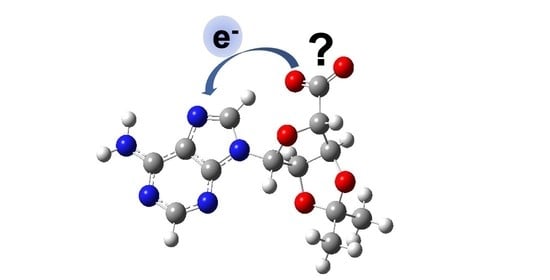
Intramolecular Photo-Oxidation as a Potential Source to Probe Biological Electron Damage: A Carboxylated Adenosine Analogue as Case Study
Abstract
1. Introduction
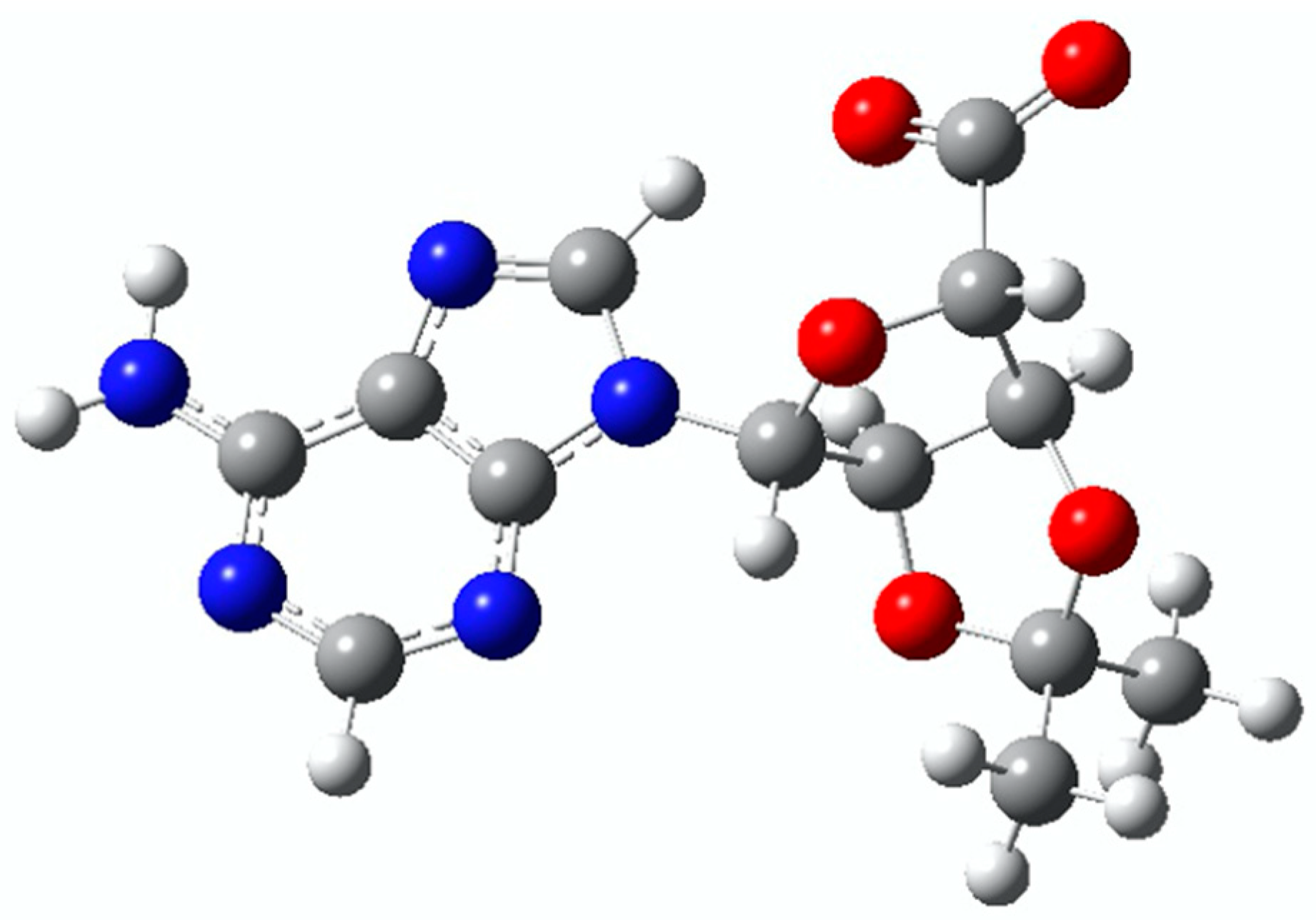
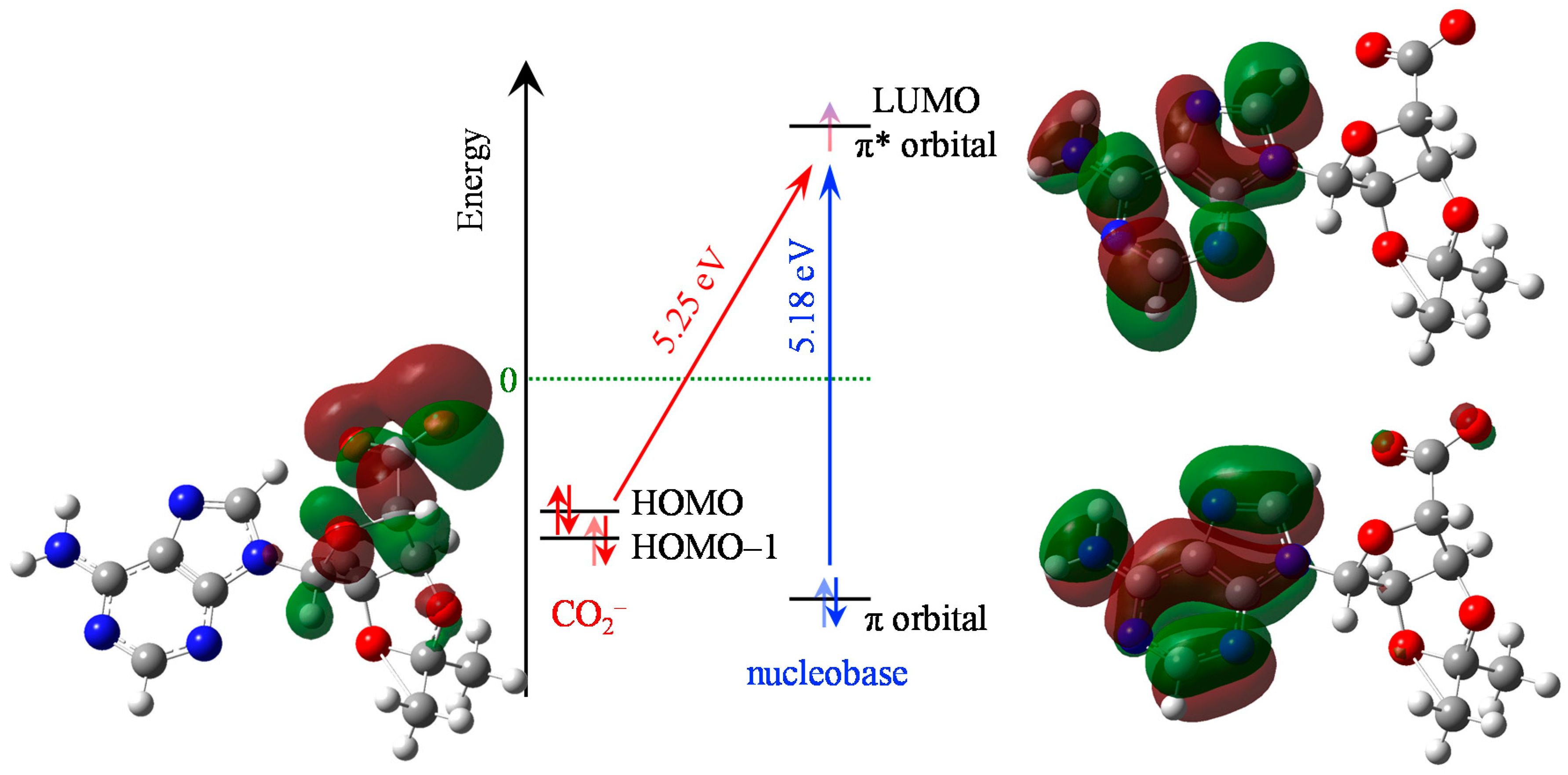
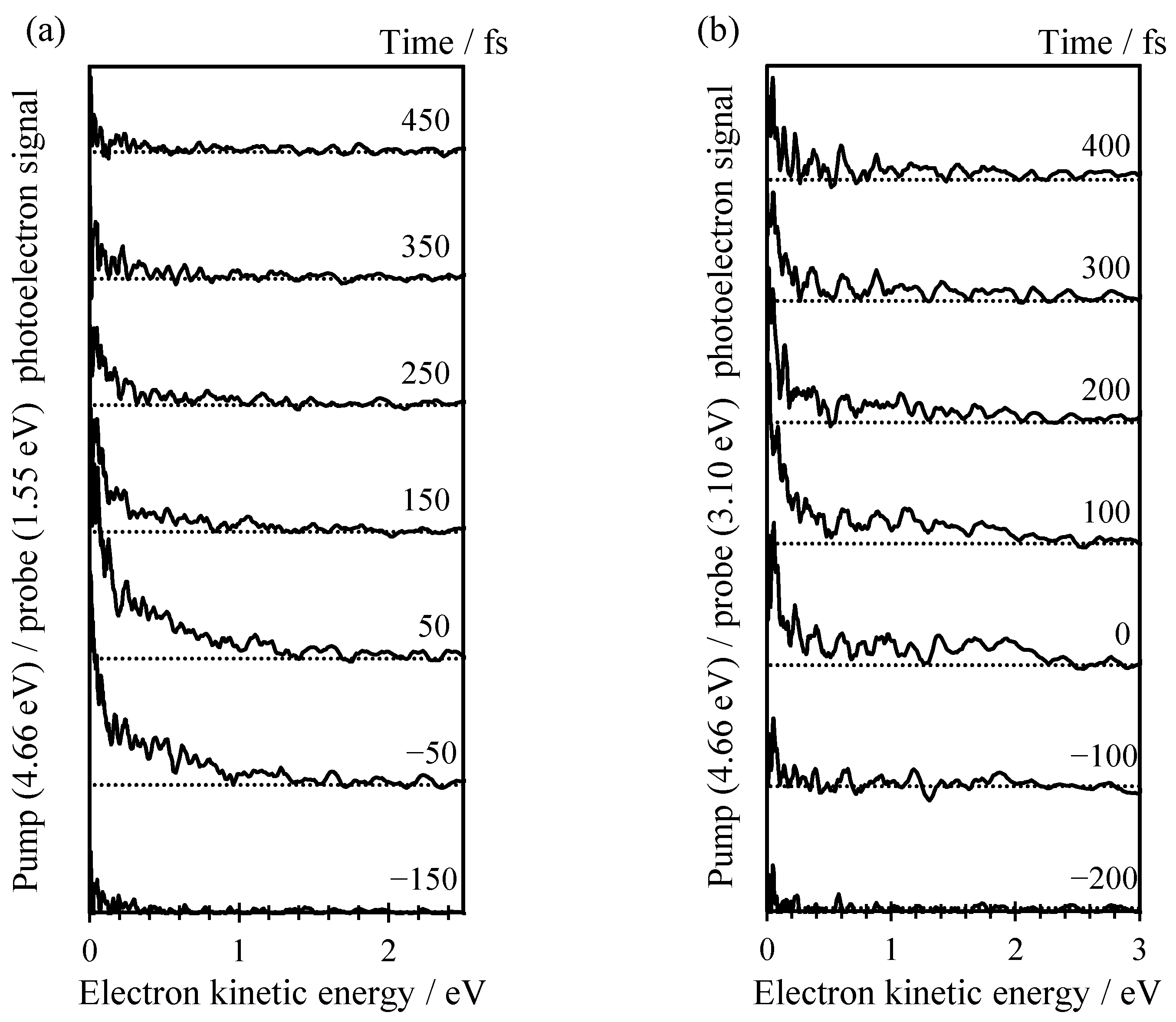
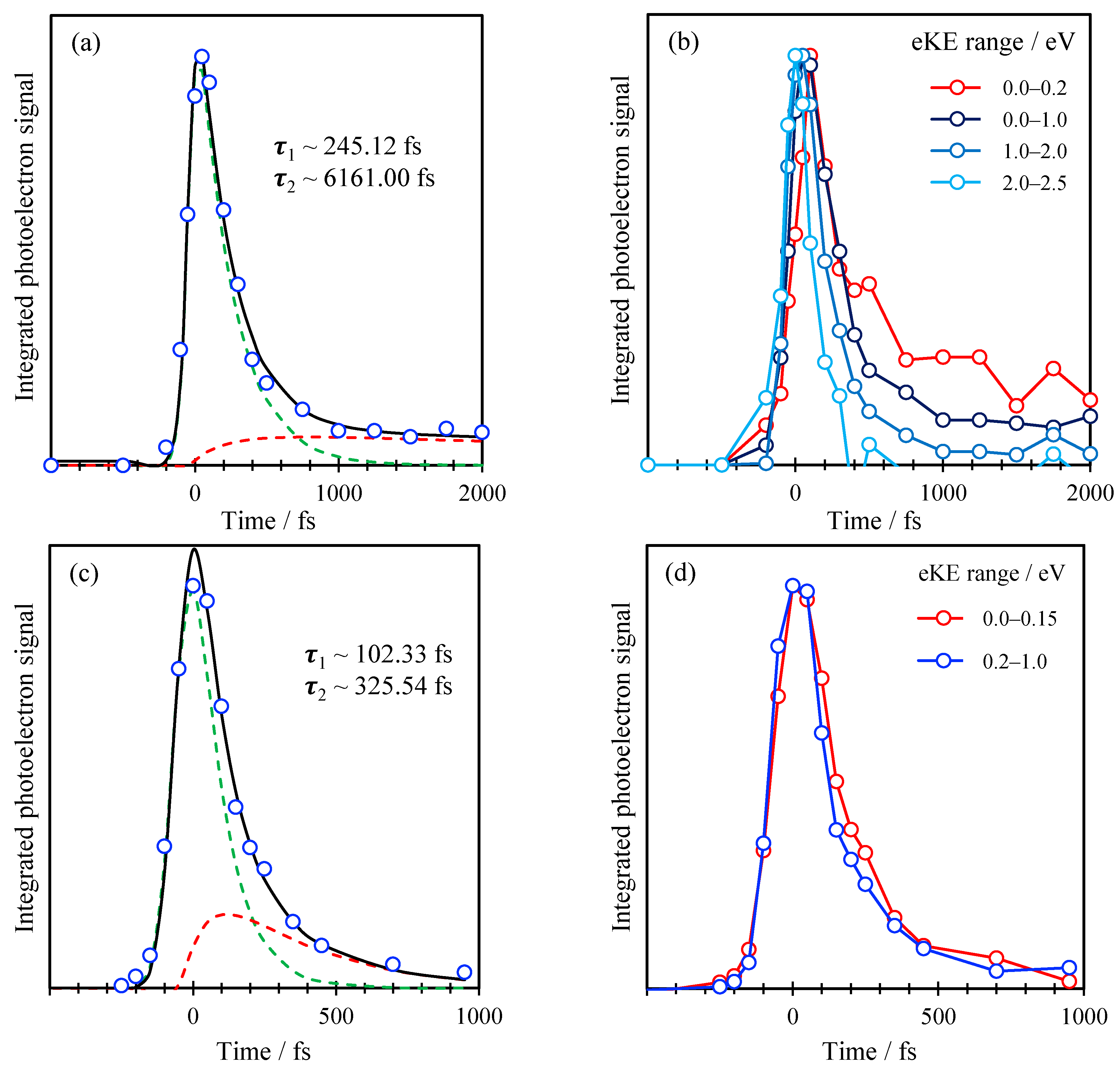
2. Results
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alberts, B. Molecular Biology of the Cell, 5th ed.; Garland Science: New York, NY, USA, 2008. [Google Scholar]
- Steenken, S. Purine bases, nucleosides, and nucleotides: Aqueous solution redox chemistry and transformation reactions of their radical cations and e- and OH adducts. Chem. Rev. 1989, 89, 503–520. [Google Scholar] [CrossRef]
- Burrows, C.J.; Muller, J.G. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 1998, 98, 1109–1151. [Google Scholar] [CrossRef]
- Simons, J. How do low-energy (0.1–2 eV) electrons cause DNA-strand breaks? Acc. Chem. Res. 2006, 39, 772–779. [Google Scholar] [CrossRef]
- Kumar, A.; Becker, D.; Adhikary, A.; Sevilla, M.D. Reaction of Electrons with DNA: Radiation Damage to Radiosensitization. Int. J. Mol. Sci. 2019, 20, 3998. [Google Scholar] [CrossRef]
- Zheng, Y.; Cloutier, P.; Hunting, D.J.; Wagner, J.R.; Sanche, L. Glycosidic bond cleavage of thymidine by low-energy electrons. J. Am. Chem. Soc. 2004, 126, 1002–1003. [Google Scholar] [CrossRef]
- Aflatooni, K.; Gallup, G.A.; Burrow, P.D. Electron attachment energies of the DNA bases. J. Phys. Chem. A 1998, 102, 6205–6207. [Google Scholar] [CrossRef]
- Ptasińska, S.; Denifl, S.; Scheier, P.; Märk, T.D. Inelastic electron interaction (attachment/ionization) with deoxyribose. J. Chem. Phys. 2004, 120, 8505–8511. [Google Scholar] [CrossRef]
