Cyanovirin-N Binds Viral Envelope Proteins at the Low-Affinity Carbohydrate Binding Site without Direct Virus Neutralization Ability
Abstract
1. Introduction
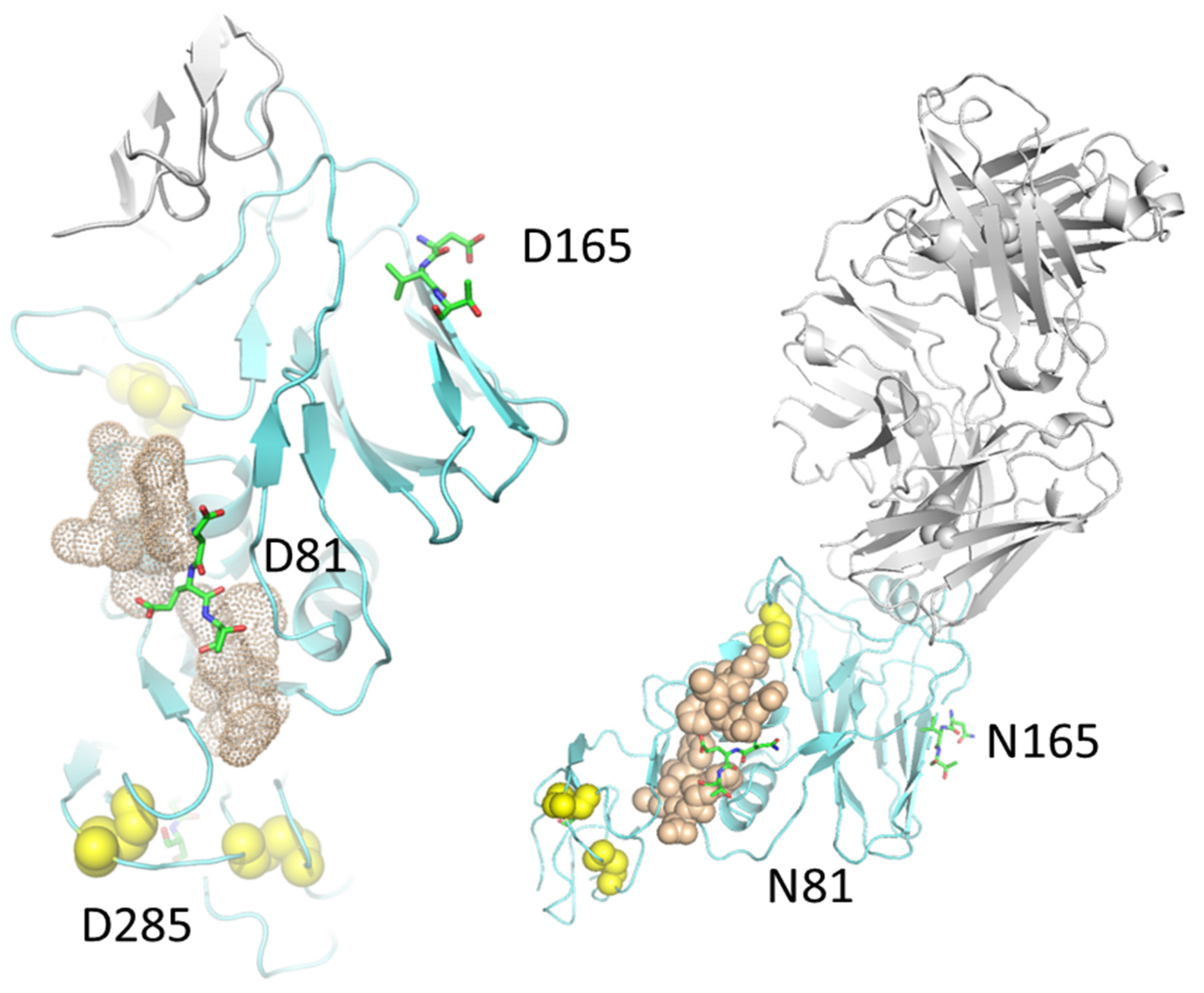
2. Results
3. Discussion
3.1. CV–N and 2G12 Binding Affinities for HIV gp120, HIV gp140, HA, and Ebola GP1,2
3.2. Avidity Correlated with the Number of CV–N Carbohydrate Binding Sites to Recognize Spike Proteins
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Expression of 2G12, HC19 Fab and HA H3 Top
4.3. HIV-1 Envelope Spike Proteins
4.4. SPR Binding Studies
4.5. Enzyme-Linked Immunosorbent Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wu, C.; Chen, W.; Chen, J.; Han, B.; Peng, Z.; Ge, F.; Wei, B.; Liu, M.; Zhang, M.; Qian, C.; et al. Preparation of monoPEGylated Cyanovirin-N’s derivative and its anti-influenza A virus bioactivity in vitro and in vivo. J. Biochem. 2015, 157, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Mazur-Marzec, H.; Cegłowska, M.; Konkel, R.; Pyrć, K. Antiviral Cyanometabolites—A Review. Biomolecules 2021, 11, 474. [Google Scholar] [CrossRef] [PubMed]
- Smee, D.F.; Bailey, K.W.; Wong, M.-H.; O’Keefe, B.R.; Gustafson, K.R.; Mishin, V.P.; Gubareva, L.V. Treatment of influenza A (H1N1) virus infections in mice and ferrets with cyanovirin-N. Antivir. Res. 2008, 80, 266–271. [Google Scholar] [CrossRef]
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nat. Cell Biol. 1981, 289, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Otterstrom, J.J.; Brandenburg, B.; Koldijk, M.H.; Juraszek, J.; Tang, C.; Mashaghi, S.; Kwaks, T.; Goudsmit, J.; Vogels, R.; Friesen, R.H.E.; et al. Relating influenza virus membrane fusion kinetics to stoichiometry of neutralizing antibodies at the single-particle level. Proc. Natl. Acad. Sci. USA 2014, 111, E5143–E5148. [Google Scholar] [CrossRef]
- Lee, P.S.; Yoshida, R.; Ekiert, D.; Sakai, N.; Suzuki, Y.; Takada, A.; Wilson, I.A. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc. Natl. Acad. Sci. USA 2012, 109, 17040–17045. [Google Scholar] [CrossRef]
- Waheed, A.A.; Gitzen, A.; Swiderski, M.; Freed, E.O. High-Mannose But Not Complex-Type Glycosylation of Tetherin Is Required for Restriction of HIV-1 Release. Viruses 2018, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-F.; Chang, C.-F.; Tsai, H.-P.; Chi, C.-Y.; Su, I.-J.; Wang, J.-R. Glycan-binding preferences and genetic evolution of human seasonal influenza A(H3N2) viruses during 1999–2007 in Taiwan. PLoS ONE 2018, 13, e0196727. [Google Scholar] [CrossRef]
