisi, V.; Kriegstein, A.R.; Kosik, K.S. A Primate lncRNA Mediates Notch Signaling during Neuronal Development by Sequestering miRNA. Neuron 2016, 90, 1174–1188. [Google Scholar] [CrossRef] [PubMed]
- Weng, R.; Lu, C.; Liu, X.; Li, G.; Lan, Y.; Qiao, J.; Bai, M.; Wang, Z.; Guo, X.; Ye, D.; et al. Long Noncoding RNA-1604 Orchestrates Neural Differentiation through the miR-200c/ZEB Axis. Stem Cells 2018, 36, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Q.; Liu, B.; Mei, L.; Ma, S.; Wang, S.; Wang, R.; Zhang, Y.; Niu, C.; Xiong, Z.; et al. The long noncoding RNA Synage regulates synapse stability and neuronal function in the cerebellum. Cell Death Differ. 2021. [Google Scholar] [CrossRef]
- Wang, D.; Chen, Y.; Liu, M.; Cao, Q.; Wang, Q.; Zhou, S.; Wang, Y.; X, S.M.; Gu, X.; Luo, Z.; et al. cro The long noncoding RNA Arrl1 inhibits neurite outgrowth by functioning as a competing endogenous RNA during neuronal regeneration in rats. J. Biol. Chem. 2020, 295, 8374–8386. [Google Scholar] [CrossRef]
- Lee, S.; Kopp, F.; Chang, T.-C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef]
- Barry, G.; Briggs, J.A.; Hwang, D.W.; Nayler, S.P.; Fortuna, P.R.J.; Jonkhout, N.; Dachet, F.; Maag, J.L.V.; Mestdagh, P.; Singh, E.M.; et al. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci. Rep. 2017, 7, 40127. [Google Scholar] [CrossRef]
- Butler, A.A.; Johnston, D.R.; Kaur, S.; Lubin, F.D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 2019, 12, eaaw9277. [Google Scholar] [CrossRef]
- Keihani, S.; Kluever, V.; Mandad, S.; Bansal, V.; Rahman, R.-U.; Fritsch, E.; Gomes, L.C.; Gärtner, A.; Kügler, S.; Urlaub, H.; et al. The long noncoding RNA neuroLNC regulates presynaptic activity by interacting with the neurodegeneration-associated protein TDP-43. Sci. Adv. 2019, 5, eaay2670. [Google Scholar] [CrossRef]
- Grinman, E.; Nakahata, Y.; Avchalumov, Y.; Espadas, I.; Swarnkar, S.; Yasuda, R.; Puthanveettil, S.V. Activity-regulated synaptic targeting of lncRNA ADEPTR mediates structural plasticity by localizing Sptn1 and AnkB in dendrites. Sci. Adv. 2021, 7, eabf0605. [Google Scholar] [CrossRef]
- Gkogkas, C.; Sonenberg, N.; Costa-Mattioli, M. Translational control mechanisms in long-lasting synaptic plasticity and memory. J. Biol. Chem. 2010, 285, 31913–31917. [Google Scholar] [CrossRef]
- Musumeci, S.A.; Bosco, P.; Calabrese, G.; Bakker, C.; De Sarro, G.B.; Elia, M.; Ferri, R.; Oostra, B.A. Audiogenic seizures susceptibility in transgenic mice with fragile X syndrome. Epilepsia 2000, 41, 19–23. [Google Scholar] [CrossRef]
- Zhong, J.; Chuang, S.-C.; Bianchi, R.; Zhao, W.; Paul, G.; Thakkar, P.; Liu, D.; Fenton, A.A.; Wong, R.K.S.; Tiedge, H. Regulatory BC1 RNA and the fragile X mental retardation protein: Convergent functionality in brain. PLoS ONE 2010, 5, e15509. [Google Scholar] [CrossRef]
- Wang, H.; Iacoangeli, A.; Popp, S.; Muslimov, I.A.; Imataka, H.; Sonenberg, N.; Lomakin, I.B.; Tiedge, H. Dendritic BC1 RNA: Functional role in regulation of translation initiation. J. Neurosci. 2002, 22, 10232–10241. [Google Scholar] [CrossRef]
- Eom, T.; Berardi, V.; Zhong, J.; Risuleo, G.; Tiedge, H. Dual nature of translational control by regulatory BC RNAs. Mol. Cell. Biol. 2011, 31, 4538–4549. [Google Scholar] [CrossRef]
