In Vitro and In Silico Anti-Arboviral Activities of Dihalogenated Phenolic Derivates of L-Tyrosine
Abstract
1. Introduction
2. Results
2.1. Dihalogenated Compounds Derived from L-Tyrosine Are Not Toxic in the In Vitro and In Silico Models
2.2. Effect of Phenolic Dihalogenated Compounds Derived from L-Tyrosine on the Production of Infectious Viral Particles in the VERO Cell Line Depends on the Type of Virus
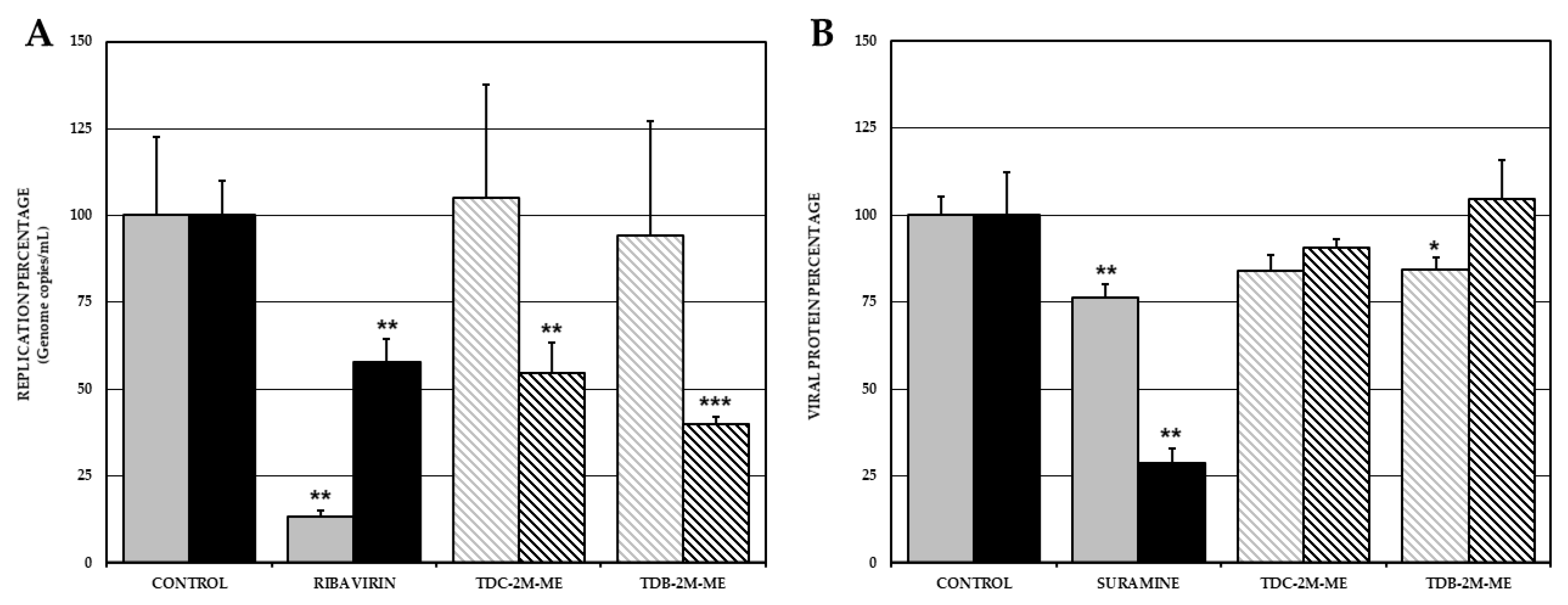
2.3. TDC-2M-ME and TDB-2M-ME Only Inhibit Viral Genome Replication in the Alphavirus Infection Model Whilst TDB-2M-ME Inhibits One Viral Protein Synthesis in the ZIKV Infection Model
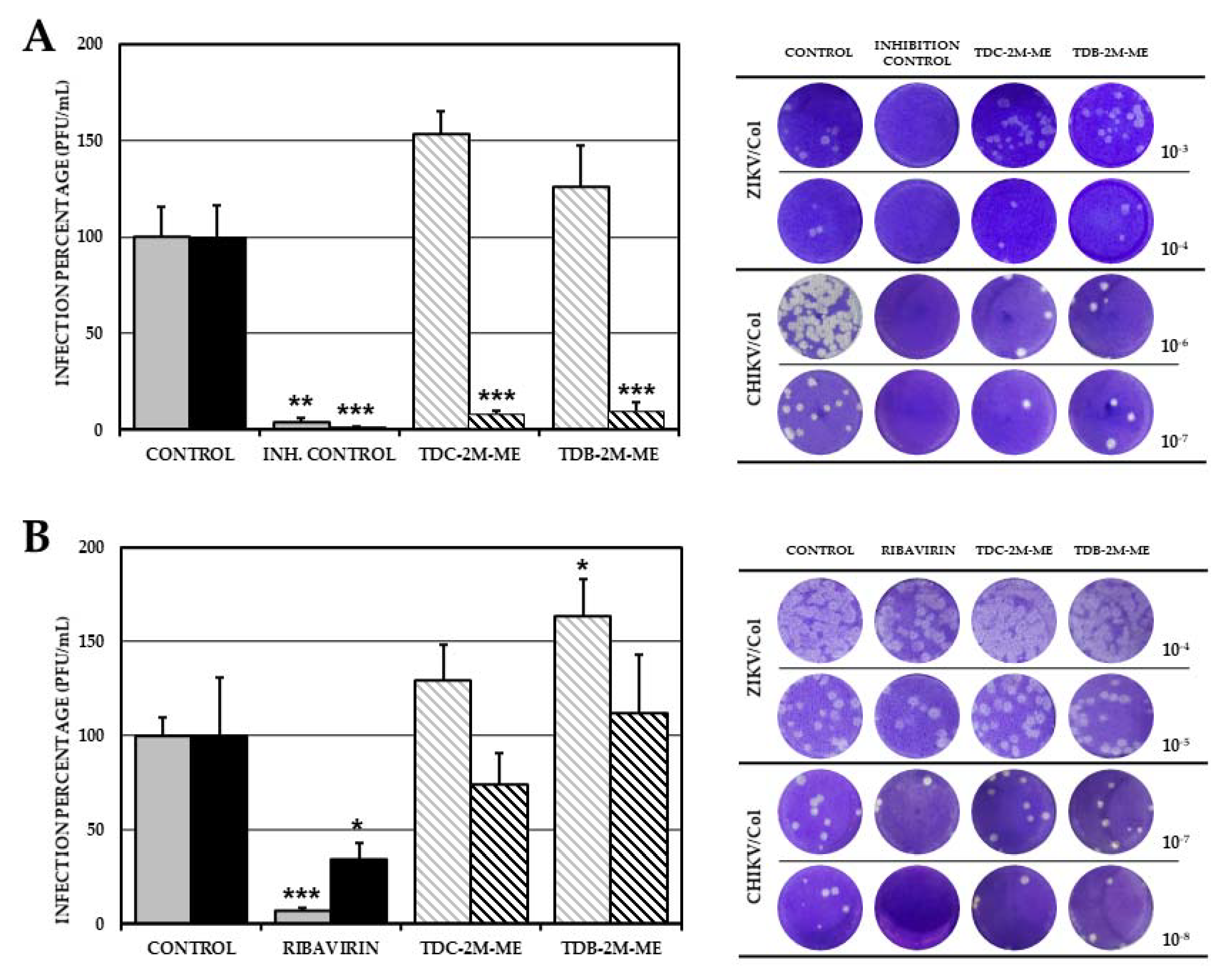
2.4. TDC-2M-ME and TDB-2M-ME Treatment Prior to Infection Inhibits the Alphavirus Model (CHIKV/Col)
2.5. TDB-2M-ME Presents Virucidal Activity in the ZIKV-Infection Model
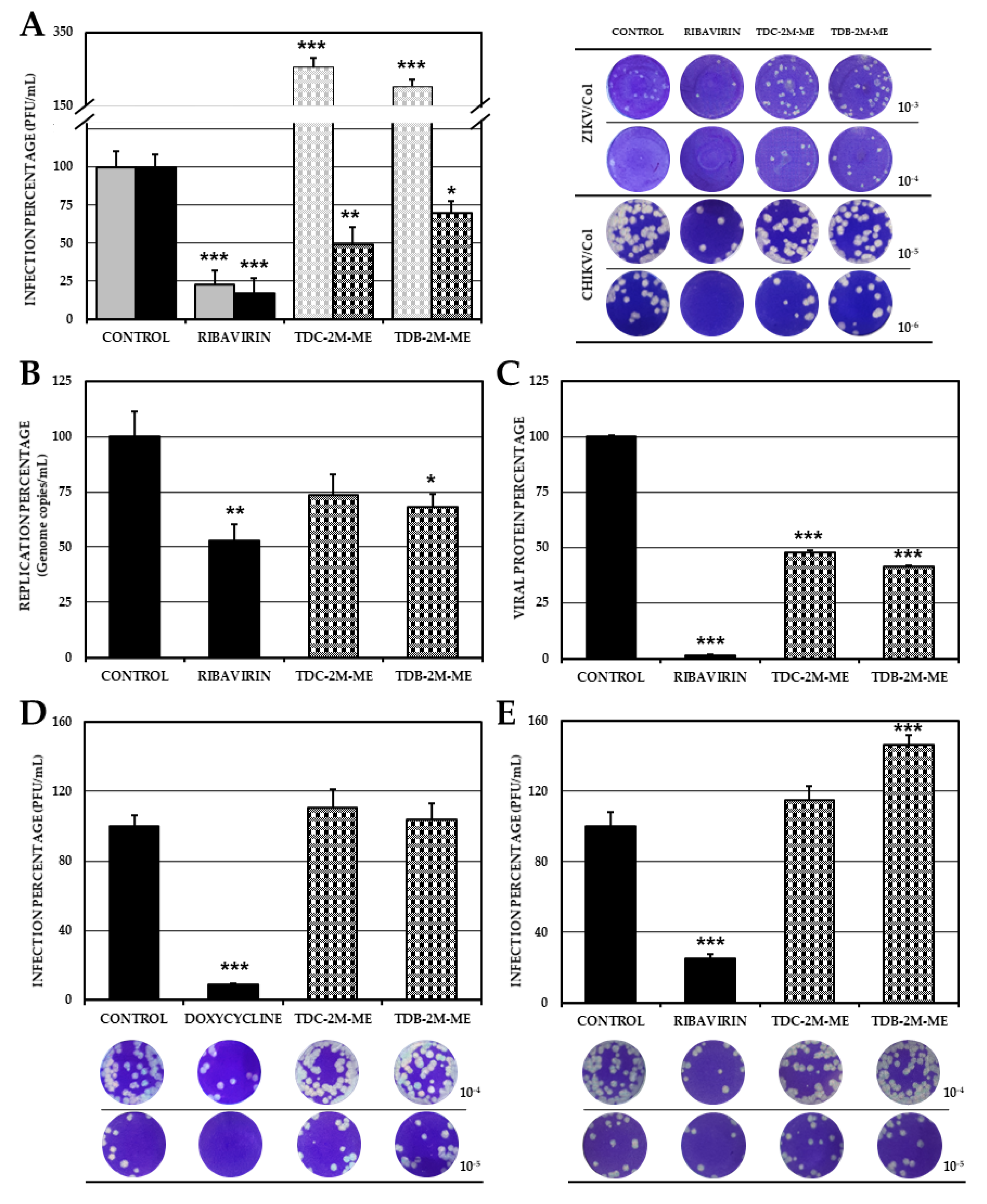
2.6. The Effect of TDB-2M-ME and TDC-2M-ME on the Production of Infectious Viral Particles Depends on the Cell Line
2.7. Phenolic Dihalogenated Compounds Derived from L-Tyrosine Present Favorable Interaction with the Viral Helicases of ZIKV and CHIKV and Some Cellular Proteins
2.8. The Interactions between Virucidal Compound, TDB-2M-ME, and ZIKV Envelope Domains Were Not Stable over the Time
3. Discussion
4. Materials and Methods
4.1. Synthesis of Phenolic Dihalogenated Compounds Derived from L-Tyrosine
4.2. Cells, Viruses and Controls
4.3. Toxicity Assay
4.3.1. Cytotoxicity
4.3.2. In Silico Toxicological Modeling
4.4. In Vitro Antiviral Strategies
4.5. Quantification of Infection
4.5.1. Titration by Plaque Formation
4.5.2. RT-qPCR
4.5.3. Cell ELISA
4.6. Statistical Analysis
4.7. In Silico Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Paixão, E.S.; Teixeira, M.G.; Rodrigues, L.C. Zika, chikungunya and dengue: The causes and threats of new and re-emerging arboviral diseases. BMJ Glob. Health 2018, 3, e000530. [Google Scholar] [CrossRef]
- Jones, R.; Kulkarni, M.A.; Davidson, T.M.; Team, R.-L.R.; Talbot, B. Arbovirus vectors of epidemiological concern in the Americas: A scoping review of entomological studies on Zika, dengue and chikungunya virus vectors. PLoS ONE 2020, 15, e0220753. [Google Scholar] [CrossRef] [PubMed]