- Li, X.; Sevilla, M.D.; Sanche, L. Density functional theory studies of electron interaction with DNA: Can zero eV electrons induce strand breaks? J. Am. Chem. Soc. 2003, 125, 13668–13669. [Google Scholar] [CrossRef]
- Scheer, A.M.; Aflatooni, K.; Gallup, G.A.; Burrow, P.D. Bond Breaking and Temporary Anion States in Uracil and Halouracils: Implications for the DNA Bases. Phys. Rev. Lett. 2004, 92, 068102. [Google Scholar] [CrossRef]
- Baccarelli, I.; Bald, I.; Gianturco, F.A.; Illenberger, E.; Kopyra, J. Electron-induced damage of DNA and its components: Experiments and theoretical models. Phys. Rep. 2011, 508, 1–44. [Google Scholar] [CrossRef]
- Sanche, L. Role of secondary low energy electrons in radiobiology and chemoradiation therapy of cancer. Chem. Phys. Lett. 2009, 474, 1–6. [Google Scholar] [CrossRef]
- Sanche, L. Low energy electron-driven damage in biomolecules. Eur. Phys. J. D 2005, 35, 367–390. [Google Scholar] [CrossRef]
- Boudaiffa, B.; Cloutier, P.; Hunting, D.; Huels, M.A.; Sanche, L. Resonant formation of DNA strand breaks by low-energy (3 to 20 eV) electrons. Science 2000, 287, 1658–1660. [Google Scholar]
- Burrow, P.D.; Gallup, G.A.; Modelli, A. Are There π* Shape Resonances in Electron Scattering from Phosphate Groups? J. Phys. Chem. A 2008, 112, 4106–4113. [Google Scholar] [CrossRef]
- Fennimore, M.A.; Matsika, S. Core-excited and shape resonances of uracil. Phys. Chem. Chem. Phys. 2016, 18, 30536–30545. [Google Scholar] [CrossRef]
- Abdoul-Carime, H.; Gohlke, S.; Illenberger, E. Site-specific dissociation of DNA bases by slow electrons at early stages of irradiation. Phys. Rev. Lett. 2004, 92, 168103. [Google Scholar] [CrossRef]
- Barrios, R.; Skurski, P.; Simons, J. Mechanism for damage to DNA by low-energy electrons. J. Phys. Chem. B 2002, 106, 7991–7994. [Google Scholar] [CrossRef]
