PLS-DA Model for the Evaluation of Attention Deficit and Hyperactivity Disorder in Children and Adolescents through Blood Serum FTIR Spectra
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Stage
2.2. Spectroscopic Stage
2.3. Statistical Stage
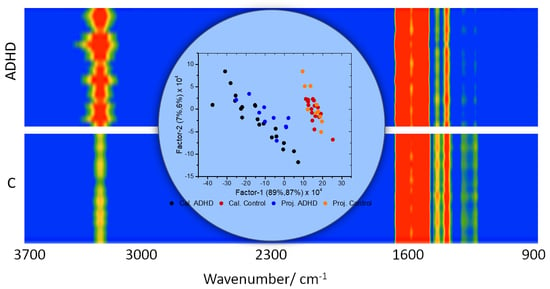
3. Results and Discussion
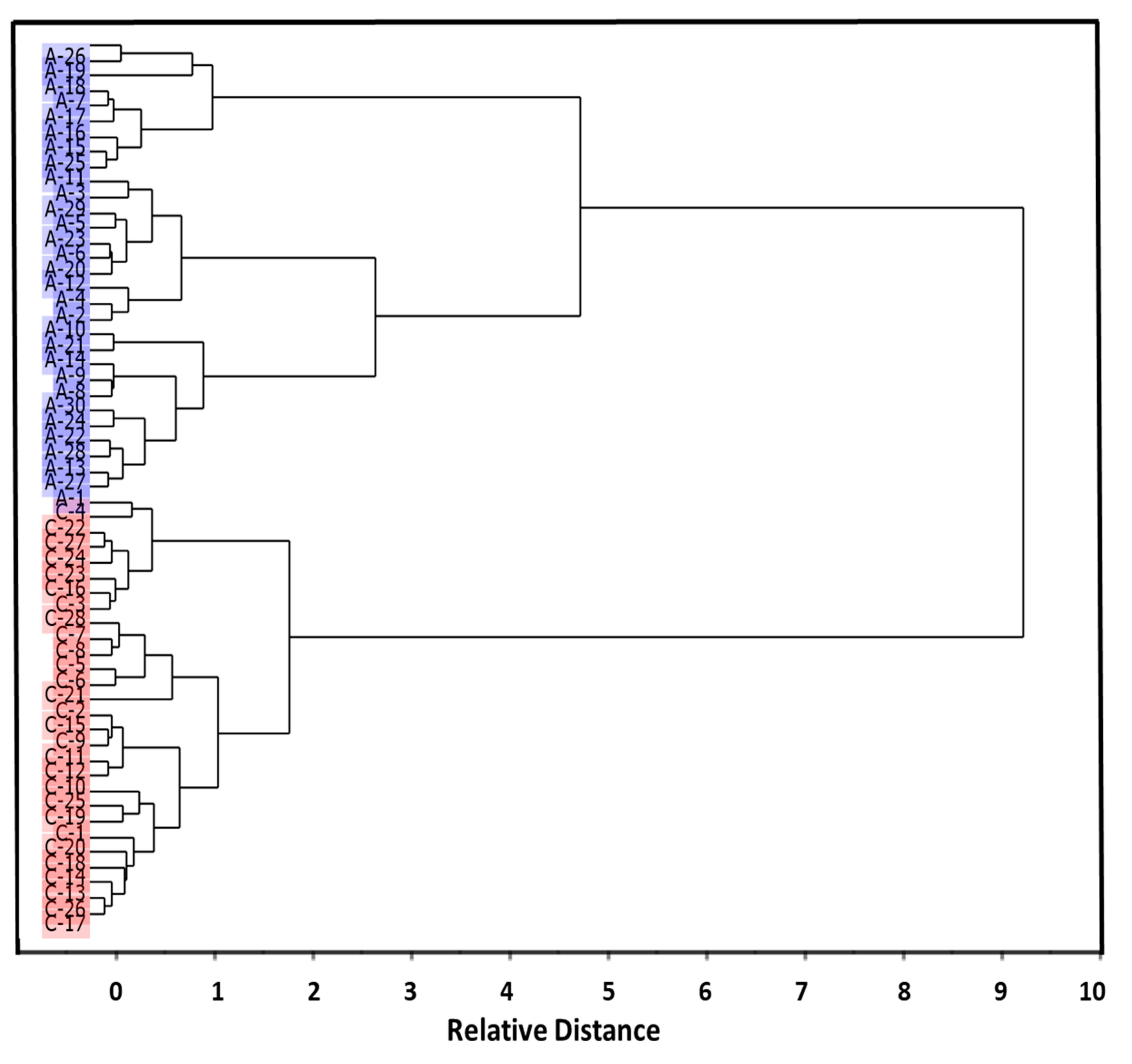
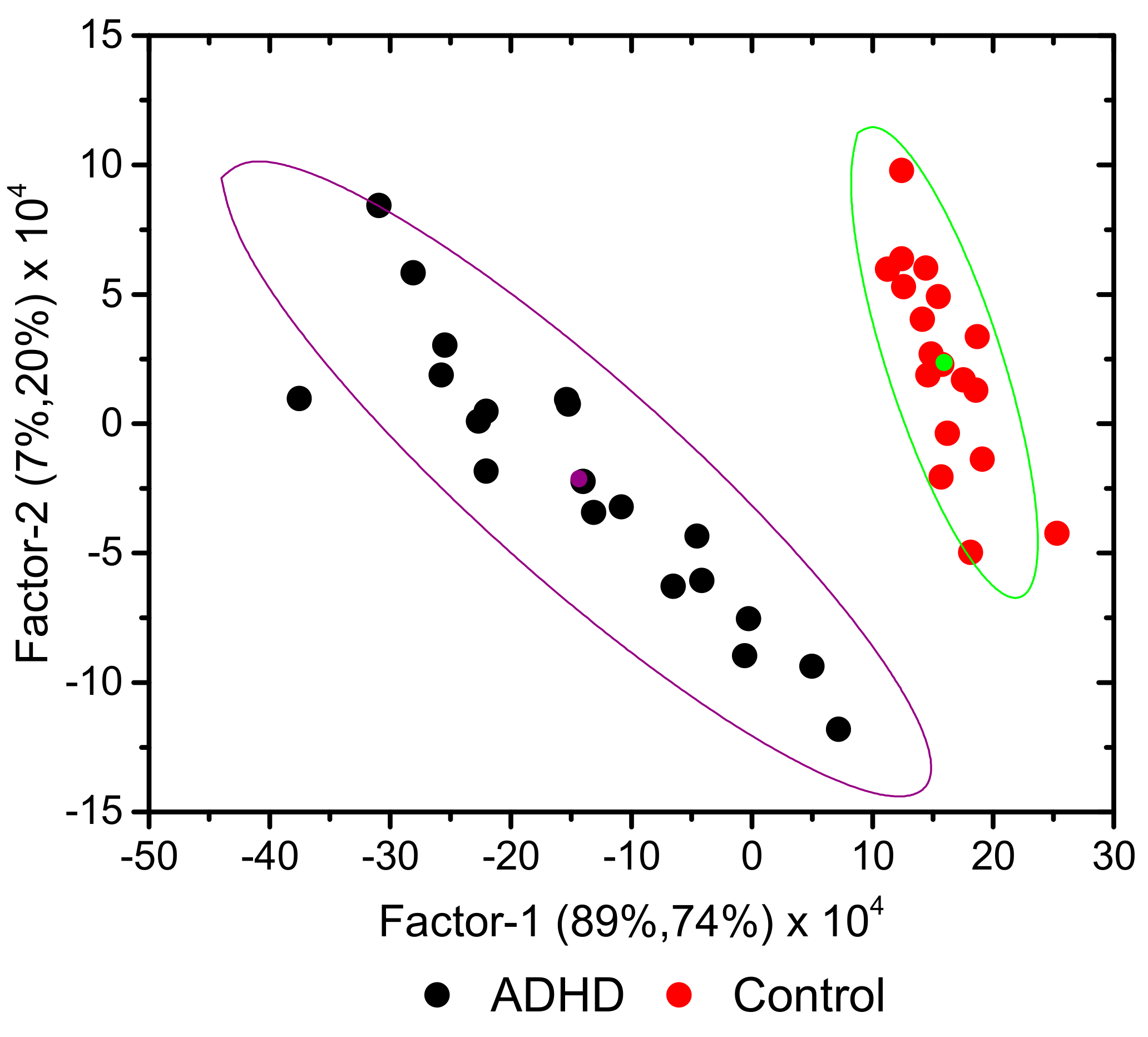
3.1. Preliminary Data Analysis
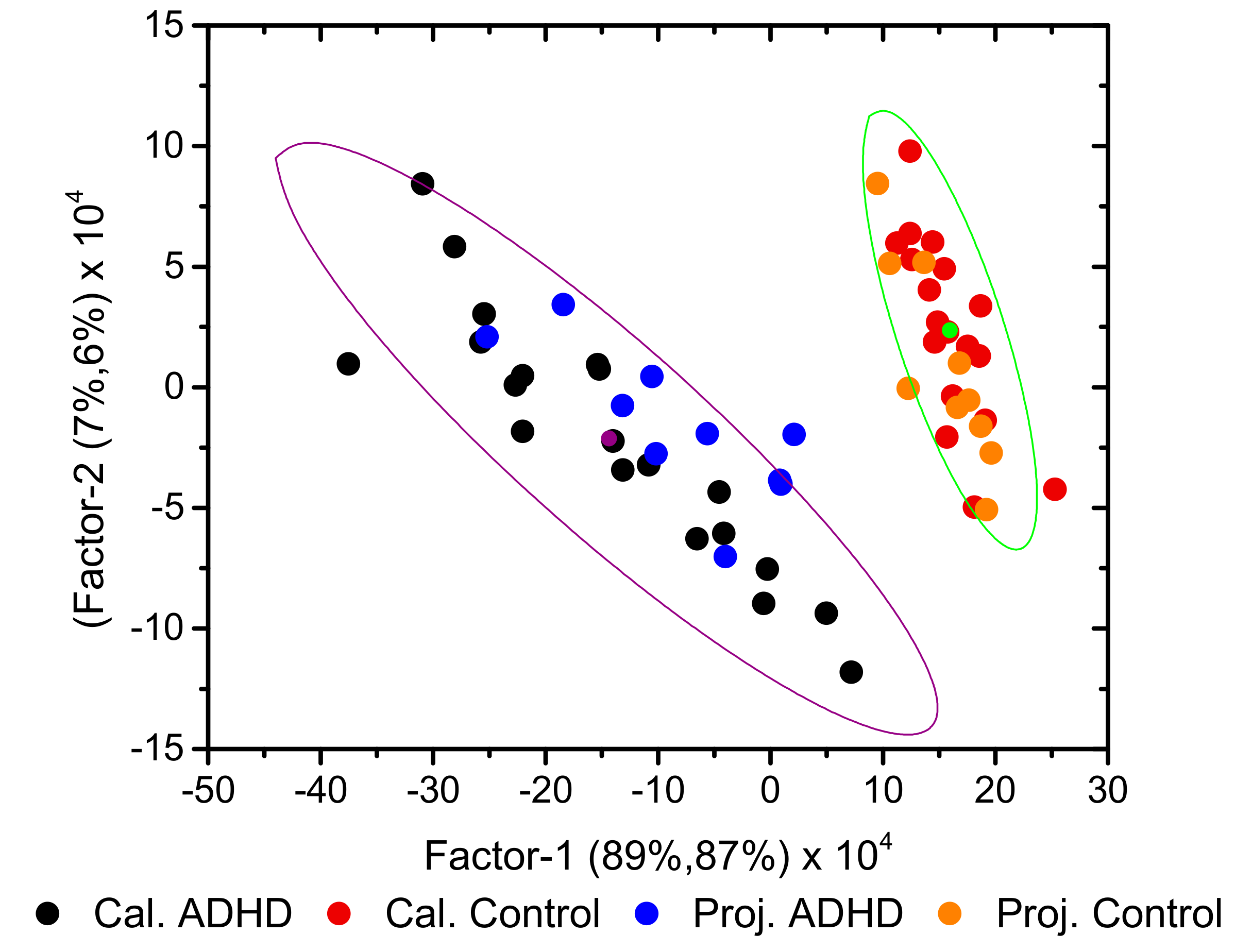
3.2. Development of Classification Model
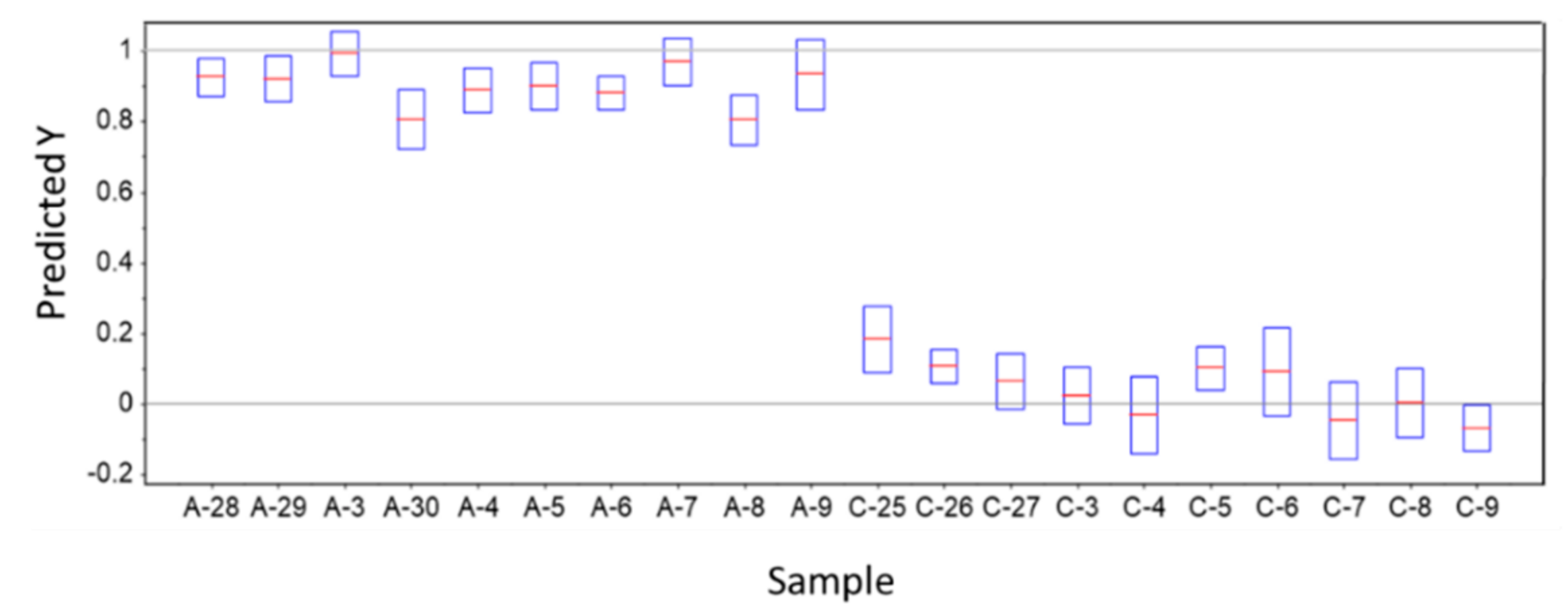
3.3. Predictions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sroubek, A.; Kelly, M.; Li, X. Inattentiveness in Attention-Deficit/Hyperactivity Disorder. Neurosci. Bull. 2013, 29, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Faraone, S.V.; Rostain, A.L.; Blader, J.; Busch, B.; Childress, A.C.; Connor, D.F.; Newcorn, J.H. Practitioner Review: Emotional Dysregulation in Attention-Deficit/Hyperactivity Disorder—Implications for Clinical Recognition and Intervention. J. Child Psychol. Psychiatry Allied Discip. 2019, 60, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Tenenbaum, R.B.; Musser, E.D.; Morris, S.; Ward, A.R.; Raiker, J.S.; Coles, E.K.; Pelham, W.E. Response Inhibition, Response Execution, and Emotion Regulation among Children with Attention-Deficit/Hyperactivity Disorder. J. Abnorm. Child Psychol. 2019, 47, 589–603. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Arlington, VA, USA, 2013. [Google Scholar]
- Ramsay, J.R. Cognitive Behavioral Therapy for Adult ADHD; Routledge: Abingdon-on-Thames, UK, 2007; pp. 25–26. [Google Scholar]
- Steinau, S. Diagnostic Criteria in Attention Deficit Hyperactivity Disorder—Changes in DSM 5. Front. Psychiatry 2013, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- National Institute for Health and Care Excellence. Attention Deficit Hyperactivity Disorder: Diagnosis and Management; National Guideline Centre: London, UK, 2019. [Google Scholar]