- Epelboin, Y.; Talaga, S.; Epelboin, L.; Dusfour, I. Zika virus: An updated review of competent or naturally infected mosquitoes. PLoS Negl. Trop. Dis. 2017, 11, e0005933. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Hernández, M.Y.; Ruiz-Saenz, J.; Villamizar, L.J.; Gómez-Rangel, S.Y.; Martínez-Gutierrez, M. Co-circulation and simultaneous co-infection of dengue, chikungunya, and zika viruses in patients with febrile syndrome at the Colombian-Venezuelan border. BMC Infect. Dis. 2018, 18, 61. [Google Scholar] [CrossRef]
- Cabral-Castro, M.J.; Cavalcanti, M.G.; Peralta, R.H.S.; Peralta, J.M. Molecular and serological techniques to detect co-circulation of DENV, ZIKV and CHIKV in suspected dengue-like syndrome patients. J. Clin. Virol. 2016, 82, 108–111. [Google Scholar] [CrossRef]
- Guo, C.; Zhou, Z.; Wen, Z.; Liu, Y.; Zeng, C.; Xiao, D.; Ou, M.; Han, Y.; Huang, S.; Liu, D. Global epidemiology of dengue outbreaks in 1990–2015: A systematic review and meta-analysis. Front. Cellul. Infect. Microbiol. 2017, 7, 317. [Google Scholar] [CrossRef]
- Russo, F.B.; Jungmann, P.; Beltrão-Braga, P.C.B. Zika infection and the development of neurological defects. Cellul. Microbiol. 2017, 19, e12744. [Google Scholar] [CrossRef]
- Suhrbier, A. Rheumatic manifestations of chikungunya: Emerging concepts and interventions. Nat. Rev. Rheumatol. 2019, 15, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Becher, B.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Mukhopadhyay, S.; Merits, A.; Bolling, B.; Nasar, F.; Coffey, L.L.; Powers, A.; Weaver, S.C. ICTV virus taxonomy profile: Togaviridae. J. Gen. Virol. 2018, 99, 761–762. [Google Scholar] [CrossRef] [PubMed]
- Chambers, T.J.; Hahn, C.S.; Galler, R.; Rice, C.M. Flavivirus genome organization, expression, and replication. Ann. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef]
- Tsetsarkin, K.A.; Chen, R.; Sherman, M.B.; Weaver, S.C. Chikungunya virus: Evolution and genetic determinants of emergence. Curr. Opin. Virol. 2011, 1, 310–317. [Google Scholar] [CrossRef]
- Kuhn, R.J.; Zhang, W.; Rossmann, M.G.; Pletnev, S.V.; Corver, J.; Lenches, E.; Jones, C.T.; Mukhopadhyay, S.; Chipman, P.R.; Strauss, E.G. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell 2002, 108, 717–725. [Google Scholar] [CrossRef]
- World Health Organization. Equipment for Vector Control. Specification Guidelines. Available online: https://bit.ly/2KzBtz5 (accessed on 25 May 2019).