- Gnirß, K.; Zmora, P.; Blazejewska, P.; Winkler, M.; Lins, A.; Nehlmeier, I.; Gärtner, S.; Moldenhauer, A.-S.; Hofmann-Winkler, H.; Wolff, T.; et al. Tetherin Sensitivity of Influenza A Viruses Is Strain Specific: Role of Hemagglutinin and Neuraminidase. J. Virol. 2015, 89, 9178–9188. [Google Scholar] [CrossRef]
- O’Keefe, B.R.; Smee, D.F.; Turpin, J.A.; Saucedo, C.J.; Gustafson, K.R.; Mori, T.; Blakeslee, D.; Buckheit, R.; Boyd, M.R. Potent Anti-Influenza Activity of Cyanovirin-N and Interactions with Viral Hemagglutinin. Antimicrob. Agents Chemother. 2003, 47, 2518–2525. [Google Scholar] [CrossRef]
- Fleury, D.; Barrère, B.; Bizebard, T.; Daniels, R.S.; Skehel, J.J.; Knossow, M. A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat. Genet. 1999, 6, 530–534. [Google Scholar] [CrossRef]
- Calarese, D.A.; Lee, H.-K.; Huang, C.-Y.; Best, M.D.; Astronomo, R.D.; Stanfield, R.L.; Katinger, H.; Burton, D.R.; Wong, C.-H.; Wilson, I.A. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. USA 2005, 102, 13372–13377. [Google Scholar] [CrossRef] [PubMed]
- Doores, K.J.; Fulton, Z.; Huber, M.; Wilson, I.A.; Burton, D.R. Antibody 2G12 Recognizes Di-Mannose Equivalently in Domain- and Nondomain-Exchanged Forms but Only Binds the HIV-1 Glycan Shield if Domain Exchanged. J. Virol. 2010, 84, 10690–10699. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.; Bhabha, G.; Elsliger, M.-A.; Friesen, R.H.E.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody Recognition of a Highly Conserved Influenza Virus Epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Friesen, R.H.; Bhabha, G.; Kwaks, T.; Jongeneelen, M.; Yu, W.; Ophorst, C.; Cox, F.; Korse, H.J.; Brandenburg, B.; et al. A Highly Conserved Neutralizing Epitope on Group 2 Influenza A Viruses. Science 2011, 333, 843–850. [Google Scholar] [CrossRef]
- Esser, M.T.; Mori, T.; Mondor, I.; Sattentau, Q.J.; Dey, B.; Berger, E.A.; Boyd, M.R.; Lifson, J.D. Cyanovirin-N Binds to gp120 To Interfere with CD4-Dependent Human Immunodeficiency Virus Type 1 Virion Binding, Fusion, and Infectivity but Does Not Affect the CD4 Binding Site on gp120 or Soluble CD4-Induced Conformational Changes in gp120. J. Virol. 1999, 73, 4360–4371. [Google Scholar] [CrossRef]
- Singh, R.S.; Walia, A.K.; Khattar, J.S.; Singh, D.P.; Kennedy, J.F. Cyanobacterial lectins characteristics and their role as antiviral agents. Int. J. Biol. Macromol. 2017, 102, 475–496. [Google Scholar] [CrossRef]
- Salunke, S.B.; Babu, N.S.; Chen, C.-T. Iron(iii) chloride as an efficient catalyst for stereoselective synthesis of glycosyl azides and a cocatalyst with Cu(0) for the subsequent click chemistry. Chem. Commun. 2011, 47, 10440–10442. [Google Scholar] [CrossRef]
