Analysis of Kojic Acid Derivatives as Competitive Inhibitors of Tyrosinase: A Molecular Modeling Approach
Abstract
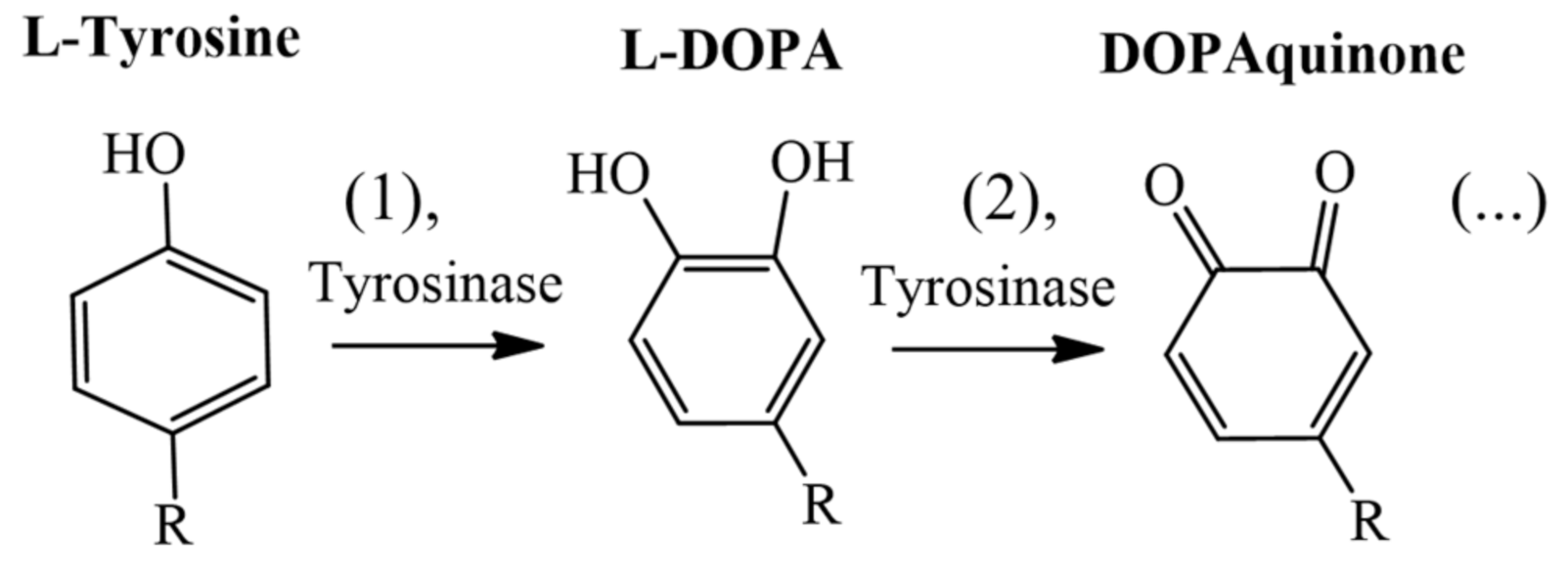
1. Introduction
2. Material and Methods
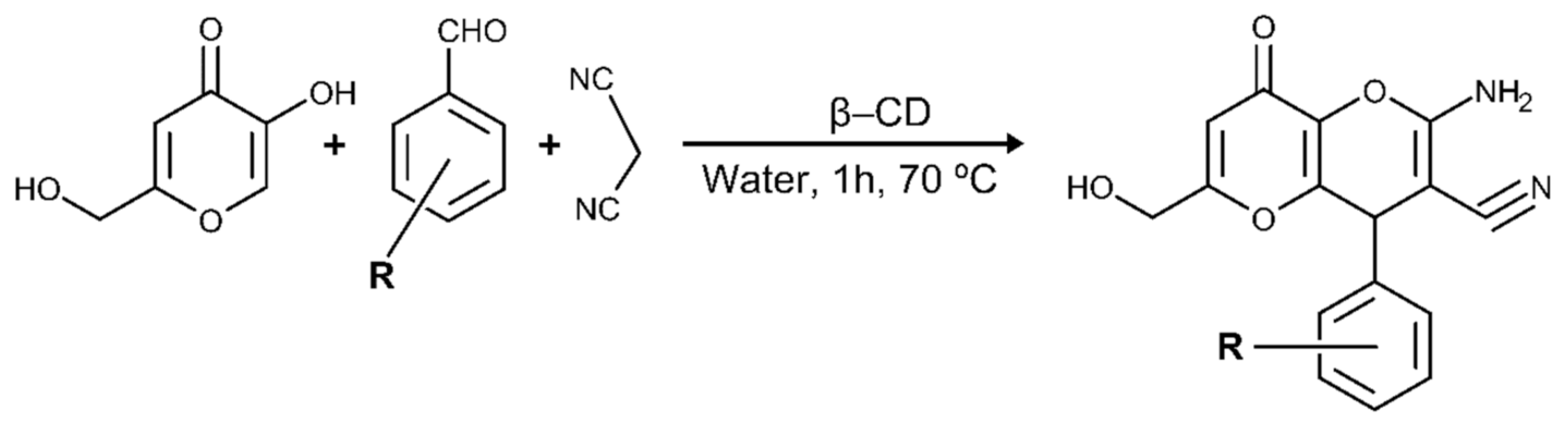
2.1. KA Derivatives Based on Aromatic Aldehydes and Malononitrile
2.2. Evaluation of Drug-like Properties