- Kim, K.-S. Current challenges in the development of vaccines and drugs against emerging vector-borne diseases. Curr. Med. Chem. 2019, 26, 2974–2986. [Google Scholar] [CrossRef]
- Godói, I.P.; Lemos, L.L.P.; de Araújo, V.E.; Bonoto, B.C.; Godman, B.; Guerra Junior, A.A. CYD-TDV dengue vaccine: Systematic review and meta-analysis of efficacy, immunogenicity and safety. J. Comp. Effect. Res. 2017, 6, 165–180. [Google Scholar] [CrossRef]
- Poland, G.A.; Kennedy, R.B.; Ovsyannikova, I.G.; Palacios, R.; Ho, P.L.; Kalil, J. Development of vaccines against Zika virus. Lancet Infect. Dis. 2018, 18, e211–e219. [Google Scholar] [CrossRef]
- Powers, A.M. Vaccine and therapeutic options to control Chikungunya virus. Clin. Microbiol. Rev. 2018, 31, e00104–e00116. [Google Scholar] [CrossRef]
- Loaiza-Cano, V.; Monsalve-Escudero, L.M.; Martinez-Gutierrez, M.; Sousa, D.P.d. Antiviral Role of Phenolic Compounds against Dengue Virus: A Review. Biomolecules 2021, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.; Kaur, M.; Minneman, K.P. Antiviral lead compounds from marine sponges. Mar. Drugs 2010, 8, 2619–2638. [Google Scholar] [CrossRef] [PubMed]
- Galeano, E.; Martínez, A.; Thomas, O.P.; Robledo, S.; Munoz, D. Antiparasitic bromotyrosine derivatives from the Caribbean marine sponge Aiolochroia crassa. Quim. Nova 2012, 35, 1189–1193. [Google Scholar] [CrossRef]
- Zapata, W.; Gomez-Archila, L.G.; Galeano, E.; Martínez, A.; Castrillón, F.J.D.; Rugeles, M.T. Bromotyrosine derivatives from marine sponges inhibit the HIV-1 replication in vitro. Vitae 2014, 21, 114–125. [Google Scholar]
- Restrepo, M.P.; Jaramillo, E.G.; Martínez, A.M.; Restrepo, S.R. Synthesis and trypanocide activity of chloro-l-tyrosine and bromo-l-tyrosine derivatives. Med. Chem. Res. 2018, 27, 2454–2465. [Google Scholar] [CrossRef]
- Blázquez, A.B.; Martín-Acebes, M.A.; Saiz, J.-C. Inhibition of West Nile virus multiplication in cell culture by anti-Parkinsonian drugs. Front. Microbiol. 2016, 7, 296. [Google Scholar] [CrossRef] [PubMed]
- Frakolaki, E.; Kalliampakou, K.I.; Kaimou, P.; Moraiti, M.; Kolaitis, N.; Boleti, H.; Koskinas, J.; Vassilacopoulou, D.; Vassilaki, N. Emerging Role of l-Dopa Decarboxylase in Flaviviridae Virus Infections. Cells 2019, 8, 837. [Google Scholar] [CrossRef] [PubMed]
