Search for Non-Protein Protease Inhibitors Constituted with an Indole and Acetylene Core
Abstract
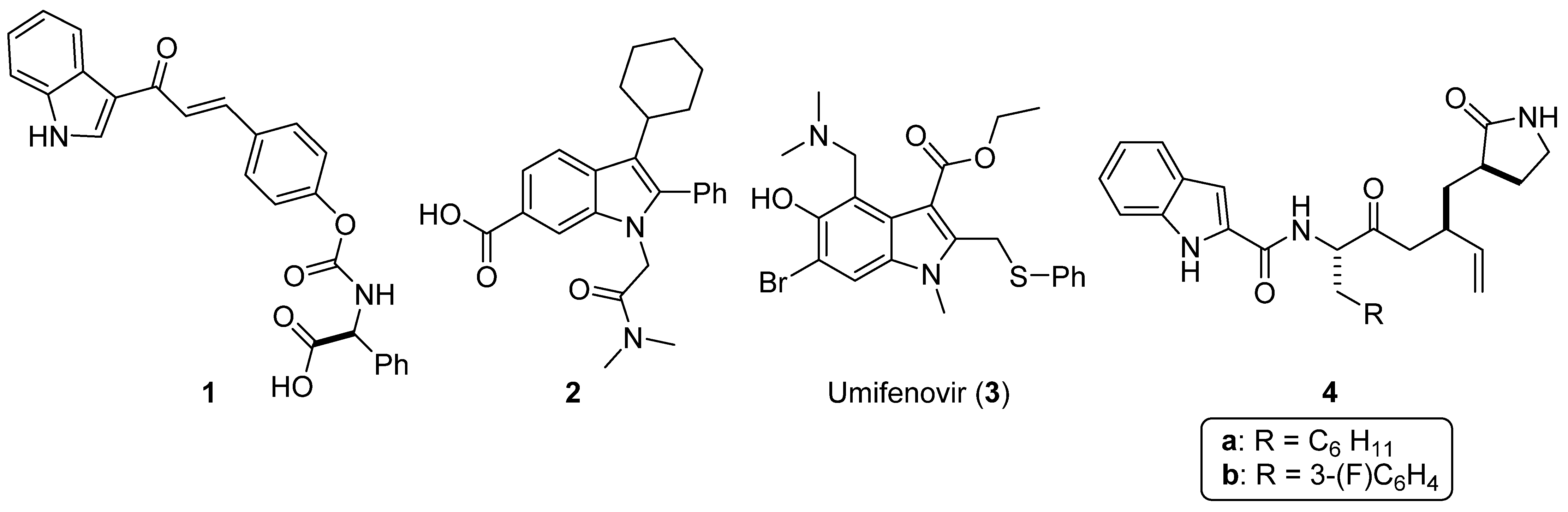
:1. Introduction
2. Results and Discussion
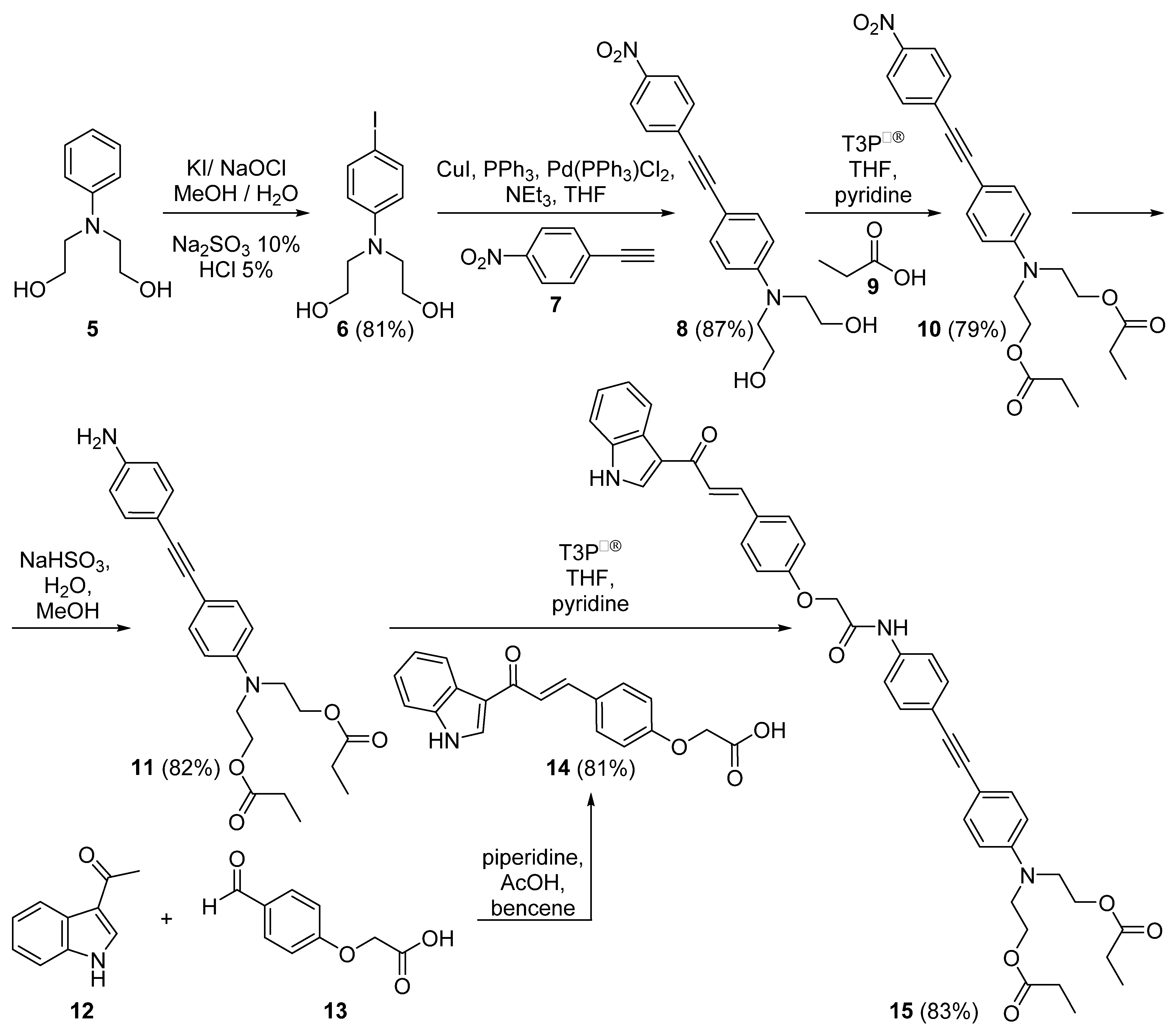
2.1. Chemical Synthesis
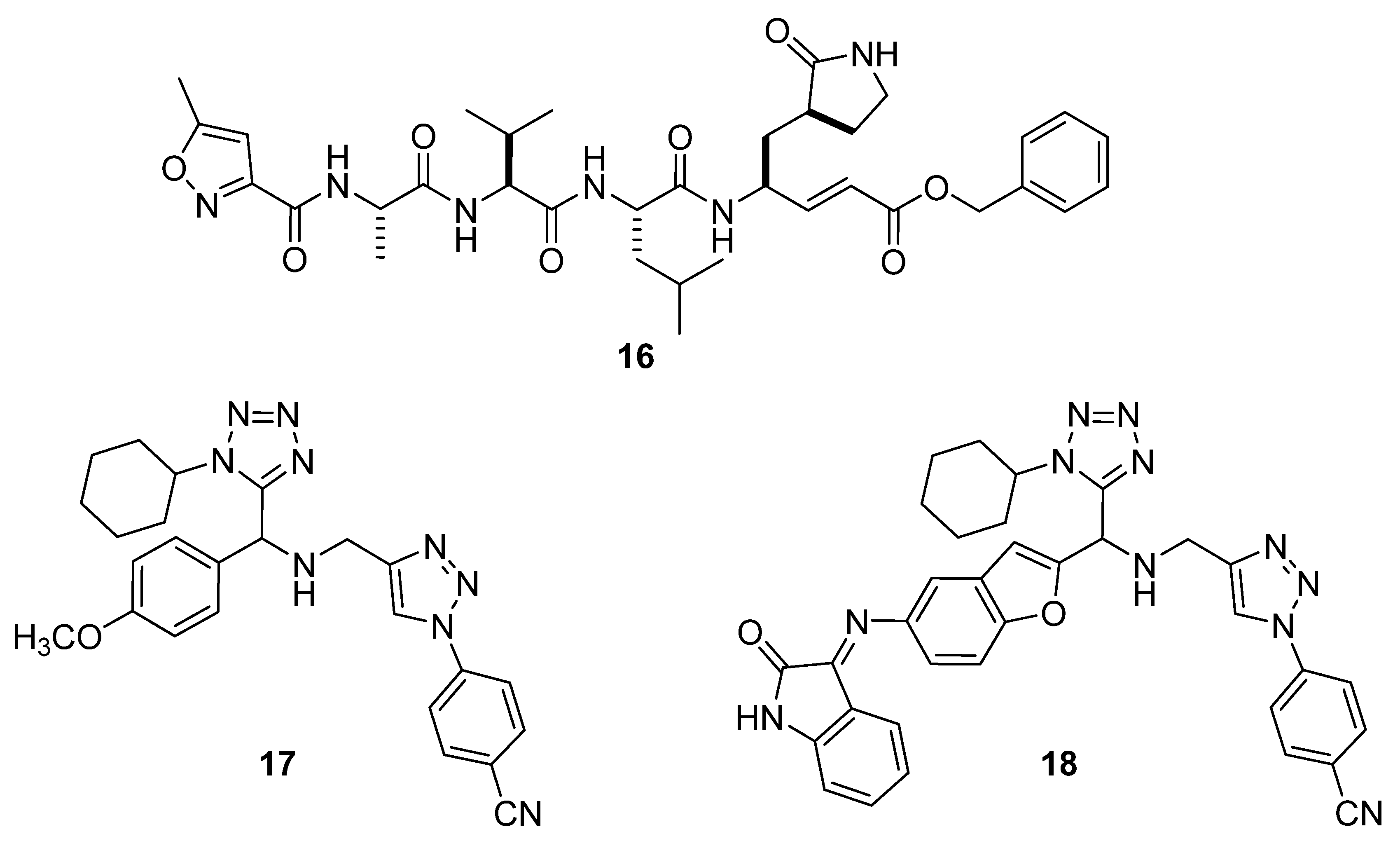
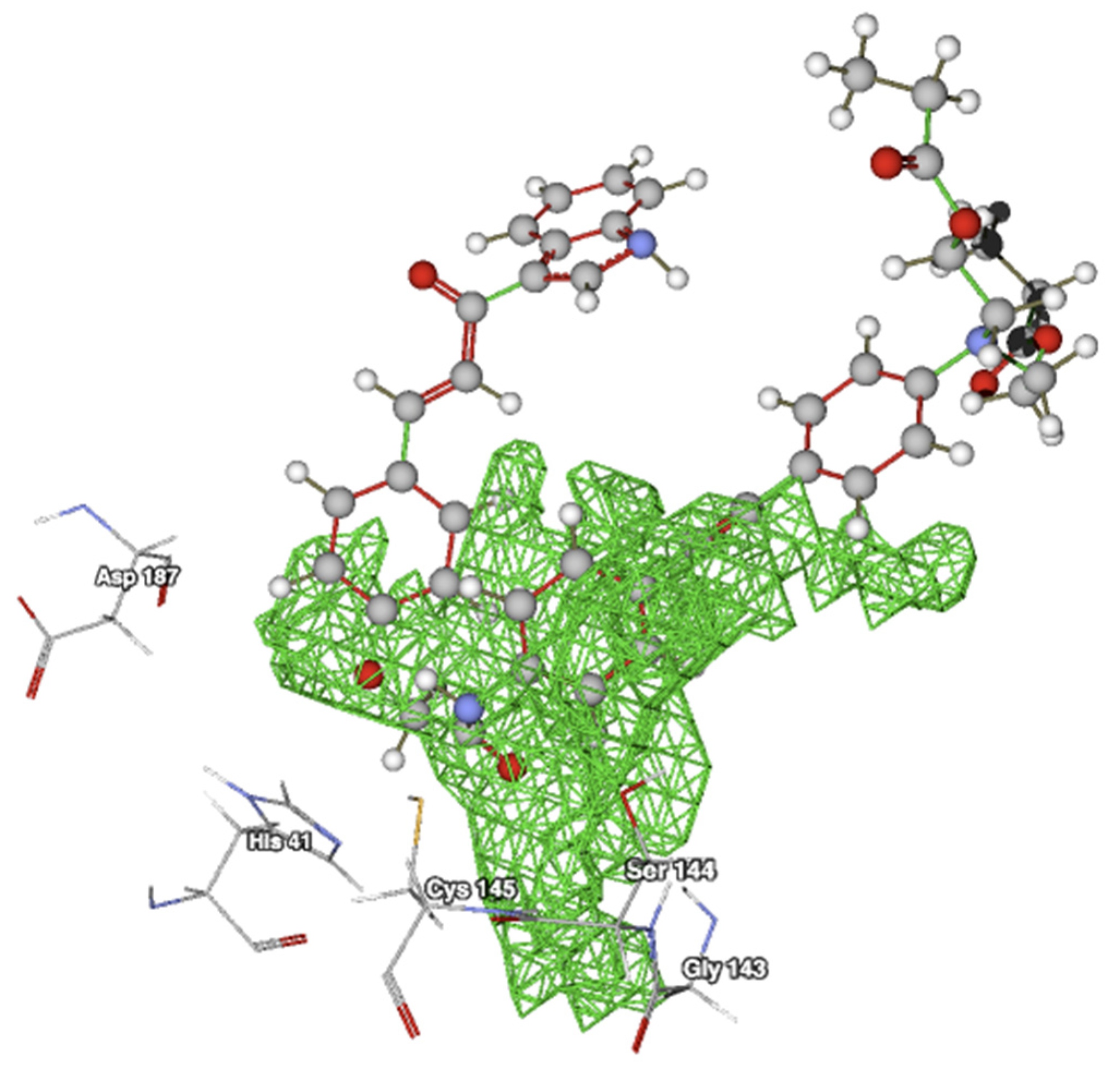
2.2. Docking Studies with SARS-CoV-2-Mpro
2.3. Docking Studies with KLK5 Protease
3. Materials and Methods
3.1. Materials and Instrumentation
3.2. Chemical Synthesis
3.2.1. 2,2′-[(4-iodophenyl)azanediyl]di(ethan-1-ol) (6)
3.2.2. 2,2′-({4-[(4-nitrophenyl)ethynyl]phenyl}azanediyl)di(ethan-1-ol) (8)
3.2.3. 2,2′-({4-[(4-nitrophenyl)ethynyl]phenyl}azanediyl)di(ethane-2,1-diyl)dipropianate (10)
3.2.4. 2,2′-({4-[(4-aminophenyl)ethynyl]phenyl}azanediyl) di(ethane-2,1-diyl)dipropianate (11)
3.2.5. {4-[(1E)-3-(1H-indol-3-yl)-3-oxoprop-1-en-1-yl]phenoxy} acetic acid (14)
3.2.6. 2,2′-[(4-{[4-({4-[(1. E)-3-(1H-indol-3-yl)-3-oxoprop-1-en-1-yl]phenoxy}acetamido)phe-nyl]ethynyl}phenyl)azanediyl]di(ethane-2,1-diyl)dipropionate (15)
3.3. Computational Assays
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abbenante, G.; Fairlie, D.P. Proteases Inhibitors in the clinic. Med. Chem. 2005, 1, 71–104. [Google Scholar] [CrossRef]
- Grzonka, Z.; Jankowska, E.; Kasprzykowski, F.; Kasprzykowska, R.; Lankiewicz, L.; Wiczk, W.; Wieczerzak, E.; Ciarkowski, J.; Drabik, P.; Janowski, R.; et al. Structural studies of cysteine proteases and their inhibitors. Acta Biochim. Pol. 2001, 48, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Fear, G.; Komarnytsky, S.; Raskin, I. Protease inhibitors and their peptidomimetic derivatives as potential drugs. Pharmacol. Ther. 2007, 113, 354–368. [Google Scholar] [CrossRef] [PubMed]
- Tong, L. Viral Proteases. Chem. Rev. 2002, 102, 4609–4626. [Google Scholar] [CrossRef] [PubMed]
- Colarusso, S.; Koch, U.; Gerlach, B.; Steinkuhler, C.; Francesco, R.D.; Altamura, S.; Matassa, V.G.; Narjes, F. Phenethyl Amides as Novel Noncovalent Inhibitors of Hepatitis C Virus NS3/4A Protease: Discovery, Initial SAR, and Molecular Modeling. J. Med. Chem. 2003, 46, 345–348. [Google Scholar] [CrossRef]