- Kooij, S.J.; Bejerot, S.; Blackwell, A.; Caci, H.; Casas-Brugué, M.; Carpentier, P.J.; Edvinsson, D.; Fayyad, J.; Foeken, K.; Fitzgerald, M.; et al. European Consensus Statement on Diagnosis and Treatment of Adult ADHD: The European Network Adult ADHD. BMC Psychiatry 2010, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Bálint, S.; Czobor, P.; Mészáros, A.; Simon, V.; Bitter, I. Neuropsychological Impairments in Adult Attention Deficit Hyperactivity Disorder: A Literature Review. Psychiatr. Hung. 2008, 23, 324–335. [Google Scholar] [PubMed]
- Ginsberg, Y.; Quintero, J.; Anand, E.; Casillas, M.; Upadhyaya, H.P. Underdiagnosis of Attention-Deficit/Hyperactivity Disorder in Adult Patients: A Review of the Literature. Prim. Care Companion CNS Disord. 2014, 16. [Google Scholar] [CrossRef] [PubMed]
- Millichap, J.G. Chapter 2: Causative Factors. In Attention Deficit Hyperactivity Disorder Handbook: A Physician’s Guide to ADHD, 2nd ed.; Springer Science: New York, NY, USA, 2010. [Google Scholar]
- Thapar, A.; Cooper, M.; Eyre, O.; Langley, K. What have we learnt about the causes of ADHD? J. Child Psychol. Psychiatry Allied Discip. 2013, 54, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Walitza, S.; Drechsler, R.; Ball, J. The School Child with ADHD. Ther. Umsch. 2012, 69, 467–473. [Google Scholar] [CrossRef]
- Wilens, T.E.; Spencer, T.J. Understanding Attention-Deficit/Hyperactivity Disorder from Childhood to Adulthood. Postgrad. Med. 2010, 122, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Scassellati, C.; Bonvicini, C.; Faraone, S.V.; Gennarelli, M. Biomarkers and Attention-Deficit/Hyperactivity Disorder: A Systematic Review and Meta-Analyses. J. Am. Acad. Child Adolesc. Psychiatry 2012, 51, 1003–1019. [Google Scholar] [CrossRef]