- Kremerskothen, J.; Zopf, D.; Walter, P.; Cheng, J.G.; Nettermann, M.; Niewerth, U.; Maraia, R.J.; Brosius, J. Heterodimer SRP9/14 is an integral part of the neural BC200 RNP in primate brain. Neurosci. Lett. 1998, 245, 123–126. [Google Scholar] [CrossRef]
- Muddashetty, R.; Khanam, T.; Kondrashov, A.; Bundman, M.; Iacoangeli, A.; Kremerskothen, J.; Duning, K.; Barnekow, A.; Hüttenhofer, A.; Tiedge, H.; et al. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 2002, 321, 433–445. [Google Scholar] [CrossRef]
- Zalfa, F.; Giorgi, M.; Primerano, B.; Moro, A.; Di Penta, A.; Reis, S.; Oostra, B.; Bagni, C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 2003, 112, 317–327. [Google Scholar] [CrossRef]
- Duning, K.; Buck, F.; Barnekow, A.; Kremerskothen, J. SYNCRIP, a component of dendritically localized mRNPs, binds to the translation regulator BC200 RNA. J. Neurochem. 2008, 105, 351–359. [Google Scholar] [CrossRef]
- Tiedge, H.; Chen, W.; Brosius, J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci. 1993, 13, 2382–2390. [Google Scholar] [CrossRef]
- Eom, T.; Muslimov, I.A.; Tsokas, P.; Berardi, V.; Zhong, J.; Sacktor, T.C.; Tiedge, H. Neuronal BC RNAs cooperate with eIF4B to mediate activity-dependent translational control. J. Cell Biol. 2014, 207, 237–252. [Google Scholar] [CrossRef]
- Iacoangeli, A.; Rozhdestvensky, T.S.; Dolzhanskaya, N.; Tournier, B.; Schütt, J.; Brosius, J.; Denman, R.B.; Khandjian, E.W.; Kindler, S.; Tiedge, H. On BC1 RNA and the fragile X mental retardation protein. Proc. Natl. Acad. Sci. USA 2008, 105, 734–739. [Google Scholar] [CrossRef]
- Darnell, J.C.; Fraser, C.E.; Mostovetsky, O.; Stefani, G.; Jones, T.A.; Eddy, S.R.; Darnell, R.B. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005, 19, 903–918. [Google Scholar] [CrossRef]
- Yan, X.; Denman, R.B. Conformational-dependent and independent RNA binding to the fragile x mental retardation protein. J. Nucleic Acids 2011, 2011, 246127. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Booy, E.P.; McRae, E.K.; Ezzati, P.; Choi, T.; Gussakovsky, D.; McKenna, S.A. Comprehensive analysis of the BC200 ribonucleoprotein reveals a reciprocal regulatory function with CSDE1/UNR. Nucleic Acids Res. 2018, 46, 11575–11591. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Chuang, S.-C.; Bianchi, R.; Zhao, W.; Lee, H.; Fenton, A.A.; Wong, R.K.S.; Tiedge, H. BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 2009, 29, 9977–9986. [Google Scholar] [CrossRef] [PubMed]
- Liau, W.-S.; Samaddar, S.; Banerjee, S.; Bredy, T.W. On the functional relevance of spatiotemporally-specific patterns of experience-dependent long noncoding RNA expression in the brain. RNA Biol. 2021, 18, 1025–1036. [Google Scholar] [CrossRef]
- Luo, J.; Qu, L.; Gao, F.; Lin, J.; Liu, J.; Lin, A. LncRNAs: Architectural scaffolds or more potential roles in phase separation. Front. Genet. 2021, 12, 626234. [Google Scholar] [CrossRef]
- Cajigas, I.; Leib, D.E.; Cochrane, J.; Luo, H.; Swyter, K.R.; Chen, S.; Clark, B.S.; Thompson, J.; Yates, J.R.; Kingston, R.E.; et al. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development 2015, 142, 2641–2652. [Google Scholar] [CrossRef]
- Tsuiji, H.; Yoshimoto, R.; Hasegawa, Y.; Furuno, M.; Yoshida, M.; Nakagawa, S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells 2011, 16, 479–490. [Google Scholar] [CrossRef]