- Ismail, N.S.M.; Hatori, M. Molecular Medeling based approach, Synthesis and in vitro assay to new indole inhibitors of hepatitis C NS3/4A Serine protease. Bioorg. Med. Chem. 2011, 19, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.S.M.; Dine, R.S.E.; Hattori, M.; Takahashi, K.; Ihara, M. Computer based design, synthesis and biological evaluation of novel indole derivatives as HCV NS3-4A serine protease inhibitors. Bioorg. Med. Chem. 2008, 16, 7877–7887. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.; Pacini, B.; Avolio, S.; Filippo, M.; Migliaccio, G.; Laufer, R.; Francesco, R.; Rowley, M.; Narjes, F. Development and Preliminary Optimization of Indole-N-Acetamide Inhibitors of Hepatitis C Virus NS5B Polymerase. J. Med. Chem. 2005, 48, 1314–1317. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.; Avolio, S.; Pacini, B.; Di Filippo, M.; Altamura, S.; Tomei, L.; Paonessa, G.; Di Marco, S.; Carfi, A.; Giuliano, C.; et al. Potent Inhibitors of Subgenomic Hepatitis C Virus RNA Replication through Optimization of Indole-N-Acetamide Allosteric Inhibitors of the Viral NS5B Polymerase. J. Med. Chem. 2005, 48, 4547–4557. [Google Scholar] [CrossRef] [PubMed]
- Stansfield, I.; Pompei, M.; Conte, I.; Ercolani, C.; Migliaccio, G.; Jairaj, M.; Giuliano, C.; Rowley, M.; Narjes, F. Development of carboxylic acid replacements in indole-N-acetamide inhibitors of hepatitis C virus NS5B polymerase. Bioorg. Med. Chem. Lett. 2007, 17, 5143–5149. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, B.; Hashimoto, K. Current status of potential therapeutic candidates for the COVID-19 crisis. Brain Behav. Immun. 2020, 87, 59–73. [Google Scholar] [CrossRef] [PubMed]
- McKee, D.L.; Sternberg, A.; Stange, U.; Laufer, S.; Naujokat, C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020, 157, 104859. [Google Scholar] [CrossRef]
- Fischer, A.; Sellner, M.; Neranjan, S.; Smieško, M.; Lill, M.A. Potential Inhibitors for Novel Coronavirus Protease Identified by Virtual Screening of 606 Million Compounds. Int. J. Mol. Sci. 2020, 21, 3626. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 main protease as drug target. Bioorganic Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef]