- Schilling, P.; Kontaxis, G.; Dragosits, M.; Schiestl, R.H.; Becker, C.F.W.; Maier, I. Mannosylated hemagglutinin peptides bind cyanovirin-N independent of disulfide-bonds in complementary binding sites. RSC Adv. 2020, 10, 11079–11087. [Google Scholar] [CrossRef]
- Fleury, D.; Wharton, S.A.; Skehel, J.J.; Knossow, M.; Bizebard, T. Antigen distortion allows influenza virus to escape neutralization. Nat. Genet. 1998, 5, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Bizebard, T.; Gigant, B.; Rigolet, P.; Rasmussen, B.; Diat, O.; Peter, B.Ã.; Wharton, S.A.; Skehel, J.J.; Knossow, M. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nat. Cell Biol. 1995, 376, 92–94. [Google Scholar] [CrossRef]
- Doores, K.J.; Fulton, Z.; Hong, V.; Patel, M.K.; Scanlan, C.N.; Wormald, M.R.; Finn, M.G.; Burton, D.R.; Wilson, I.A.; Davis, B.G. A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity. Proc. Natl. Acad. Sci. USA 2010, 107, 17107–17112. [Google Scholar] [CrossRef] [PubMed]
- Hoorelbeke, B.; Huskens, D.; Férir, G.; François, K.O.; Takahashi, A.; Van Laethem, K.; Schols, D.; Tanaka, H.; Balzarini, J. Actinohivin, a Broadly Neutralizing Prokaryotic Lectin, Inhibits HIV-1 Infection by Specifically Targeting High-Mannose-Type Glycans on the gp120 Envelope. Antimicrob. Agents Chemother. 2010, 54, 3287–3301. [Google Scholar] [CrossRef]
- Barrientos, L.G.; Matei, E.; LaSala, F.; Delgado, R.; Gronenborn, A.M. Dissecting carbohydrate–Cyanovirin-N binding by structure-guided mutagenesis: Functional implications for viral entry inhibition. Protein Eng. Des. Sel. 2006, 19, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Bolmstedt, A.J.; O’Keefe, B.R.; Shenoy, S.R.; McMahon, J.B.; Boyd, M.R. Cyanovirin-N Defines a New Class of Antiviral Agent Targeting N-Linked, High-Mannose Glycans in an Oligosaccharide-Specific Manner. Mol. Pharmacol. 2001, 59, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Keeffe, J.R.; Gnanapragasam, P.N.P.; Gillespie, S.K.; Yong, J.; Bjorkman, P.J.; Mayo, S.L. Designed oligomers of cyanovirin-N show enhanced HIV neutralization. Proc. Natl. Acad. Sci. USA 2011, 108, 14079–14084. [Google Scholar] [CrossRef] [PubMed]
- Matei, E.; Zheng, A.; Furey, W.; Rose, J.; Aiken, C.; Gronenborn, A.M. Anti-HIV Activity of Defective Cyanovirin-N Mutants Is Restored by Dimerization. J. Biol. Chem. 2010, 285, 13057–13065. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, L.G.; O’Keefe, B.R.; Bray, M.; Sanchez, A.; Gronenborn, A.M.; Boyd, M.R. Cyanovirin-N binds to the viral surface glycoprotein, GP1,2 and inhibits infectivity of Ebola virus. Antivir. Res. 2003, 58, 47–56. [Google Scholar] [CrossRef]