- Abdullahi, A.; Wang, V.; Auger, C.; Patsouris, D.; Amini-Nik, S.; Jeschke, M.G. Catecholamines Induce Endoplasmic Reticulum Stress via Both Alpha and Beta Receptors. Shock Augusta Ga 2020, 53, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Galzigna, L.; Zanatta, L.; Esposito, N. Toxicity of dopamine and dopaminochrome on cultured cells. Neurotox. Res. 1999, 1, 149–152. [Google Scholar] [CrossRef] [PubMed]
- El-Saadi, M.W.; Williams-Hart, T.; Salvatore, B.A.; Mahdavian, E. Use of in-silico assays to characterize the ADMET profile and identify potential therapeutic targets of fusarochromanone, a novel anti-cancer agent. Silico Pharmacol. 2015, 3, 6. [Google Scholar] [CrossRef]
- Paemanee, A.; Hitakarun, A.; Roytrakul, S.; Smith, D.R. Screening of melatonin, α-tocopherol, folic acid, acetyl-L-carnitine and resveratrol for anti-dengue 2 virus activity. BMC Res. Notes 2018, 11, 307. [Google Scholar] [CrossRef]
- Hitakarun, A.; Khongwichit, S.; Wikan, N.; Roytrakul, S.; Yoksan, S.; Rajakam, S.; Davidson, A.D.; Smith, D.R. Evaluation of the antiviral activity of orlistat (tetrahydrolipstatin) against dengue virus, Japanese encephalitis virus, Zika virus and chikungunya virus. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Gomez-Calderon, C.; Mesa-Castro, C.; Robledo, S.; Gomez, S.; Bolivar-Avila, S.; Diaz-Castillo, F.; Martinez-Gutierrez, M. Antiviral effect of compounds derived from the seeds of Mammea americana and Tabernaemontana cymosa on Dengue and Chikungunya virus infections. BMC Complementary Altern. Med. 2017, 17. [Google Scholar] [CrossRef]
- Rupp, J.C.; Sokoloski, K.J.; Gebhart, N.N.; Hardy, R.W. Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 2015, 96, 2483. [Google Scholar] [CrossRef] [PubMed]
- Mehrbod, P.; Ande, S.R.; Alizadeh, J.; Rahimizadeh, S.; Shariati, A.; Malek, H.; Hashemi, M.; Glover, K.K.; Sher, A.A.; Coombs, K.M. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence 2019, 10, 376–413. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Srivastava, R.; Howell, S.H.; Bassham, D.C. Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 2016, 85, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.P.; Ng, M.-L.; Vasudevan, S.G. Differential unfolded protein response during Chikungunya and Sindbis virus infection: CHIKV nsP4 suppresses eIF2α phosphorylation. Virol. J. 2013, 10, 1–15. [Google Scholar] [CrossRef]
- Ferraz, A.C.; Moraes, T.d.F.S.; da Cruz Nizer, W.S.; dos Santos, M.; Tótola, A.H.; Ferreira, J.M.S.; Vieira-Filho, S.A.; Rodrigues, V.G.; Duarte, L.P.; de Brito Magalhães, C.L. Virucidal activity of proanthocyanidin against Mayaro virus. Antivir. Res. 2019, 168, 76–81. [Google Scholar] [CrossRef]