- Zhou, Z.-F.; Menna, M.; Cai, Y.-S.; Guo, Y.-W. Polyacetylenes of Marine Origin: Chemistry and Bioactivity. Chem. Rev. 2015, 115, 1543–1596. [Google Scholar] [CrossRef]
- Minto, R.E.; Blacklock, B.J. Biosynthesis and function of polyacetylenes and allied natural products. Prog. Lipid Res. 2008, 47, 233–306. [Google Scholar] [CrossRef] [Green Version]
- Dembitsky, V.M. Anticancer Activity of Natural and Synthetic Acetylenic Lipids. Lipids 2006, 41, 883–924. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, W.; Zhang, B.; Jiang, X.-M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F.; et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianconi, V.; Violi, F.; Fallarino, F.; Pignatelli, P.; Sahebkar, A.; Pirro, M. Is Acetylsalicylic Acid a Safe and Potentially Useful Choice for Adult Patients with COVID-19? Drugs 2020, 80, 1383–1396. [Google Scholar] [CrossRef]
- Yang, L.; Liu, W.; Yu, X.; Wu, M.; Reichert, J.M.; Ho, M. COVID-19 antibody therapeutics tracker: A global online database of antibody therapeutics for the prevention and treatment of COVID-19. Antibody Ther. 2020, 3, 205–212. [Google Scholar] [CrossRef]
- Shivanika, C.; Deepak, K.S.; Ragunathan, V.; Tiwari, P.; Sumitha, A.; Brindha, D.P. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J. Biomol. Struct. Dyn. 2020, 8, 1–27. [Google Scholar] [CrossRef]
- Joshi, R.S.; Jagdale, S.S.; Bansode, S.B.; Shankar, S.S.; Tellis, M.B.; Pandya, V.K.; Chugh, A.; Giri, A.P.; Kulkarni, M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020, 5, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.; Saha, T.; Islam, K.J.; Suman, R.H.; Biswas, S.; Rahat, E.U.; Hossen, R.; Islam, R.; Hossain, N.; Al Mamun, A.; et al. Virtual screening, molecular dynamics and structure–activity relationship studies to identify potent approved drugs for Covid-19 treatment. J. Biomol. Struct. Dyn. 2020, 21, 1–11. [Google Scholar] [CrossRef]
- Umesh; Kundu, D.; Selvaraj, C.; Singh, S.K.; Dubey, V.K. Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. J. Biomol. Struct. Dyn. 2020, 2, 1–9. [Google Scholar] [CrossRef]
- Mazzini, S.; Musso, L.; Dallavalle, S.; Artali, R. Putative SARS-CoV-2 M Inhibitors from an In-House Library of Natural and Nature-Inspired Products: A Virtual Screening and Molecular Docking Study. Molecules 2020, 25, 3745. [Google Scholar] [CrossRef] [PubMed]