- Berdys, J.; Anusiewicz, I.; Skurski, P.; Simons, J. Damage to Model DNA Fragments from Very Low-Energy (<1 eV) Electrons. J. Am. Chem. Soc. 2004, 126, 6441–6447. [Google Scholar] [CrossRef]
- Bhaskaran, R.; Sarma, M. The role of the shape resonance state in low energy electron induced single strand break in 2′-deoxycytidine-5′-monophosphate. Phys. Chem. Chem. Phys. 2015, 17, 15250–15257. [Google Scholar] [CrossRef] [PubMed]
- Khorsandgolchin, G.; Sanche, L.; Cloutier, P.; Wagner, J.R. Strand Breaks Induced by Very Low Energy Electrons: Product Analysis and Mechanistic Insight into the Reaction with TpT. J. Am. Chem. Soc. 2019, 141, 10315–10323. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Cloutier, P.; Hunting, D.; Sanche, L. Dissociative Electron Attachment to DNA. Phys. Rev. Lett. 2003, 90, 208102. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Burrow, P.D.; Cai, Z.L.; Cloutier, P.; Hunting, D.; Sanche, L. DNA strand breaks induced by 0-4 eV electrons: The role of shape resonances. Phys. Rev. Lett. 2004, 93, 068101. [Google Scholar] [CrossRef]
- Zheng, Y.; Cloutier, P.; Hunting, D.J.; Wagner, J.R.; Sanche, L. Phosphodiester and N-glycosidic bond cleavage in DNA induced by 4-15 eV electrons. J. Chem. Phys. 2006, 124, 064710. [Google Scholar] [CrossRef]
- Li, Z.; Cloutier, P.; Sanche, L.; Wagner, J.R. Low-energy electron-induced DNA damage: Effect of base sequence in oligonucleotide trimers. J. Am. Chem. Soc. 2010, 132, 5422–5427. [Google Scholar] [CrossRef]
- Horke, D.A.; Li, Q.; Blancafort, L.; Verlet, J.R. Ultrafast above-threshold dynamics of the radical anion of a prototypical quinone electron-acceptor. Nat. Chem. 2013, 5, 711–717. [Google Scholar] [CrossRef] [PubMed]