- Simchovitz, A.; Hanan, M.; Yayon, N.; Lee, S.; Bennett, E.R.; Greenberg, D.S.; Kadener, S.; Soreq, H. A lncRNA survey finds increases in neuroprotective LINC-PINT in Parkinson’s disease substantia nigra. Aging Cell 2020, 19, 1–13. [Google Scholar] [CrossRef]
- Ang, C.E.; Ma, Q.; Wapinski, O.L.; Fan, S.; Flynn, R.A.; Lee, Q.Y.; Coe, B.; Onoguchi, M.; Olmos, V.H.; Do, B.T.; et al. The novel lncRNA lnc-NR2F1 is pro-neurogenic and mutated in human neurodevelopmental disorders. Elife 2019, 8, e41770. [Google Scholar] [CrossRef]
- van de Vondervoort, I.I.G.M.; Gordebeke, P.M.; Khoshab, N.; Tiesinga, P.H.E.; Buitelaar, J.K.; Kozicz, T.; Aschrafi, A.; Glennon, J.C. Long non-coding RNAs in neurodevelopmental disorders. Front. Mol. Neurosci. 2013, 6, 53. [Google Scholar] [CrossRef]
- Zhang, S.-F.; Gao, J.; Liu, C.-M. The role of non-coding RNAs in neurodevelopmental disorders. Front. Genet. 2019, 10, 1033. [Google Scholar] [CrossRef]
- Tong, Z.; Zhou, Y.; Wang, J. Identification and functional analysis of long non-coding RNAs in autism spectrum disorders. Front. Genet. 2020, 11, 849. [Google Scholar] [CrossRef]
- Bilinovich, S.M.; Lewis, K.; Grepo, N.; Campbell, D.B. The long noncoding rna rps10p2-as1 is implicated in autism spectrum disorder risk and modulates gene expression in human neuronal progenitor cells. Front. Genet. 2019, 10, 970. [Google Scholar] [CrossRef]
- Luo, T.; Ou, J.; Cao, L.; Peng, X.; Li, Y.; Tian, Y. The Autism-related lncRNA MSNP1AS regulates moesin protein to influence the RhoA, Rac1, and PI3K/Akt pathways and regulate the structure and survival of neurons. Autism Res. 2020, 13, 2073–2082. [Google Scholar] [CrossRef]
- Oliver, R.J.; Mandyam, C.D. Regulation of adult neurogenesis by non-coding RNAs: Implications for substance use disorders. Front. Neurosci. 2018, 12, 849. [Google Scholar] [CrossRef]
- Barros, I.I.; Leão, V.; Santis, J.O.; Rosa, R.C.A.; Brotto, D.B.; Storti, C.B.; Siena, Á.D.D.; Molfetta, G.A.; Silva, W.A. Non-syndromic intellectual disability and its pathways: A long noncoding RNA perspective. Non-coding RNA 2021, 7, 22. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, J. LncRNAs: Macromolecules with big roles in neurobiology and neurological diseases. Metab. Brain Dis. 2017, 32, 281–291. [Google Scholar] [CrossRef]
- Wu, Y.-Y.; Kuo, H.-C. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J. Biomed. Sci. 2020, 27, 49. [Google Scholar] [CrossRef]
- Yao, Z.-H.; Wang, J.; Shen, B.-Z.; Li, Y.-T.; Yao, X.-L.; Zhang, S.-F.; Zhang, Y.; Hu, J.-C.; Xie, Y.-C. Identification of a hippocampal lncRNA-regulating network in cognitive dysfunction caused by chronic cerebral hypoperfusion. Aging 2020, 12, 19520–19538. [Google Scholar] [CrossRef]
- Yang, S.; Lim, K.H.; Kim, S.H.; Joo, J.Y. Molecular landscape of long noncoding RNAs in brain disorders. Mol. Psychiatry 2021, 26, 1060–1074. [Google Scholar] [CrossRef]
- Qi, P.; Zhou, X.-Y.; Du, X. Circulating long non-coding RNAs in cancer: Current status and future perspectives. Mol. Cancer 2016, 15, 39. [Google Scholar] [CrossRef]
- Musiek, E.S.; Holtzman, D.M. Three dimensions of the amyloid hypothesis: Time, space and “wingmen”. Nat. Neurosci. 2015, 18, 800–806. [Google Scholar] [CrossRef]