- Cortés-García, C.J.; Chacón-García, L.; Mejia-Benavides, J.E.; Díaz-Cervantes, E. Tackling the SARS-CoV-2 main protease using hybrid derivatives of 1,5-disubstituted tetrazole-1,2,3-triazoles: An in silico assay. PeerJ Phys. Chem. 2020, 2, e10. [Google Scholar] [CrossRef]
- Sotiropoulou, G.; Pampalakis, G.; Diamandis, E.P. Functional Roles of Human Kallikrein-related Peptidases. J. Biol. Chem. 2009, 284, 32989–32994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avgeris, M.; Papachristopoulou, G.; Polychronis, A.; Scorilas, A. Down-regulation of kallikrein-related peptidase 5 (KLK5) expression in breast cancer patients: A biomarker for the differential diagnosis of breast lesions. Clin. Proteom. 2011, 8, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.M.; Bae, K.S. Inhibitory Effects of Indole-3-Acetonitrile-4- Methoxy-2-S-Β-D-Glucopyranoside From Nasturtium officinale R. Br on Human Kallikrein 5 and 7 Protease. Nat. Prod. Commun. 2020, 15, 1–4. [Google Scholar] [CrossRef]
- Díaz-Cervantes, E.; Robles, J.; Aguilera-Granja, F. Understanding the structure, electronic properties, solubility in water, and protein interactions of three novel nano-devices against ovarian cancer: A computational study. J. Nanopart. Res. 2018, 20, 266. [Google Scholar] [CrossRef]
- Han, S.-Y.; Qiao, J.-Q.; Zhang, Y.-Y.; Lian, H.-Z.; Ge, X. Determination of n-octanol/water partition coefficients of weak ionizable solutes by RP-HPLC with neutral model compounds. Talanta 2012, 97, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Testa, B.; Fahr, A. Lipophilicity and Its Relationship with Passive Drug Permeation. Pharm Res. 2011, 28, 962–977. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, A.J.; Franson, A.D.; Cassals, A.G.; Donald, K.A.; Yu, A.J.; Garimallaprabhakaran, A.K.; Lynda, A.; Morrison, L.A.; Murelli, R.P. Importance of lipophilicity for potent anti-herpes simplex virus-1 activity of α-hydroxytropolones. Med. Chem. Commun. 2019, 10, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, B.; Jin, Z.; Yang, H.; Rao, Z.; Liu, X.; Zhang, B.; Jin, Z.; Yang, H.; Rao, Z. The Crystal Structure of COVID-19 Main Protease in Complex with an Inhibitor N3; Worldwide Protein Data Bank: London, UK, 2020. [Google Scholar] [CrossRef]