- Cheuk, D.K.L.; Wong, V. Attention-Deficit Hyperactivity Disorder and Blood Mercury Level: A Case-Control Study in Chinese Children. Neuropediatrics 2006, 37, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Jiamei, L.; Yunbo, M.; Min, L. Blood and Hair Zinc Levels in Children with Attention Deficit Hyperactivity Disorder: A Meta-Analysis. Asian J. Psychiatry 2020, 47, 101805. [Google Scholar]
- Yang, R.; Zhan, Y.; Gao, W.; Lin, N.; Li, R.; Zhao, Z. Blood Levels of Trace Elements in Children with Attention-Deficit Hyperactivity Disorder: Results from a Case-Control Study. Biol. Trace Elem. Res. 2006, 187, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, X.; Yang, B.; Ma, F.; Wang, S.; Tang, M.; Hao, M. Case–Control Study of Blood Lead Levels and Attention Deficit Hyperactivity Disorder in Chinese Children. Environ. Health Perspect. 2008, 116, 1401–1406. [Google Scholar] [CrossRef] [PubMed]
- Viktorinova, A.; Ursinyova, M.; Trebaticka, J.; Uhnakova, I.; Durackova, Z.; Masanova, V. Changed Plasma Levels of Zinc and Copper to Zinc Ratio and Their Possible Associations with Parent and Teacher Rated Symptoms in Children with Attention Deficit Hyperactivity Disorder. Biol. Trace Elem. Res. 2016, 169, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.R.; Hsu, S.F.; Hsu, C.D.; Hwang, L.H.; Yang, S.C. Dietary Patterns and Blood Fatty Acid Composition in Children with Attention-Deficit Hyperactivity Disorder in Taiwan. J. Nutr. Biochem. 2004, 15, 467–472. [Google Scholar] [CrossRef]
- Wang, M.; Hu, Z.; Liu, L.; Li, H.; Qian, Q.; Niu, H. Disrupted Functional Brain Connectivity Networks in Children with Attention-Deficit/Hyperactivity Disorder: Evidence from Resting-State Functional Near-Infrared Spectroscopy. Neurophotonics 2020, 7, 015012. [Google Scholar] [CrossRef] [PubMed]