- West, C.W.; Bull, J.N.; Antonkov, E.; Verlet, J.R. Anion resonances of para-benzoquinone probed by frequency-resolved photoelectron imaging. J. Phys. Chem. A 2014, 118, 11346–11354. [Google Scholar] [CrossRef]
- Dessent, C.E.H.; Kim, J.; Johnson, M.A. Photochemistry of Halide Ion−Molecule Clusters: Dipole-Bound Excited States and the Case for Asymmetric Solvation. Acc. Chem. Res. 1998, 31, 527–534. [Google Scholar] [CrossRef]
- Serxner, D.; Dessent, C.E.H.; Johnson, M.A. Precursor of the Iaq− charge-transfer-to-solvent (CTTS) band in I—(H2O)n clusters. J. Chem. Phys. 1996, 105, 7231–7234. [Google Scholar] [CrossRef]
- Talbot, J.J.; Yang, N.; Huang, M.; Duong, C.H.; McCoy, A.B.; Steele, R.P.; Johnson, M.A. Spectroscopic Signatures of Mode-Dependent Tunnel Splitting in the Iodide–Water Binary Complex. J. Phys. Chem. A 2020, 124, 2991–3001. [Google Scholar] [CrossRef]
- Dessent, C.E.H.; Bailey, C.G.; Johnson, M.A. Dipole-bound excited states of the I−CH3CN and I−(CH3CN)2 ion–molecule complexes: Evidence for asymmetric solvation. J. Chem. Phys. 1995, 103, 2006–2015. [Google Scholar] [CrossRef]
- Dessent, C.E.H.; Kim, J.; Johnson, M.A. Spectroscopic observation of vibrational Feshbach resonances in near-threshold photoexcitation of X−·CH3NO2 (X− = I− and Br−). Faraday Discuss. 2000, 115, 395–406. [Google Scholar] [CrossRef]
- Dessent, C.E.H.; Bailey, C.G.; Johnson, M.A. Observation of the dipole-bound excited state of the I−⋅acetone ion-molecule complex. J. Chem. Phys. 1995, 102, 6335–6338. [Google Scholar] [CrossRef]
- Kunin, A.; Neumark, D.M. Time-resolved radiation chemistry: Femtosecond photoelectron spectroscopy of electron attachment and photodissociation dynamics in iodide-nucleobase clusters. Phys. Chem. Chem. Phys. 2019, 21, 7239–7255. [Google Scholar] [CrossRef] [PubMed]