- Mody, V.; Ho, J.; Wills, S.; Mawri, A.; Lawson, L.; Ebert, M.C.C.J.C.; Fortin, G.M.; Rayalam, S.; Taval, S. Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. Commun. Biol. 2021, 4, 93. [Google Scholar] [CrossRef]
- Castanon-Alonso, S.L.; Morales-Saavedra, O.G.; Baez-Pimiento, S.; Ortega-Martinez, R.; Antonio, A.; Rodriguez-Rosales, A.A.; Hernandez-Rojas, M.E. Synthesis and overall nonlinear optical characterization of poly(hexa-2,4-diynylen-1,6-dioxydibenzoate) containing 2,2-(4-((4-nitrophenyl)ethynyl)phenylazanediyl)diethanol. Mater. Chem. Phys. 2012, 133, 528–540. [Google Scholar] [CrossRef]
- Jablonkai, E.; Henyecz, R.; Milen, M.; Kóti, J.; Keglevich, G. T3P®-assisted esterification and amidation of phosphinic acids. Tetrahedron 2014, 70, 8280–8285. [Google Scholar] [CrossRef]
- Jablonkai, E.; Milen, M.; Drahos, L.; Keglevich, G. Esterification of five-membered cyclic phosphinic acids under mild conditions using propylphosphonic anhydride (T3P®). Tetrahedron Lett. 2013, 54, 5873–5875. [Google Scholar] [CrossRef]
- Dunetz, J.R.; Xiang, Y.; Baldwin, A.; Ringling, J. General and Scalable Amide Bond Formation with Epimerization-Prone Substrates Using T3P and Pyridine. Org. Lett. 2011, 13, 5048–5051. [Google Scholar] [CrossRef]
- Randall, A.; Scheuerman, R.; Tumelty, D. The reduction of aromatic nitro groups on solid supports using sodium hydrosulfite (Na2S2O4). Tetrahedron Lett. 2000, 41, 6531–6535. [Google Scholar] [CrossRef]
- Williamson, K.L. Reduction of Indigo: Sodium Hydrosulfite as a Reducing Agent. J. Chem. Educ. 1989, 66, 359. [Google Scholar] [CrossRef] [Green Version]
- iMODS. Available online: http://imods.chaconlab.org/ (accessed on 2 March 2021).
- Tetko, I.V.; Tanchuk, V.Y. Application of associative neural networks for prediction of lipophilicity in ALOGPS 2.1 program. J. Chem. Inf. Comput. Sci. 2002, 42, 1136–1145. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Wallingford CT, 2009. Available online: https://gaussian.com/g09citation/ (accessed on 2 March 2021).