- Güven, A.; Altınkaynak, M.; Dolu, N. Combining Functional Near-Infrared Spectroscopy and EEG Measurements for the Diagnosis of Attention-Deficit Hyperactivity Disorder. Neural Comput. Appl. 2020, 32, 8367–8380. [Google Scholar] [CrossRef]
- Chen, H.; Chen, W.; Song, Y.; Sun, L.; Li, X. EEG Characteristics of Children with Attention-Deficit/Hyperactivity Disorder. Neuroscience 2019, 406, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Nigg, J.T.; Knottnerus, G.M.; Martel, M.M.; Nikolas, M.; Cavanagh, K.; Karmaus, W.; Rappley, M.D. Low Blood Lead Levels Associated with Clinically Diagnosed Attention-Deficit/Hyperactivity Disorder and Mediated by Weak Cognitive Control. Biol. Psychiatry 2008, 63, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.D.; Shelton, R.C.; Duman, R.S. Functional Biomarkers of Depression: Diagnosis, Treatment, and Pathophysiology. Neuropsychopharmacology 2011, 36, 2375–2394. [Google Scholar] [CrossRef] [PubMed]
- Ogruc Ildiz, G.; Bayari, S.; Yorguner, N.; Fausto, R. Blood Serum Infrared Spectra Based Chemometric Models for Auxiliary Diagnosis of Autism Spectrum Disorder. In Autism Spectrum Disorder: Diagnosis and Treatment; El-Bazm, A.S., Mahmoud, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; in the press. [Google Scholar]
- Shaw, R.A.; Mantsch, H.H. Infrared Spectroscopy of Biological Fluids in Clinical and Diagnostic Analysis. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2006; Volume 2, pp. 1–24. [Google Scholar]
- Ogruc Ildiz, G.; Bayari, S.; Karadag, A.; Kaygisiz, E.; Fausto, R. Fourier Transform Infrared Spectroscopy Based Complementary Diagnosis Tool for Autism Spectrum Disorder in Children and Adolescents. Molecules 2020, 25, 2079. [Google Scholar] [CrossRef] [PubMed]
- Ogruc Ildiz, G.; Bayari, S.; Aksoy, U.M.; Yorguner, N.; Bulut, H.; Yilmaz, S.S.; Halimoglu, G.; Nur Kabuk, H.; Yavuz, G.; Fausto, R. Auxiliary Differential Diagnosis of Schizophrenia and Phases of Bipolar Disorder Based on the Blood Serum Raman Spectra. J. Raman Spectrosc. 2020, 51, 2233–2244. [Google Scholar] [CrossRef]