- King, S.B.; Yandell, M.A.; Stephansen, A.B.; Neumark, D.M. Time-resolved radiation chemistry: Dynamics of electron attachment to uracil following UV excitation of iodide-uracil complexes. J. Chem. Phys. 2014, 141, 224310. [Google Scholar] [CrossRef] [PubMed]
- King, S.B.; Yandell, M.A.; Neumark, D.M. Time-resolved photoelectron imaging of the iodide–thymine and iodide–uracil binary cluster systems. Faraday Discuss. 2013, 163, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Yandell, M.A.; King, S.B.; Neumark, D.M. Time-Resolved Radiation Chemistry: Photoelectron Imaging of Transient Negative Ions of Nucleobases. J. Am. Chem. Soc. 2013, 135, 2128–2131. [Google Scholar] [CrossRef]
- King, S.B.; Stephansen, A.B.; Yokoi, Y.; Yandell, M.A.; Kunin, A.; Takayanagi, T.; Neumark, D.M. Electron accommodation dynamics in the DNA base thymine. J. Chem. Phys. 2015, 143, 024312. [Google Scholar] [CrossRef]
- Stephansen, A.B.; King, S.B.; Yokoi, Y.; Minoshima, Y.; Li, W.-L.; Kunin, A.; Takayanagi, T.; Neumark, D.M. Dynamics of dipole- and valence bound anions in iodide-adenine binary complexes: A time-resolved photoelectron imaging and quantum mechanical investigation. J. Chem. Phys. 2015, 143, 104308. [Google Scholar] [CrossRef]
- Rogers, J.P.; Anstoter, C.S.; Verlet, J.R.R. Ultrafast dynamics of low-energy electron attachment via a non-valence correlation-bound state. Nat. Chem. 2018, 10, 341–346. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.-B.; Vorpagel, E.R.; Wang, L.-S. Direct experimental observation of the low ionization potentials of guanine in free oligonucleotides by using photoelectron spectroscopy. Proc. Natl. Acad. Sci. USA 2004, 101, 17588–17592. [Google Scholar] [CrossRef]