- Stewart, J.J.P. Optimization of parameters for semiempirical methods V: Modification of NDDO approximations and application to 70 elements. J. Mol. Mod. 2007, 13, 1173–1213. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Chen, C. GEMDOCK: A generic evolutionary method for molecular docking. Proteins 2004, 55, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, R.; Christensen, M.H. MolDock: A New Technique for High-Accuracy Molecular Docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef] [PubMed]
- Koes, D.R.; Camacho, C.J. ZINCPharmer: Pharmacophore search of the ZINC database. Nucleic Acids Res. 2012, 40, W409–W414. [Google Scholar] [CrossRef]



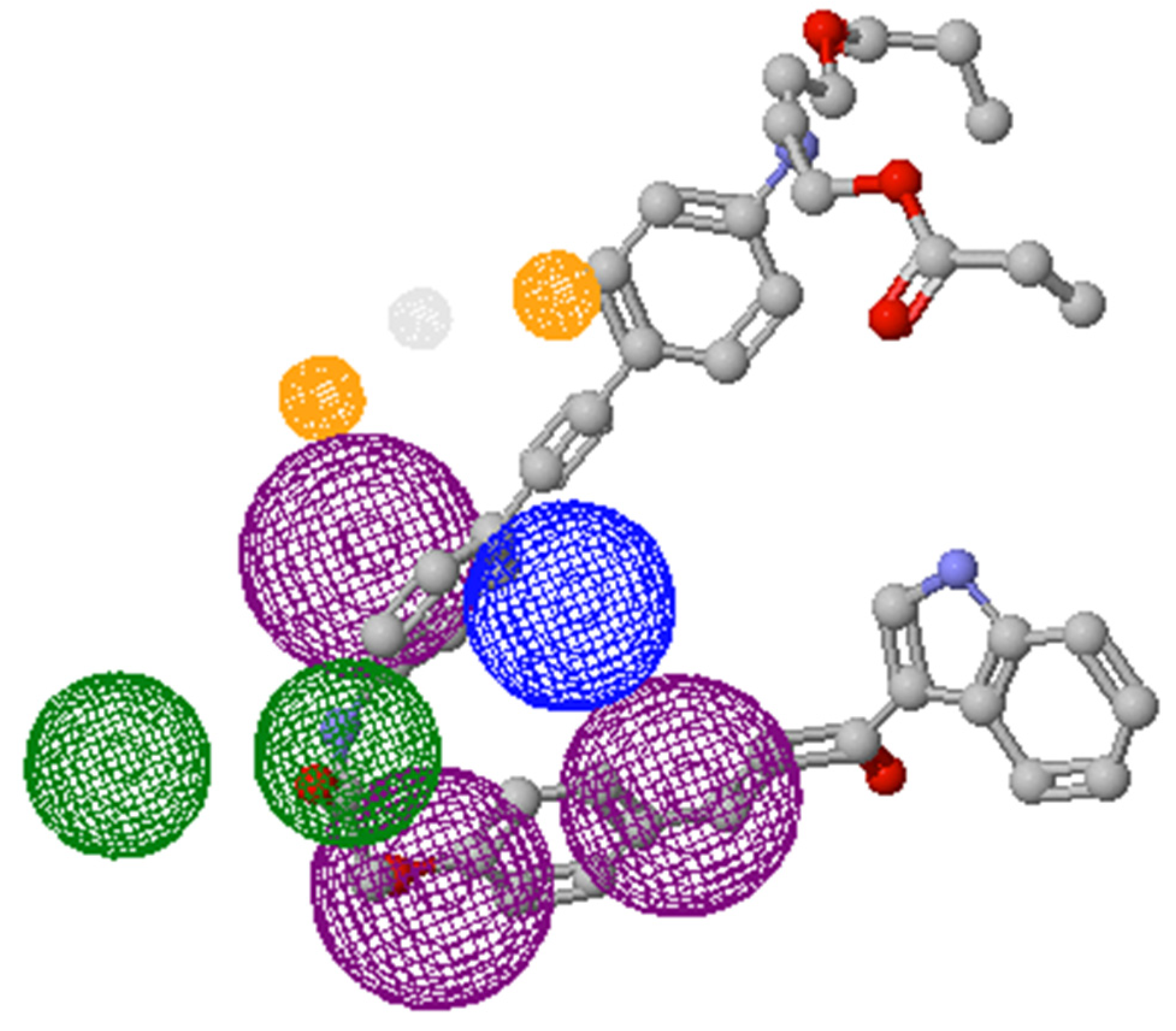
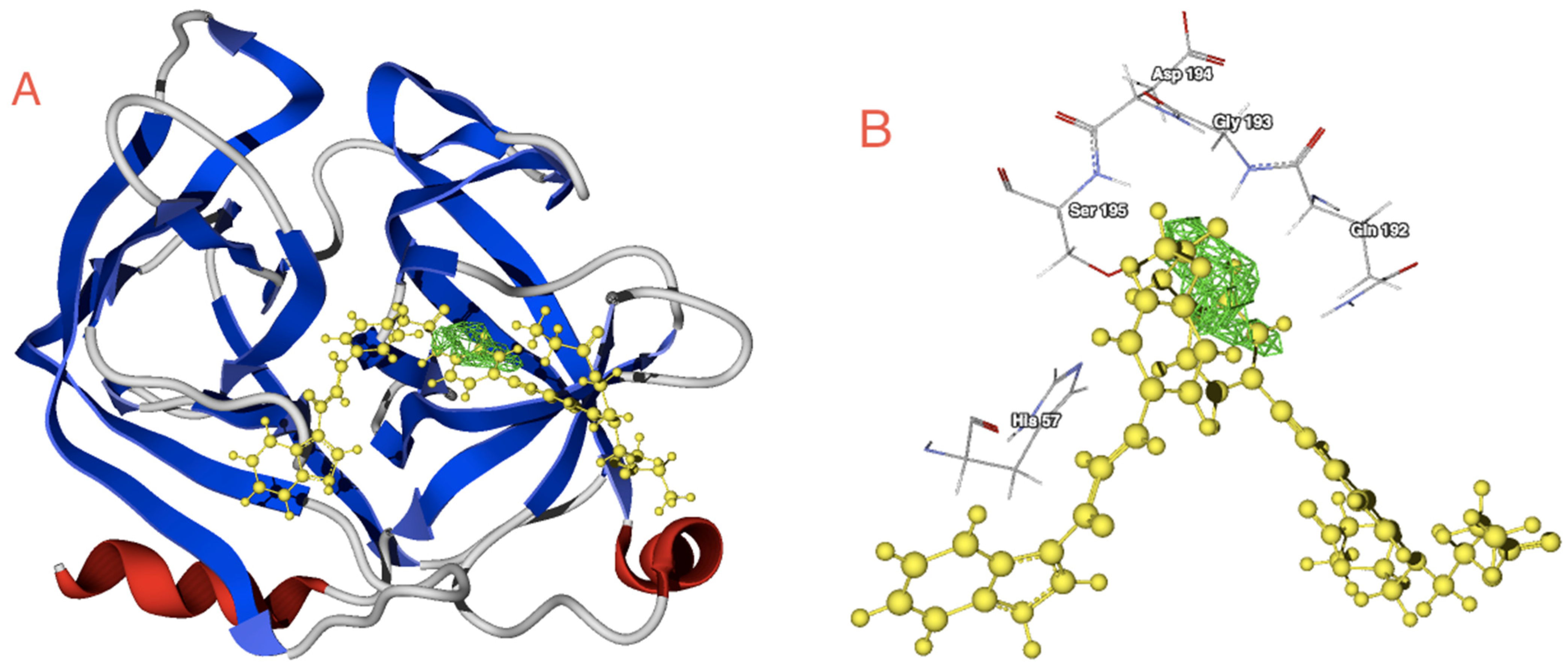
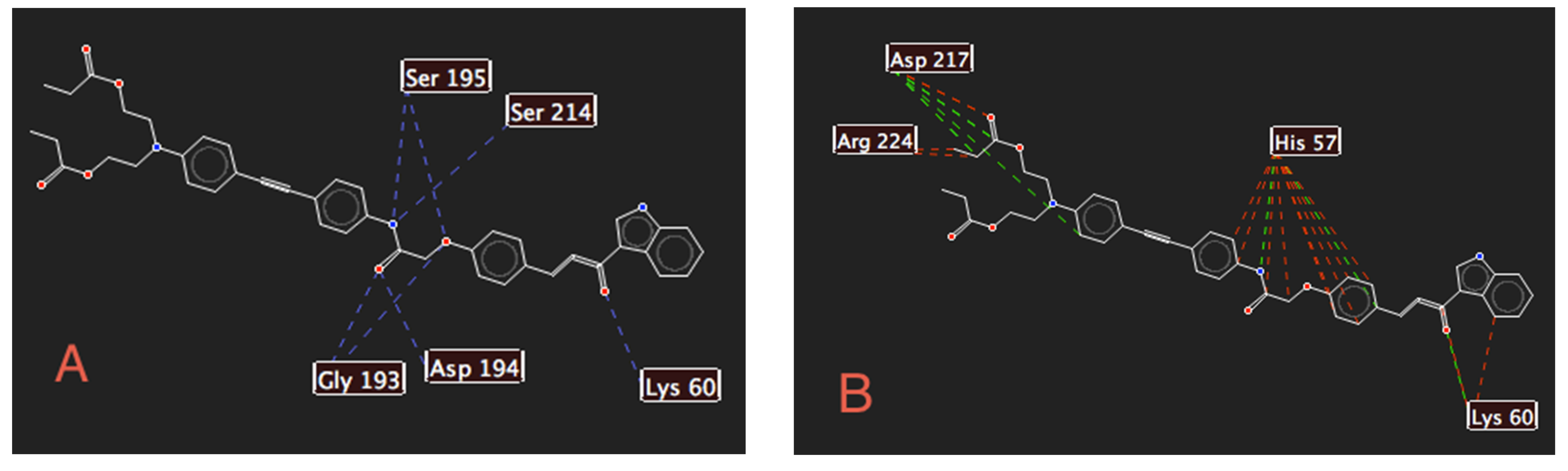
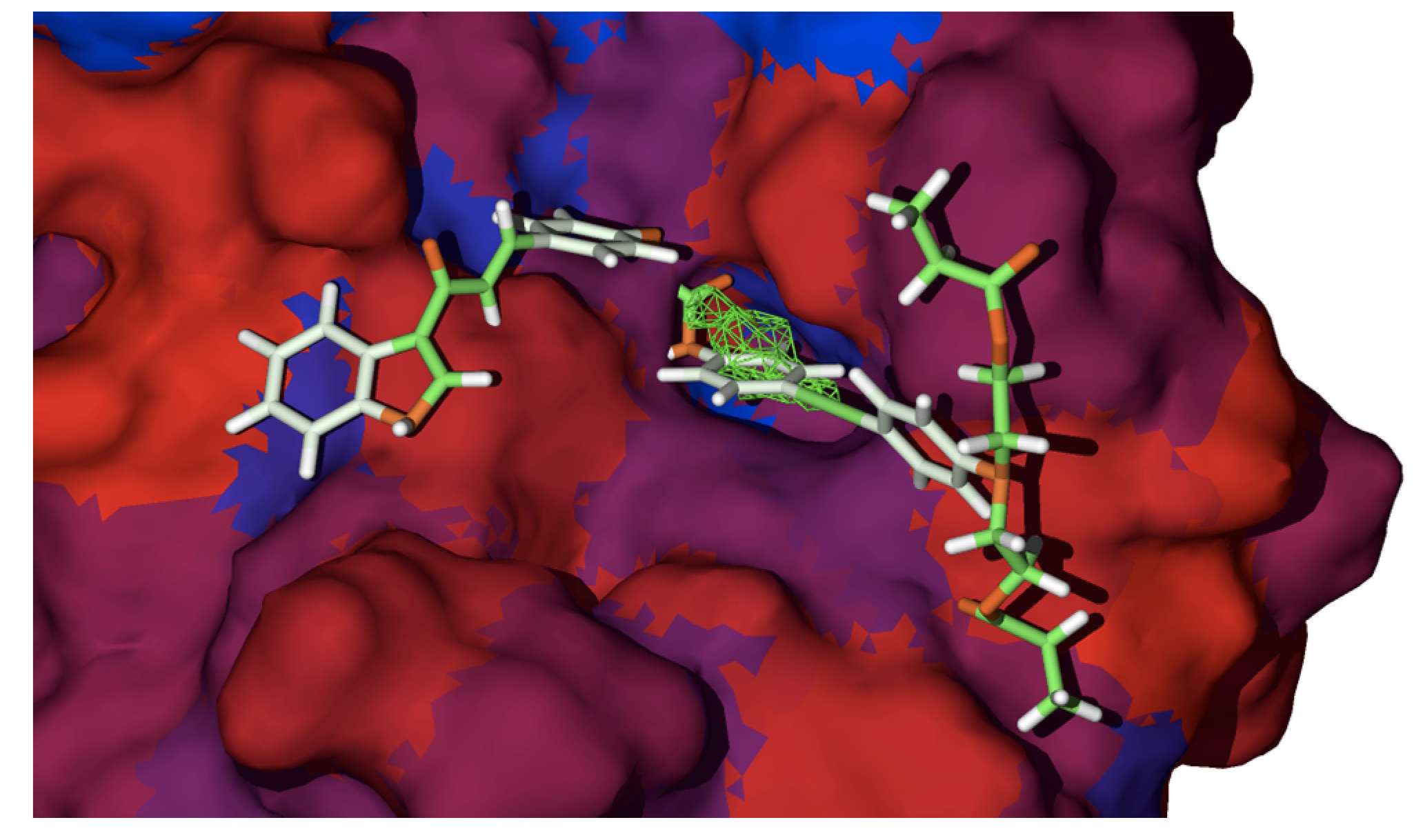
| Molecule | E | HBond | Elstat | VdW | LE |
|---|---|---|---|---|---|
| 1 | −191.25 | −6.11 | −0.64 | −52.36 | −5.63 |
| 2 | −186.18 | −2.72 | −0.80 | −49.59 | −6.21 |
| 3 | −177.44 | −5.56 | −0.73 | 20.92 | −6.12 |