- Ahmed, S.S.S.J.; Santosh, W.; Kumar, S.; Christlet, T.H.T. Neural Network Algorithm for the Early Detection of Parkinson’s Disease from Blood Plasma by FTIR Micro-Spectroscopy. Vib. Spectrosc. 2010, 53, 181–188. [Google Scholar] [CrossRef]
- Deleris, G.; Petibois, C. Applications of FT-IR Spectrometry to Plasma Contents Analysis and Monitoring. Vib. Spectrosc. 2003, 32, 129–136. [Google Scholar] [CrossRef]
- Khanmohammadi, M.; Ghasemi, K.; Garmarudi, A.B.; Ramin, M. Diagnostic Prediction of Renal Failure from Blood Serum Analysis by FTIR Spectrometry and Chemometrics. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 136, 1782–1785. [Google Scholar] [CrossRef] [PubMed]
- Mostaco-Guidolin, L.B.; Bachmann, L. Application of FTIR Spectroscopy for Identification of Blood and Leukemia Biomarkers: A Review over the Past 15 Years. Appl. Spectrosc. Rev. 2011, 46, 388–404. [Google Scholar] [CrossRef]
- Erukhimovitch, V.; Talyshinsky, M.; Souprun, Y.; Huleihel, M. FTIR Spectroscopy Examination of Leukemia Patients Plasma. Vibrat. Spectrosc. 2006, 40, 40–46. [Google Scholar] [CrossRef]
- Lewis, P.D.; Lewis, K.E.; Ghosal, R.; Bayliss, S.; Lloyd, A.J.; Wills, J.; Godfrey, R.; Kloer, P.; Mur, L.A.J. Evaluation of FTIR Spectroscopy as a Diagnostic Tool for Lung Cancer Using Sputum. BMC Cancer 2010, 10, 640. [Google Scholar] [CrossRef] [PubMed]
- Mordechai, S.; Shufan, E.; Katz Porat, B.S.; Salman, A. Early Diagnosis of Alzheimer’s Disease Using Infrared Spectroscopy of Isolated Blood Samples Followed by Multivariate Analyses. Analyst 2017, 142, 1276–1284. [Google Scholar] [CrossRef]
- Krimm, S.; Bandekar, J. Vibrational Spectroscopy and Conformation of Peptides, Polypeptides and Proteins. Adv. Protein Chem. 1986, 38, 181–364. [Google Scholar] [PubMed]