- Moscona, A. Oseltamivir resistance—disabling our influenza defenses. N. Engl. J. Med. 2005, 353, 2633–2636. [Google Scholar] [CrossRef]
- Gutiérrez, I.S.; Lin, F.-Y.; Vanommeslaeghe, K.; Lemkul, J.A.; Armacost, K.A.; Brooks, C.L., III; MacKerell, A.D., Jr. Parametrization of halogen bonds in the CHARMM general force field: Improved treatment of ligand–protein interactions. Bioorg. Med. Chem. 2016, 24, 4812–4825. [Google Scholar] [CrossRef]
- Danelius, E.; Andersson, H.; Jarvoll, P.; Lood, K.; Grafenstein, J.; Erdelyi, M.T. Halogen bonding: A powerful tool for modulation of peptide conformation. Biochemistry 2017, 56, 3265–3272. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, R.; Brown, D.T.; Paredes, A. Structural differences observed in arboviruses of the alphavirus and flavivirus genera. Adv. Virol. 2014, 2014. [Google Scholar] [CrossRef]
- Nikitina, E.; Larionova, I.; Choinzonov, E.; Kzhyshkowska, J. Monocytes and macrophages as viral targets and reservoirs. Int. J. Mol. Sci. 2018, 19, 2821. [Google Scholar] [CrossRef]
- Mumtaz, N.; Jimmerson, L.C.; Bushman, L.R.; Kiser, J.J.; Aron, G.; Reusken, C.B.; Koopmans, M.P.; van Kampen, J.J. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Ant. Res. 2017, 146, 161–163. [Google Scholar] [CrossRef]
- Martínez-Betancur, V.; Martínez-Gutierrez, M. Proteomic profile of human monocytic cells infected with dengue virus. Asian Pac. J. Trop. Biomed. 2016, 6, 914–923. [Google Scholar] [CrossRef]
- Martínez Betancur, V.; Marín Villa, M.; Martinez Gutierrez, M. Infection of epithelial cells with dengue virus promotes the expression of proteins favoring the replication of certain viral strains. J. Med. Virol. 2014, 86, 1448–1458. [Google Scholar] [CrossRef]
- Thaker, S.K.; Chapa, T.; Garcia, G., Jr.; Gong, D.; Schmid, E.W.; Arumugaswami, V.; Sun, R.; Christofk, H.R. Differential Metabolic Reprogramming by Zika Virus Promotes Cell Death in Human versus Mosquito Cells. Cell Metabol. 2019, 29, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Bera, A.K.; Kuhn, R.J.; Smith, J.L. Structure of the Flavivirus helicase: Implications for catalytic activity, protein interactions, and proteolytic processing. J. Virol. 2005, 79, 10268–10277. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Prosser, O.; Kumar, P.; Tuplin, A.; Giri, R. Small molecule inhibitors possibly targeting the rearrangement of Zika virus envelope protein. Antiv. Res. 2020, 182, 104876. [Google Scholar] [CrossRef]