| 4a | −209.02 | −6.92 | −1.00 | −51.58 | −6.33 |
| 4b | −221.35 | −3.08 | 0.24 | −27.07 | −6.51 |
| 15 (this work) | −224.29 | −5.48 | −0.87 | −54.97 | −4.23 |
| Co-Crystal (16) | −198.93 | −4.05 | −1.30 | −53.08 | −4.06 |
| 17 | −181.75 | −2.52 | −0.65 | −52.43 | −5.19 |
| 18 | −255.79 | −7.99 | −0.11 | −60.73 | −5.44 |
| Molecule | E | HBond | Electro | VdW | LE |
|---|---|---|---|---|---|
| 15 | −168.87 | −4.97 | −1.32 | 88.53 | −3.19 |
| Tube-altre [32] | −518.38 | −0.58 | −2.55 | 21.72 | −2.41 |
| Molecule | MW | LogP a |
|---|---|---|
| 1 | 440.46 | 4.97 |
| 2 | 404.51 | 4.91 |
| 3 | 477.42 | 5.14 |
| 4a | 449.60 | 4.08 |
| 4b | 461.54 | 2.93 |
| 15 | 711.82 | 6.70 |
| Co-Crystal (16) | 680.80 | 2.11 |
| 17 | 469.55 | 2.79 |
| 18 | 623.68 | 3.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almaraz-Girón, M.A.; Calderón-Jaimes, E.; Carrillo, A.S.; Díaz-Cervantes, E.; Alonso, E.C.; Islas-Jácome, A.; Domínguez-Ortiz, A.; Castañón-Alonso, S.L. Search for Non-Protein Protease Inhibitors Constituted with an Indole and Acetylene Core. Molecules 2021, 26, 3817. https://doi.org/10.3390/molecules26133817