- Liu, K.Z.; Shi, M.H.; Mantsch, H.H. Molecular and Chemical Characterization of Blood Cells by Infrared Spectroscopy: A New Optical Tool in Hematology. Blood Cells Mol. Dis. 2005, 35, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Pearson, K. On Lines and Planes of Closest Fit to Systems of Points in Space. Philos. Mag. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a Complex of Statistical Variables into Principal Components. J. Educ. Psychol. 1933, 24, 417–441. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a Complex of Statistical Variables into Principal Components. J. Educ. Psychol. 1933, 24, 498–520. [Google Scholar] [CrossRef]
- Hotelling, H. Relations Between Two Sets of Variates. Biometrika 1936, 28, 321–377. [Google Scholar] [CrossRef]
- Wold, H. Quantitative Sociology: International Perspectives on Mathematical and Statistical Model Building; Academic Press: New York, NY, USA, 1975; pp. 307–357. [Google Scholar]
- CAMO Software Inc. The UnscramblerTM Version 10.5; CAMO A/S: Trondheim, Norway, 2018. [Google Scholar]
- Ward, J.H.J. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-Regression: A Basic Tool of Chemometrics. Chemom. Intell. Lab. Sys. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Wold, H. Estimation of Principal Components and Related Models by Iterative Least Squares. In Multivariate Analysis; Krishnaiah, P.R., Ed.; Academic Press: New York, NY, USA, 1966; pp. 391–420. [Google Scholar]
- Bair, E.; Hastie, T.; Paul, D.; Tibshirani, R. Prediction by Supervised Principal Components. J. Am. Stat. Assoc. 2006, 101, 119–137. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.-Y.; Jemain, A.A. Partial Least Squares-Discriminant Analysis (PLS-DA) for Classification of High-Dimensional (HD) Data: A Review of Contemporary Practice Strategies and Knowledge Gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef] [PubMed]