- Ma, N.; Tie, C.; Yu, B.; Zhang, W.; Wan, J. Identifying lncRNA-miRNA-mRNA networks to investigate Alzheimer’s disease pathogenesis and therapy strategy. Aging 2020, 12, 2897–2920. [Google Scholar] [CrossRef]
- Mus, E.; Hof, P.R.; Tiedge, H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2007, 104, 10679–10684. [Google Scholar] [CrossRef]
- Massone, S.; Vassallo, I.; Fiorino, G.; Castelnuovo, M.; Barbieri, F.; Borghi, R.; Tabaton, M.; Robello, M.; Gatta, E.; Russo, C.; et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 2011, 41, 308–317. [Google Scholar] [CrossRef]
- Ciarlo, E.; Massone, S.; Penna, I.; Nizzari, M.; Gigoni, A.; Dieci, G.; Russo, C.; Florio, T.; Cancedda, R.; Pagano, A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. Dis. Model. Mech. 2013, 6, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Arisi, I.; D’Onofrio, M.; Brandi, R.; Felsani, A.; Capsoni, S.; Drovandi, G.; Felici, G.; Weitschek, E.; Bertolazzi, P.; Cattaneo, A. Gene expression biomarkers in the brain of a mouse model for Alzheimer’s disease: Mining of microarray data by logic classification and feature selection. J. Alzheimers. Dis. 2011, 24, 721–738. [Google Scholar] [CrossRef] [PubMed]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Carrieri, C.; Forrest, A.R.R.; Santoro, C.; Persichetti, F.; Carninci, P.; Zucchelli, S.; Gustincich, S. Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front. Cell. Neurosci. 2015, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Jangir, D.K.; Verma, G.; Shekhar, S.; Hanpude, P.; Kumar, S.; Kumari, R.; Singh, N.; Sarovar Bhavesh, N.; Ranjan Jana, N.; et al. S-nitrosylation of UCHL1 induces its structural instability and promotes α-synuclein aggregation. Sci. Rep. 2017, 7, 44558. [Google Scholar] [CrossRef]
- Barrachina, M.; Castaño, E.; Dalfó, E.; Maes, T.; Buesa, C.; Ferrer, I. Reduced ubiquitin C-terminal hydrolase-1 expression levels in dementia with Lewy bodies. Neurobiol. Dis. 2006, 22, 265–273. [Google Scholar] [CrossRef]
- Kraus, T.F.J.; Haider, M.; Spanner, J.; Steinmaurer, M.; Dietinger, V.; Kretzschmar, H.A. Altered Long Noncoding RNA Expression Precedes the Course of Parkinson’s Disease-a Preliminary Report. Mol. Neurobiol. 2017, 54, 2869–2877. [Google Scholar] [CrossRef]
- Liu, S.; Cui, B.; Dai, Z.; Shi, P.; Wang, Z.; Guo, Y. Long Non-coding RNA HOTAIR Promotes Parkinson’s Disease Induced by MPTP Through up-regulating the Expression of LRRK2. Curr. Neurovasc. Res. 2016, 13, 115–120. [Google Scholar] [CrossRef]
- Cai, L.J.; Tu, L.; Huang, X.M.; Huang, J.; Qiu, N.; Xie, G.H.; Liao, J.X.; Du, W.; Zhang, Y.Y.; Tian, J.Y. LncRNA MALAT1 facilitates inflammasome activation via epigenetic suppression of Nrf2 in Parkinson’s disease. Mol. Brain 2020, 13, 130. [Google Scholar] [CrossRef]
- Lin, Q.; Hou, S.; Dai, Y.; Jiang, N.; Lin, Y. LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson’s disease through RAB3IP. Biol. Chem. 2019, 400, 1217–1228. [Google Scholar] [CrossRef]
- Sunwoo, J.-S.; Lee, S.-T.; Im, W.; Lee, M.; Byun, J.-I.; Jung, K.-H.; Park, K.-I.; Jung, K.-Y.; Lee, S.K.; Chu, K.; et al. Altered Expression of the Long Noncoding RNA NEAT1 in Huntington’s Disease. Mol. Neurobiol. 2017, 54, 1577–1586. [Google Scholar] [CrossRef]