- Chatterley, A.S.; Johns, A.S.; Stavros, V.G.; Verlet, J.R.R. Base-Specific Ionization of Deprotonated Nucleotides by Resonance Enhanced Two-Photon Detachment. J. Phys. Chem. A 2013, 117, 5299–5305. [Google Scholar] [CrossRef]
- Epp, J.B.; Widlanski, T.S. Facile Preparation of Nucleoside-5‘-carboxylic Acids. J. Org. Chem. 1999, 64, 293–295. [Google Scholar] [CrossRef] [PubMed]
- Chatterley, A.S.; West, C.W.; Roberts, G.M.; Stavros, V.G.; Verlet, J.R.R. Mapping the Ultrafast Dynamics of Adenine onto Its Nucleotide and Oligonucleotides by Time-Resolved Photoelectron Imaging. J. Phys. Chem. Lett. 2014, 5, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Stavros, V.G.; Verlet, J.R.R. Gas-Phase Femtosecond Particle Spectroscopy: A Bottom-Up Approach to Nucleotide Dynamics. Annu. Rev. Phys. Chem. 2016, 67, 211–232. [Google Scholar] [CrossRef]
- Castellani, M.E.; Avagliano, D.; Verlet, J.R.R. Ultrafast Dynamics of the Isolated Adenosine-5′-triphosphate Dianion Probed by Time-Resolved Photoelectron Imaging. J. Phys. Chem. A 2021. [Google Scholar] [CrossRef]
- Castellani, M.E.; Avagliano, D.; González, L.; Verlet, J.R.R. Site-Specific Photo-oxidation of the Isolated Adenosine-5′-triphosphate Dianion Determined by Photoelectron Imaging. J. Phys. Chem. Lett. 2020, 11, 8195–8201. [Google Scholar] [CrossRef]
- Chatterley, A.S.; West, C.W.; Stavros, V.G.; Verlet, J.R.R. Time-resolved photoelectron imaging of the isolated deprotonated nucleotides. Chem. Sci. 2014, 5, 3963–3975. [Google Scholar] [CrossRef]
- Vysotskii, Y.B. Effect of chemical substitution on the ionization potentials and electron affinity of systems with conjugated bonds. Theor. Exp. Chem. 1982, 17, 363–371. [Google Scholar] [CrossRef]
- Tully, J.C.; Preston, R.K. Trajectory Surface Hopping Approach to Nonadiabatic Molecular Collisions: The Reaction of H+ with D2. J. Chem. Phys. 1971, 55, 562–572. [Google Scholar] [CrossRef]