- Barker, M.; Rayens, W. Partial Least Squares for Discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Wiest, M.M.; German, J.B.; Harvey, D.J.; Watkins, S.M.; Hertz-Picciotto, I. Plasma Fatty Acid Profiles in Autism: A Case-Control Study. Prostaglandins Leukot. Essent. Fat. Acids 2009, 80, 221–227. [Google Scholar] [CrossRef]
- Levy, S.E.; Souders, M.C.; Ittenbach, R.F.; Giarelli, E.; Mulberg, A.E.; Pinto-Martin, J.A. Relationship of Dietary Intake to Gastrointestinal Symptoms in Children with Autistic Spectrum Disorders. Biol. Psychiatry 2007, 61, 492–497. [Google Scholar] [CrossRef]
- Wilkinson, L.; Friendly, M. The History of the Cluster Heat Map. Am. Stat. 2009, 63, 179–184. [Google Scholar] [CrossRef]
- Krilov, D.; Balarin, M.; Kosovic, M.; Gamulin, O.; Brnjas-Kralkevic, J. FT-IR Spectroscopy of Lipoproteins—A Comparative Study. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2009, 73, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Paraskevaidi, M.; Morais, C.L.M.; Lima, K.M.G.; Snowden, J.S.; Saxon, J.A.; Richardson, A.M.T.; Jones, M.; Mann, D.M.A.; Allsop, D.; Martin-Hirsch, P.L.; et al. Differential Diagnosis of Alzheimer’s Disease Using Spectrochemical Analysis of Blood. Proc. Natl. Acad. Sci. USA 2017, E7929–E7938. [Google Scholar] [CrossRef]
- Sheng, D.; Liu, X.; Li, W.; Yang, Y.; Chen, X.; Wang, X. Distinction of Leukemia Patients’ and Healthy Persons’ Serum Using FTIR Spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 101, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Infrared Spectrum of Tyrosine, Coblentz Society Collection, Copyright 2018 by the U.S. Secretary of Commerce on Behalf of the United States of America, Ref 3614. Available online: https://webbook.nist.gov/cgi/cbook.cgi?ID=C60184&Mask=80#IR-Spec (accessed on 5 April 2021).