- Godoy, A.S.; Lima, G.M.; Oliveira, K.I.; Torres, N.U.; Maluf, F.V.; Guido, R.V.; Oliva, G. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat. Commun. 2017, 8, 1–6. [Google Scholar] [CrossRef]
- Puranik, N.V.; Rani, R.; Singh, V.A.; Tomar, S.; Puntambekar, H.M.; Srivastava, P. Evaluation of the antiviral potential of halogenated dihydrorugosaflavonoids and molecular modeling with nsP3 protein of Chikungunya virus (CHIKV). ACS Omega 2019, 4, 20335–20345. [Google Scholar] [CrossRef]
- Carrillo-Hernandez, M.Y.; Ruiz-Saenz, J.; Jaimes-Villamizar, L.; Robledo-Restrepo, S.M.; Martinez-Gutierrez, M. Phylogenetic and evolutionary analysis of dengue virus serotypes circulating at the Colombian–Venezuelan border during 2015–2016 and 2018–2019. PLoS ONE 2021, 16, e0252379. [Google Scholar] [CrossRef]
- Balm, M.N.; Lee, C.K.; Lee, H.K.; Chiu, L.; Koay, E.S.; Tang, J.W. A diagnostic polymerase chain reaction assay for Zika virus. J. Med. Virol. 2012, 84, 1501–1505. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232. [Google Scholar] [CrossRef]
- Faye, O.; Faye, O.; Dupressoir, A.; Weidmann, M.; Ndiaye, M.; Sall, A.A. One-step RT-PCR for detection of Zika virus. J. Clin. Virol. 2008, 43, 96–101. [Google Scholar] [CrossRef]
- Coronado, M.A.; Eberle, R.J.; Bleffert, N.; Feuerstein, S.; Olivier, D.S.; de Moraes, F.R.; Willbold, D.; Arni, R.K. Zika virus NS2B/NS3 proteinase: A new target for an old drug-Suramin a lead compound for NS2B/NS3 proteinase inhibition. Antiv. Res. 2018, 160, 118–125. [Google Scholar] [CrossRef]
- Albulescu, I.C.; Van Hoolwerff, M.; Wolters, L.A.; Bottaro, E.; Nastruzzi, C.; Yang, S.C.; Tsay, S.-C.; Hwu, J.R.; Snijder, E.J.; Van Hemert, M.J. Suramin inhibits chikungunya virus replication through multiple mechanisms. Antiv. Res. 2015, 121, 39–46. [Google Scholar] [CrossRef]
- Rothan, H.A.; Bahrani, H.; Mohamed, Z.; Teoh, T.C.; Shankar, E.M.; Rahman, N.A.; Yusof, R. A combination of doxycycline and ribavirin alleviated chikungunya infection. PLoS ONE 2015, 10, e0126360. [Google Scholar] [CrossRef] [PubMed]