- Hammes-Schiffer, S.; Tully, J.C. Proton transfer in solution: Molecular dynamics with quantum transitions. J. Chem. Phys. 1994, 101, 4657–4667. [Google Scholar] [CrossRef]
- Mitrić, R.; Petersen, J.; Bonačić-Koutecký, V. Laser-field-induced surface-hopping method for the simulation and control of ultrafast photodynamics. Phys. Rev. A 2009, 79, 053416. [Google Scholar] [CrossRef]
- Mavri, J. Molecular Dynamics with Nonadiabatic Transitions: A Comparison of Methods. Mol. Simul. 2000, 23, 389–411. [Google Scholar] [CrossRef]
- Lecointre, J.; Roberts, G.M.; Horke, D.A.; Verlet, J.R.R. Ultrafast Relaxation Dynamics Observed Through Time-Resolved Photoelectron Angular Distributions. J. Phys. Chem. A 2010, 114, 11216–11224. [Google Scholar] [CrossRef]
- Stanley, L.H.; Anstöter, C.S.; Verlet, J.R.R. Resonances of the anthracenyl anion probed by frequency-resolved photoelectron imaging of collision-induced dissociated anthracene carboxylic acid. Chem. Sci. 2017, 8, 3054–3061. [Google Scholar] [CrossRef]
- Wiley, W.C.; McLaren, I.H. Time-of-Flight Mass Spectrometer with Improved Resolution. Rev. Sci. Instrum. 1955, 26, 1150–1157. [Google Scholar] [CrossRef]
- Horke, D.A.; Roberts, G.M.; Lecointre, J.; Verlet, J.R.R. Velocity-map imaging at low extraction fields. Rev. Sci. Instrum. 2012, 83, 063101. [Google Scholar] [CrossRef] [PubMed]
- Roberts, G.M.; Nixon, J.L.; Lecointre, J.; Wrede, E.; Verlet, J.R.R. Toward real-time charged-particle image reconstruction using polar onion-peeling. Rev. Sci. Instrum. 2009, 80, 053104. [Google Scholar] [CrossRef]
- Fingerman, S. CRC Handbook of Chemistry and Physics: A Ready-Reference Book of Chemical and Physical Data, 87th ed.; Sci-Tech News; CRC Press: Boca Raton, FL, USA, 2007; Volume 61, p. 38. [Google Scholar]