- Berry, M.D. The Potential of Trace Amines and Their Receptors for Treating Neurological and Psychiatric Diseases. Rev. Recent Clinic. Trials 2007, 2, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Zametkin, A.J.; Karoum, F.; Rapoport, J.L.; Brown, G.L.; Wyatt, R.J. Phenylethylamine Excretion in Attention Deficit Disorder. J. Am. Acad. Child Psychiatry 1984, 23, 310–314. [Google Scholar] [CrossRef]
- El Baz, F.M.; Zaki, M.M.; Youssef, A.M.; El Dorry, G.F.; Elalfy, D.Y. Study of Plasma Amino Acid Levels in Children with Autism: An Egyptian Sample. Egypt. J. Med. Hum. Genet. 2014, 15, 181–186. [Google Scholar] [CrossRef][Green Version]
- Tu, W.J.; Chen, H.; He, J. Application of LC-MS/M Analysis of Plasma Amino Acids Profiles in Children with Autism. J. Clin. Biochem. Nutr. 2012, 5, 1248–1249. [Google Scholar] [CrossRef] [PubMed]
- Tirouvanziam, R.; Obukhanych, T.V.; Laval, J.; Aronov, P.A.; Libove, R.; Banerjee, A.G.; Parker, K.J.; Ohara, R.; Herzenberg, L.A.; Herzenberg, L.A.; et al. Distinct Plasma Profile of Polar Neutral Amino Acids, Leucine, and Glutamate in Children with Autism Spectrum Disorders. J. Autism Dev. Disord. 2011, 42, 827–836. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogruc Ildiz, G.; Karadag, A.; Kaygisiz, E.; Fausto, R. PLS-DA Model for the Evaluation of Attention Deficit and Hyperactivity Disorder in Children and Adolescents through Blood Serum FTIR Spectra. Molecules 2021, 26, 3400. https://doi.org/10.3390/molecules26113400
Ogruc Ildiz G, Karadag A, Kaygisiz E, Fausto R. PLS-DA Model for the Evaluation of Attention Deficit and Hyperactivity Disorder in Children and Adolescents through Blood Serum FTIR Spectra. Molecules. 2021; 26(11):3400. https://doi.org/10.3390/molecules26113400
Chicago/Turabian StyleOgruc Ildiz, Gulce, Ahmet Karadag, Ersin Kaygisiz, and Rui Fausto. 2021. "PLS-DA Model for the Evaluation of Attention Deficit and Hyperactivity Disorder in Children and Adolescents through Blood Serum FTIR Spectra" Molecules 26, no. 11: 3400. https://doi.org/10.3390/molecules26113400
APA StyleOgruc Ildiz, G., Karadag, A., Kaygisiz, E., & Fausto, R. (2021). PLS-DA Model for the Evaluation of Attention Deficit and Hyperactivity Disorder in Children and Adolescents through Blood Serum FTIR Spectra. Molecules, 26(11), 3400. https://doi.org/10.3390/molecules26113400