- Lee, J.E.; Fusco, M.L.; Hessell, A.J.; Oswald, W.B.; Burton, D.R.; Saphire, E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nat. Cell Biol. 2008, 454, 177–182. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Hope, T.J.; Young, J.A. Differential requirements for clathrin endocytic pathway components in cellular entry by Ebola and Marburg glycoprotein pseudovirions. Virology 2011, 419, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Warfield, K.L.; Ruthel, G.; Bavari, S.; Aman, M.J.; Hope, T.J. Ebola virus uses clathrin-mediated endocytosis as an entry pathway. Virology 2010, 401, 18–28. [Google Scholar] [CrossRef]
- Mathew, M.P.; Donaldson, J.G. Glycosylation and glycan interactions can serve as extracellular machinery facilitating clathrin-independent endocytosis. Traffic 2019, 20, 295–300. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Hope, T.J. Full-length Ebola glycoprotein accumulates in the endoplasmic reticulum. Virol. J. 2011, 8, 11. [Google Scholar] [CrossRef]
- Chang, L.C.; Bewley, C.A. Potent Inhibition of HIV-1 Fusion by Cyanovirin-N Requires Only a Single High Affinity Carbohydrate Binding Site: Characterization of Low Affinity Carbohydrate Binding Site Knockout Mutants. J. Mol. Biol. 2002, 318, 1–8. [Google Scholar] [CrossRef]
- Bewley, C.A. Solution structure of a cyanovirin-N:Man alpha 1-2Man alpha complex: Structural basis for high-affinity carbohydrate-mediated binding to gp120. Structure 2001, 9, 931–940. [Google Scholar] [CrossRef]
- Yang, F.; Bewley, C.A.; Louis, J.M.; Gustafson, K.R.; Boyd, M.R.; Gronenborn, A.M.; Clore, G.M.; Wlodawer, A. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J. Mol. Biol. 1999, 288, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Wang, Z.; Glickman, R.L.; Iorio, R.M. Glycosylation within an Antigenic Site on the HN Glycoprotein of Newcastle Disease Virus Interferes with Its Role in the Promotion of Membrane Fusion. Virology 1994, 204, 17–26. [Google Scholar] [CrossRef]
- Lingwood, D.; McTamney, P.M.; Yassine, H.M.; Whittle, J.R.; Guo, X.; Boyington, J.C.; Wei, C.-J.; Nabel, G.J. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 2012, 489, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.A.; Giddens, J.; Pincetic, A.; Lomino, J.V.; Ravetch, J.V.; Wang, L.-X.; Bjorkman, P.J. Structural Characterization of Anti-Inflammatory Immunoglobulin G Fc Proteins. J. Mol. Biol. 2014, 426, 3166–3179. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, W.P.; Crameri, A.; Ha, K.D.; Brennan, T.M.; Heyneker, H.L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 1995, 164, 49–53. [Google Scholar] [CrossRef]