- Matsukawa, K.; Kukharsky, M.S.; Park, S.-K.; Park, S.; Watanabe, N.; Iwatsubo, T.; Hashimoto, T.; Liebman, S.W.; Shelkovnikova, T.A. Long non-coding RNA NEAT1_1 ameliorates TDP-43 toxicity in in vivo models of TDP-43 proteinopathy. RNA Biol. 2021, 1–9. [Google Scholar] [CrossRef]
- Johnson, R.; Richter, N.; Jauch, R.; Gaughwin, P.M.; Zuccato, C.; Cattaneo, E.; Stanton, L.W. Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol. Genomics 2010, 41, 269–274. [Google Scholar] [CrossRef]
- Johnson, R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 2012, 46, 245–254. [Google Scholar] [CrossRef]
- Vangoor, V.R.; Gomes-Duarte, A.; Pasterkamp, R.J. Long non-coding RNAs in motor neuron development and disease. J. Neurochem. 2021, 156, 777–801. [Google Scholar] [CrossRef]
- Chen, K.W.; Chen, J.A. Functional Roles of Long Non-coding RNAs in Motor Neuron Development and Disease. J. Biomed. Sci. 2020, 27, 1–14. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Nakagawa, S.; Hirose, T.; Okano, H.J.; Takao, M.; Shibata, S.; Suyama, S.; Kuwako, K.-I.; Imai, T.; Murayama, S.; et al. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 2013, 6, 31. [Google Scholar] [CrossRef]
- An, H.; Skelt, L.; Notaro, A.; Highley, J.R.; Fox, A.H.; La Bella, V.; Buchman, V.L.; Shelkovnikova, T.A. ALS-linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles. Acta Neuropathol. Commun. 2019, 7, 7. [Google Scholar] [CrossRef]
- Li, P.P.; Sun, X.; Xia, G.; Arbez, N.; Paul, S.; Zhu, S.; Peng, H.B.; Ross, C.A.; Koeppen, A.H.; Margolis, R.L.; et al. ATXN2-AS, a gene antisense to ATXN2, is associated with spinocerebellar ataxia type 2 and amyotrophic lateral sclerosis. Ann. Neurol. 2016, 80, 600–615. [Google Scholar] [CrossRef]
- Gagliardi, S.; Zucca, S.; Pandini, C.; Diamanti, L.; Bordoni, M.; Sproviero, D.; Arigoni, M.; Olivero, M.; Pansarasa, O.; Ceroni, M.; et al. Long non-coding and coding RNAs characterization in Peripheral Blood Mononuclear Cells and Spinal Cord from Amyotrophic Lateral Sclerosis patients. Sci. Rep. 2018, 8, 2378. [Google Scholar] [CrossRef]
- Han, C.-L.; Ge, M.; Liu, Y.-P.; Zhao, X.-M.; Wang, K.-L.; Chen, N.; Hu, W.; Zhang, J.-G.; Li, L.; Meng, F.-G. Long non-coding RNA H19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell Death Dis. 2018, 9, 617. [Google Scholar] [CrossRef] [PubMed]
- Fairchild, C.L.A.; Cheema, S.K.; Wong, J.; Hino, K.; Simó, S.; La Torre, A. Let-7 regulates cell cycle dynamics in the developing cerebral cortex and retina. Sci. Rep. 2019, 9, 15336. [Google Scholar] [CrossRef] [PubMed]
- Nakata, K.; Lipska, B.K.; Hyde, T.M.; Ye, T.; Newburn, E.N.; Morita, Y.; Vakkalanka, R.; Barenboim, M.; Sei, Y.; Weinberger, D.R.; et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc. Natl. Acad. Sci. USA 2009, 106, 15873–15878. [Google Scholar] [CrossRef] [PubMed]
- Law, A.J.; Kleinman, J.E.; Weinberger, D.R.; Weickert, C.S. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum. Mol. Genet. 2007, 16, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Potash, J.B.; Zandi, P.P.; Willour, V.L.; Lan, T.-H.; Huo, Y.; Avramopoulos, D.; Shugart, Y.Y.; MacKinnon, D.F.; Simpson, S.G.; McMahon, F.J.; et al. Suggestive linkage to chromosomal regions 13q31 and 22q12 in families with psychotic bipolar disorder. Am. J. Psychiatry 2003, 160, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, G.; Brynjolfsson, J.; Butler, R.; Sherrington, R.; Curtis, D.; Sigmundsson, T.; Read, T.; Murphy, P.; Sharma, T.; Petursson, H. Linkage analysis of chromosome 22q12-13 in a United Kingdom/Icelandic sample of 23 multiplex schizophrenia families. Am. J. Med. Genet. 1995, 60, 298–301. [Google Scholar] [CrossRef]