- Horke, D.A. Femtosecond Photoelectron Imaging of Anions; Durham University: Durham, UK, 2012. [Google Scholar]
- Frisch, M.J.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Krishnan, R.; Binkley, J.S.; Seeger, R.; Pople, J.A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 1980, 72, 650–654. [Google Scholar] [CrossRef]
- McLean, A.D.; Chandler, G.S. Contracted Gaussian basis sets for molecular calculations. I. Second row atoms, Z = 11–18. J. Chem. Phys. 1980, 72, 5639–5648. [Google Scholar] [CrossRef]
- Yanai, T.; Tew, D.P.; Handy, N.C. A new hybrid exchange–correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem. Phys. Lett. 2004, 393, 51–57. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castellani, M.E.; Verlet, J.R.R. Intramolecular Photo-Oxidation as a Potential Source to Probe Biological Electron Damage: A Carboxylated Adenosine Analogue as Case Study. Molecules 2021, 26, 2877. https://doi.org/10.3390/molecules26102877
Castellani ME, Verlet JRR. Intramolecular Photo-Oxidation as a Potential Source to Probe Biological Electron Damage: A Carboxylated Adenosine Analogue as Case Study. Molecules. 2021; 26(10):2877. https://doi.org/10.3390/molecules26102877
Chicago/Turabian StyleCastellani, Maria Elena, and Jan R. R. Verlet. 2021. "Intramolecular Photo-Oxidation as a Potential Source to Probe Biological Electron Damage: A Carboxylated Adenosine Analogue as Case Study" Molecules 26, no. 10: 2877. https://doi.org/10.3390/molecules26102877
APA StyleCastellani, M. E., & Verlet, J. R. R. (2021). Intramolecular Photo-Oxidation as a Potential Source to Probe Biological Electron Damage: A Carboxylated Adenosine Analogue as Case Study. Molecules, 26(10), 2877. https://doi.org/10.3390/molecules26102877