- Sanchez, A.; Trappier, S.G.; Mahy, B.W.; Peters, C.J.; Nichol, S.T. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. USA 1996, 93, 3602–3607. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Duckers, H.J.; Sullivan, N.J.; Sanchez, A.; Nabel, E.G.; Nabel, G.J. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 2000, 6, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Frey, G.; Peng, H.; Rits-Volloch, S.; Morelli, M.; Cheng, Y.; Chen, B. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc. Natl. Acad. Sci. USA 2008, 105, 3739–3744. [Google Scholar] [CrossRef]
- West, A.P.; Galimidi, R.P.; Foglesong, C.P.; Gnanapragasam, P.N.P.; Huey-Tubman, K.E.; Klein, J.S.; Suzuki, M.D.; Tiangco, N.E.; Vielmetter, J.; Bjorkman, P.J. Design and Expression of a Dimeric Form of Human Immunodeficiency Virus Type 1 Antibody 2G12 with Increased Neutralization Potency. J. Virol. 2008, 83, 98–104. [Google Scholar] [CrossRef] [PubMed][Green Version]
| HIV-1 KD (M) | Influenza KD (M) | Ebola KD (M) | |||
|---|---|---|---|---|---|
| Monomer | Trimer-Folded | ||||
| WT CV–N | 1H + 1L | 2.6 × 10−7 | 1.2 × 10−7 | 5.7 × 10−9 | 3.4 × 10−8 |
| CVN2L0-B | 2H + 1L | 4.3 × 10−8 | 7.2 × 10−8 | 2.7 × 10−9 | 2.6 × 10−8 |
| CVN2L0-N | 2L | 1.8 × 10−7 | 3.5 × 10−7 | 6.5 × 10−8 | 3.6 × 10−7 |
| CVN E | 1H | 4.5 × 10−7 | 7.2 × 10−7 | 2.0 × 10−7 | 7.2 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maier, I.; Schiestl, R.H.; Kontaxis, G. Cyanovirin-N Binds Viral Envelope Proteins at the Low-Affinity Carbohydrate Binding Site without Direct Virus Neutralization Ability. Molecules 2021, 26, 3621. https://doi.org/10.3390/molecules26123621
Maier I, Schiestl RH, Kontaxis G. Cyanovirin-N Binds Viral Envelope Proteins at the Low-Affinity Carbohydrate Binding Site without Direct Virus Neutralization Ability. Molecules. 2021; 26(12):3621. https://doi.org/10.3390/molecules26123621
Chicago/Turabian StyleMaier, Irene, Robert H. Schiestl, and Georg Kontaxis. 2021. "Cyanovirin-N Binds Viral Envelope Proteins at the Low-Affinity Carbohydrate Binding Site without Direct Virus Neutralization Ability" Molecules 26, no. 12: 3621. https://doi.org/10.3390/molecules26123621
APA StyleMaier, I., Schiestl, R. H., & Kontaxis, G. (2021). Cyanovirin-N Binds Viral Envelope Proteins at the Low-Affinity Carbohydrate Binding Site without Direct Virus Neutralization Ability. Molecules, 26(12), 3621. https://doi.org/10.3390/molecules26123621