- Rattanaburee, T.; Junking, M.; Panya, A.; Sawasdee, N.; Songprakhon, P.; Suttitheptumrong, A.; Limjindaporn, T.; Haegeman, G.; Yenchitsomanus, P.-T. Inhibition of dengue virus production and cytokine/chemokine expression by ribavirin and compound A. Antiv. Res. 2015, 124, 83–92. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Panella, A.J.; Velez, J.O.; Lambert, A.J.; Campbell, G.L. Chikungunya virus in US travelers returning from India, 2006. Emerg. Infect. Dis. 2007, 13, 764. [Google Scholar] [CrossRef]
- Quintero-Gil, D.C.; Uribe-Yepes, A.; Ospina, M.; Díaz, F.J.; Martinez-Gutierrez, M. Differences in the replicative capacities of clinical isolates of dengue virus in C6/36 cells and in urban populations of Aedes aegypti from Colombia, South America. Braz. J. Infect. Dis. 2018, 22, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Monsalve-Escudero, L.M.; Hernández-Mira, E.; Loaiza-Cano, V.; Zapata-Cardona, M.I.; Quintero-Gil, D.C.; Pájaro, Y.; Diaz-Castillo, F.; Quiñones, W.; Robledo, S.M.; Martinez-Gutierrez, M. The antiviral and virucidal activities of voacangine and structural analogs extracted from Tabernaemontana cymosa depend on the Dengue virus strain. Plants 2021, in press. [Google Scholar]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar] [PubMed]
- Trujillo-Correa, A.I.; Quintero-Gil, D.C.; Diaz-Castillo, F.; Quiñones, W.; Robledo, S.M.; Martinez-Gutierrez, M. In vitro and in silico anti-dengue activity of compounds obtained from Psidium guajava through bioprospecting. BMC Complement. Altern. Med. 2019, 19, 298. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Select. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Rakhshani, H.; Dehghanian, E.; Rahati, A. Enhanced GROMACS: Toward a better numerical simulation framework. J. Mol. Model. 2019, 25, 355. [Google Scholar] [CrossRef] [PubMed]
- Thielemann, H.C.; Cardellini, A.; Fasano, M.; Bergamasco, L.; Alberghini, M.; Ciorra, G.; Chiavazzo, E.; Asinari, P. From GROMACS to LAMMPS: GRO2LAM: A converter for molecular dynamics software. J. Mol. Model. 2019, 25, 147. [Google Scholar] [CrossRef] [PubMed]
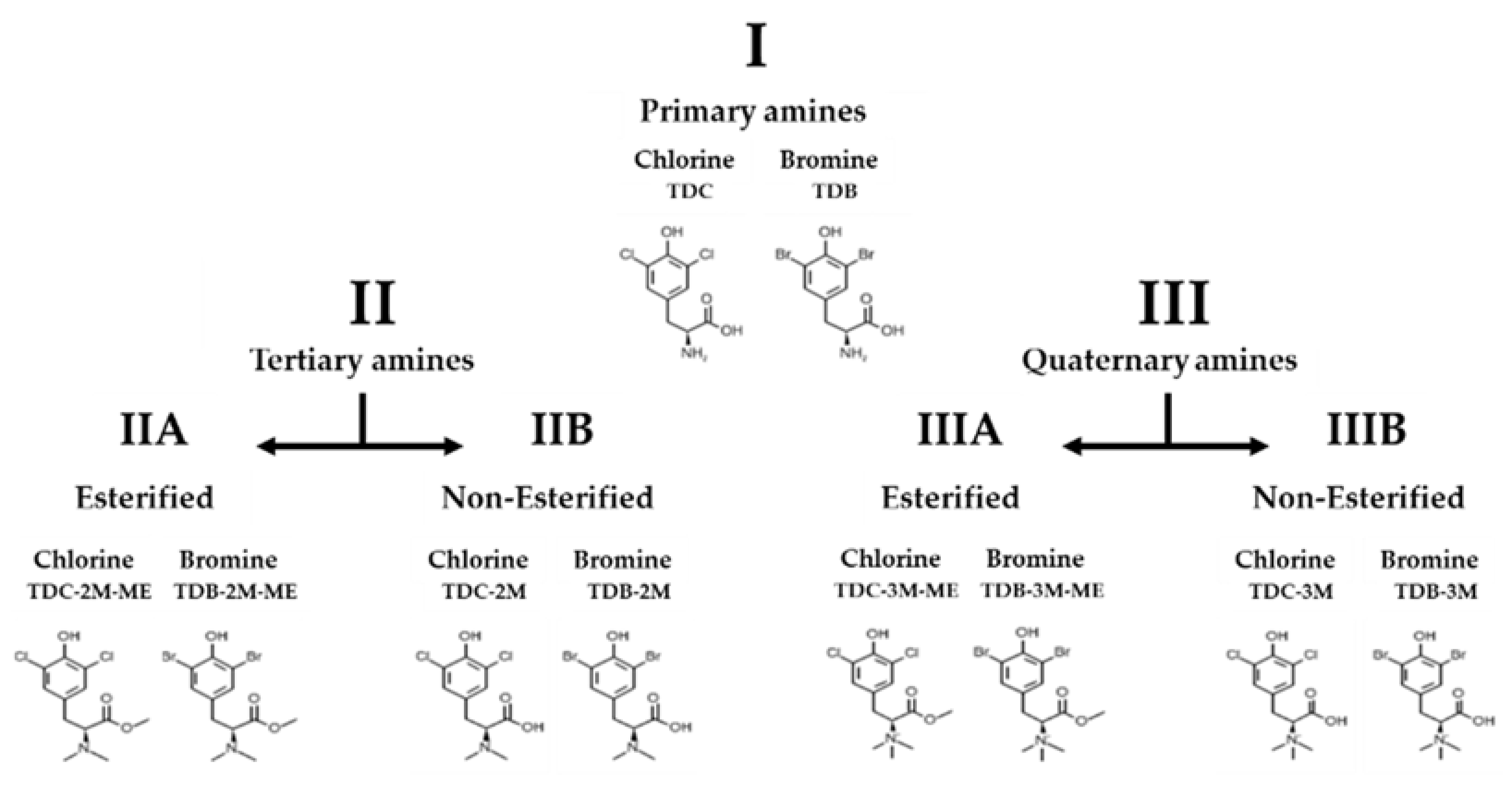




| Primary Amines | Tertiary Amines | Quaternary Amines | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Evaluated Toxicity | TDC | TDB | TDC-2M-ME | TDB-2M-ME | TDC-2M | TDB-2M | TDC-3M-ME | TDB-3M-ME | TDC-3M | TDB-3M |