Almaraz-Girón MA, Calderón-Jaimes E, Carrillo AS, Díaz-Cervantes E, Alonso EC, Islas-Jácome A, Domínguez-Ortiz A, Castañón-Alonso SL. Search for Non-Protein Protease Inhibitors Constituted with an Indole and Acetylene Core. Molecules. 2021; 26(13):3817. https://doi.org/10.3390/molecules26133817
Chicago/Turabian StyleAlmaraz-Girón, Marco A., Ernesto Calderón-Jaimes, Adrián Sánchez Carrillo, Erik Díaz-Cervantes, Edith Castañón Alonso, Alejandro Islas-Jácome, Armando Domínguez-Ortiz, and Sandra L. Castañón-Alonso. 2021. "Search for Non-Protein Protease Inhibitors Constituted with an Indole and Acetylene Core" Molecules 26, no. 13: 3817. https://doi.org/10.3390/molecules26133817
APA StyleAlmaraz-Girón, M. A., Calderón-Jaimes, E., Carrillo, A. S., Díaz-Cervantes, E., Alonso, E. C., Islas-Jácome, A., Domínguez-Ortiz, A., & Castañón-Alonso, S. L. (2021). Search for Non-Protein Protease Inhibitors Constituted with an Indole and Acetylene Core. Molecules, 26(13), 3817. https://doi.org/10.3390/molecules26133817