- Rao, S.-Q.; Hu, H.-L.; Ye, N.; Shen, Y.; Xu, Q. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr. Res. 2015, 166, 125–130. [Google Scholar] [CrossRef]
- Wang, D.Q.; Fu, P.; Yao, C.; Zhu, L.S.; Hou, T.Y.; Chen, J.G.; Lu, Y.; Liu, D.; Zhu, L.Q. Long Non-coding RNAs, Novel Culprits, or Bodyguards in Neurodegenerative Diseases. Mol. Ther. Nucleic Acids 2018, 10, 269–276. [Google Scholar] [CrossRef]
- Czermiński, J.T.; Lawrence, J.B. Silencing Trisomy 21 with XIST in Neural Stem Cells Promotes Neuronal Differentiation. Dev. Cell 2020, 52, 294–308.e3. [Google Scholar] [CrossRef]
- Vitrac, A.; Pons, S.; Balkota, M.; Lemière, N.; Raïs, C.; Bourgeois, J.-P.; Maskos, U.; Bourgeron, T.; Cloëz-Tayarani, I. A chimeric mouse model to study human iPSC-derived neurons: The case of a truncating SHANK3 mutation. Sci. Rep. 2020, 10, 13315. [Google Scholar] [CrossRef]
- Quinn, J.J.; Zhang, Q.C.; Georgiev, P.; Ilik, I.A.; Akhtar, A.; Chang, H.Y. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev. 2016, 30, 191–207. [Google Scholar] [CrossRef]
- Ramilowski, J.A.; Yip, C.W.; Agrawal, S.; Chang, J.-C.; Ciani, Y.; Kulakovskiy, I.V.; Mendez, M.; Ooi, J.L.C.; Ouyang, J.F.; Parkinson, N.; et al. Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res. 2020, 30, 1060–1072. [Google Scholar] [CrossRef]
- Ma, M.; Xiong, W.; Hu, F.; Deng, M.F.; Huang, X.; Chen, J.G.; Man, H.Y.; Lu, Y.; Liu, D.; Zhu, L.Q. A novel pathway regulates social hierarchy via lncRNA AtLAS and postsynaptic synapsin IIb. Cell Res. 2020, 30, 105–118. [Google Scholar] [CrossRef]
- Li, Y.; Chen, H.; Pan, T.; Jiang, C.; Zhao, Z.; Wang, Z.; Zhang, J.; Xu, J.; Li, X. LncRNA ontology: Inferring lncRNA functions based on chromatin states and expression patterns. Oncotarget 2015, 6, 39793–39805. [Google Scholar] [CrossRef]
- Iwakiri, J.; Hamada, M.; Asai, K. Bioinformatics tools for lncRNA research. Biochim. Biophys. Acta 2016, 1859, 23–30. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Liu, L.; Zhou, Y.; Zhang, S.; Chouvardas, P.; Johnson, R.; Li, J. LnCompare: Gene set feature analysis for human long non-coding RNAs. Nucleic Acids Res. 2019, 47, W523–W529. [Google Scholar] [CrossRef]
- Kato, M.; Carninci, P. Genome-wide technologies to study RNA-chromatin interactions. Non-coding RNA 2020, 6, 20. [Google Scholar] [CrossRef]
- Elkouris, M.; Kouroupi, G.; Vourvoukelis, A.; Papagiannakis, N.; Kaltezioti, V.; Matsas, R.; Stefanis, L.; Xilouri, M.; Politis, P.K. Long non-coding RNAs associated with neurodegeneration-linked genes are reduced in Parkinson’s disease patients. Front. Cell. Neurosci. 2019, 13, 58. [Google Scholar] [CrossRef]
- Perry, R.B.T.; Hezroni, H.; Goldrich, M.J.; Ulitsky, I. Regulation of neuroregeneration by long noncoding RNAs. Mol. Cell 2018, 72, 553–567.e5. [Google Scholar] [CrossRef]
- Jin, L.; Song, Q.; Zhang, W.; Geng, B.; Cai, J. Roles of long noncoding RNAs in aging and aging complications. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1763–1771. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).