| Chromosomal Aberrations | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Skin Sensitization | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| Respiratory Sensitization | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Neurotoxicity (Phospholipidosis) | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Cardiac Toxicity | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Endocrine Toxicity (estrogen receptor) | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Endocrine Toxicity (androgen receptor) | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Alkaline Phosphatase increase | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GGT increase | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| LDH increase | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| SGOT increase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SGPT increase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Reproductive toxicity | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Accumulated Toxicity | 2 | 1 | 3 | 4 | 1 | 3 | 5 | 4 | 2 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loaiza-Cano, V.; Monsalve-Escudero, L.M.; Restrepo, M.P.; Quintero-Gil, D.C.; Pulido Muñoz, S.A.; Galeano, E.; Zapata, W.; Martinez-Gutierrez, M. In Vitro and In Silico Anti-Arboviral Activities of Dihalogenated Phenolic Derivates of L-Tyrosine. Molecules 2021, 26, 3430. https://doi.org/10.3390/molecules26113430
Loaiza-Cano V, Monsalve-Escudero LM, Restrepo MP, Quintero-Gil DC, Pulido Muñoz SA, Galeano E, Zapata W, Martinez-Gutierrez M. In Vitro and In Silico Anti-Arboviral Activities of Dihalogenated Phenolic Derivates of L-Tyrosine. Molecules. 2021; 26(11):3430. https://doi.org/10.3390/molecules26113430
Chicago/Turabian StyleLoaiza-Cano, Vanessa, Laura Milena Monsalve-Escudero, Manuel Pastrana Restrepo, Diana Carolina Quintero-Gil, Sergio Andres Pulido Muñoz, Elkin Galeano, Wildeman Zapata, and Marlen Martinez-Gutierrez. 2021. "In Vitro and In Silico Anti-Arboviral Activities of Dihalogenated Phenolic Derivates of L-Tyrosine" Molecules 26, no. 11: 3430. https://doi.org/10.3390/molecules26113430
APA StyleLoaiza-Cano, V., Monsalve-Escudero, L. M., Restrepo, M. P., Quintero-Gil, D. C., Pulido Muñoz, S. A., Galeano, E., Zapata, W., & Martinez-Gutierrez, M. (2021). In Vitro and In Silico Anti-Arboviral Activities of Dihalogenated Phenolic Derivates of L-Tyrosine. Molecules, 26(11), 3430. https://doi.org/10.3390/molecules26113430