2.3. Molecular Docking
2.4. Molecular Dynamics (MD) Simulation
2.5. Binding Free Energy Calculations
3. Results and Discussion
3.1. Analysis of Selectivity of the KA Derivatives to the Tyrosinase Binding Site
3.2. Analysis of the Interaction of KA Derivatives with Tyrosinase Binding Pocket
3.3. Analysis of Interactions between the Histidine Residues and the Copper Ions
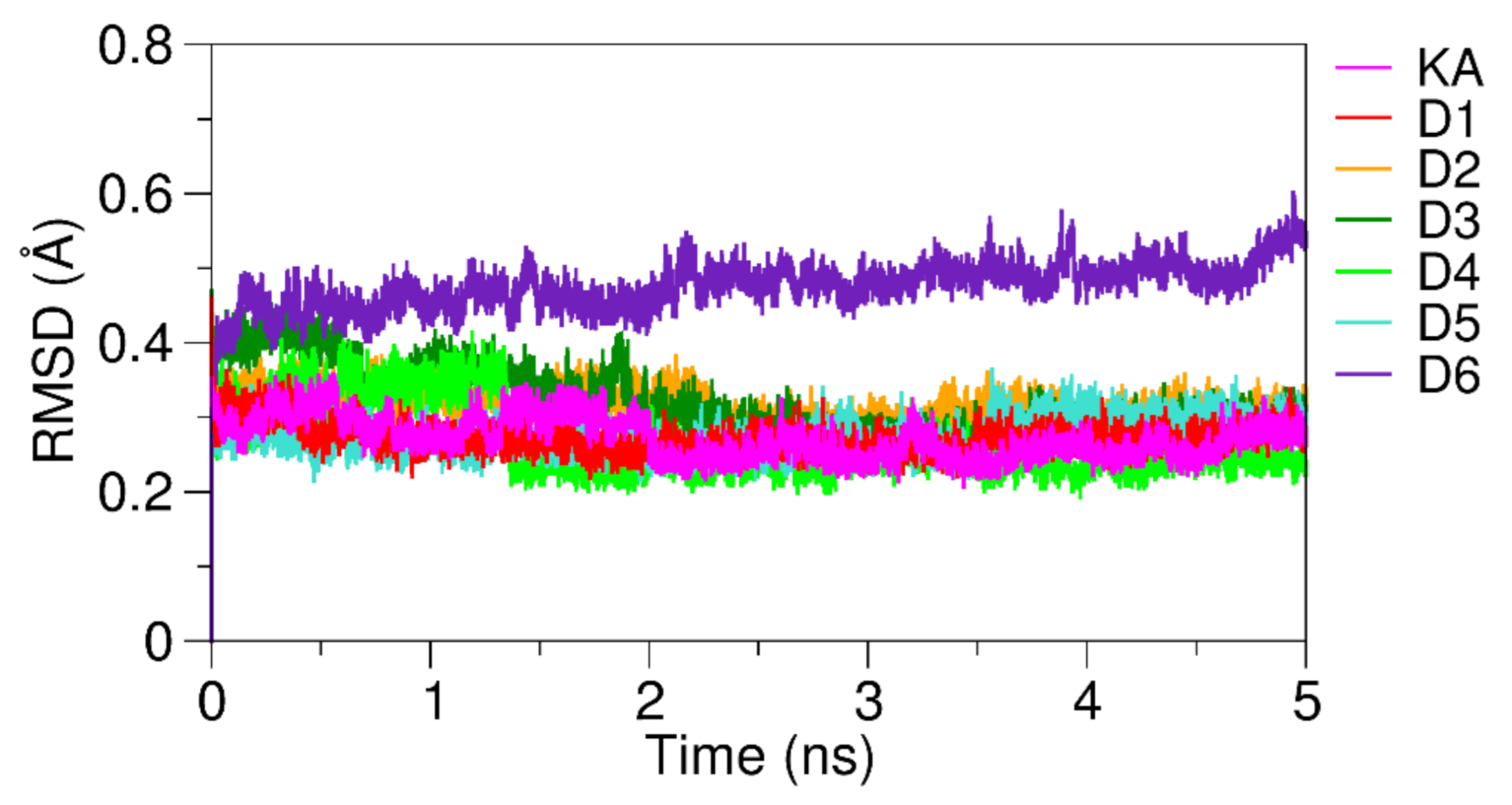
3.4. Analysis of KA Derivatives Complexed with Tyrosinase over MD Simulations
3.5. Analysis of Chelation of Copper Ions
3.6. Binding Affinity of KA Derivatives to the Tyrosinase Active Site
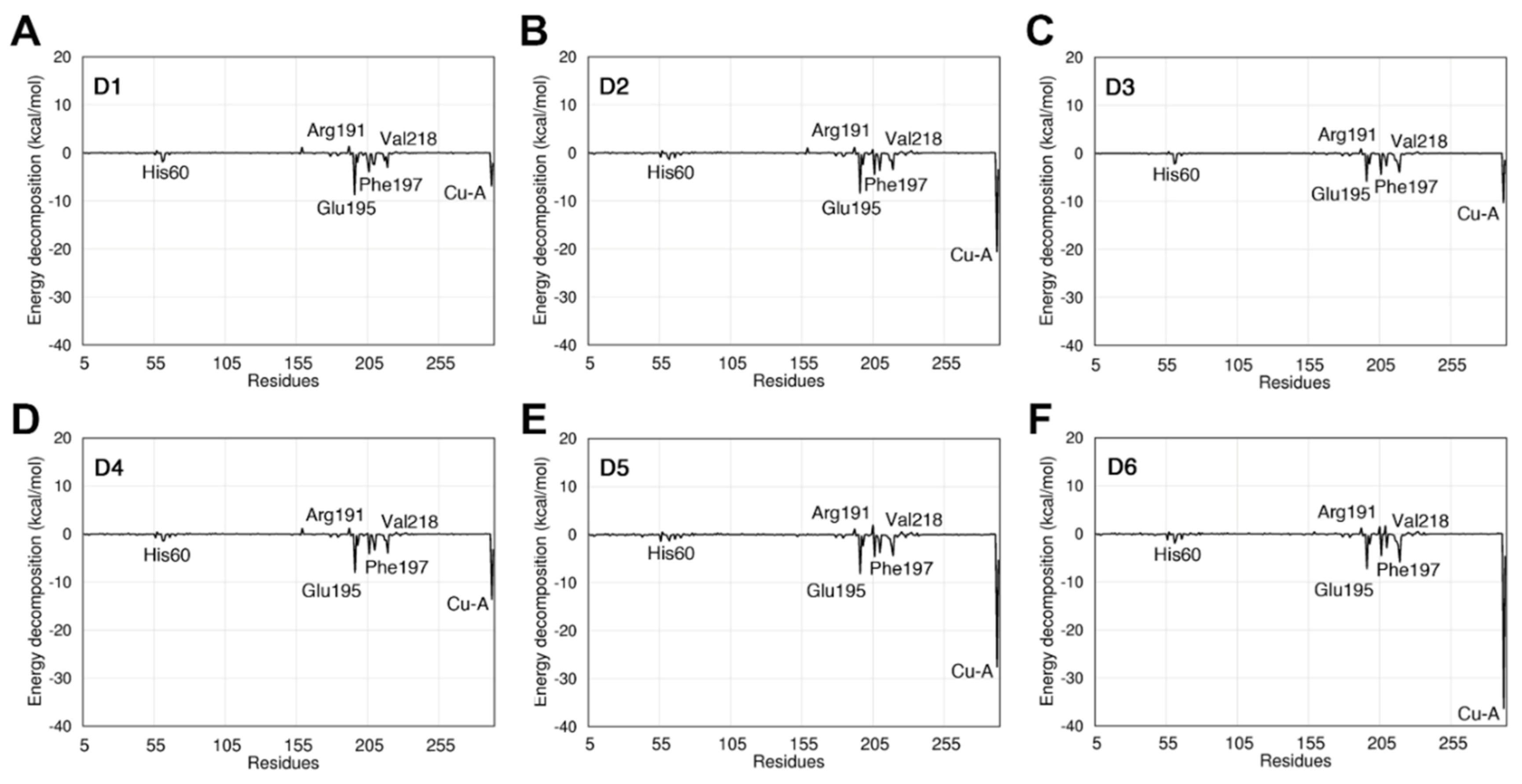
3.7. Analysis of Pairwise Energy Decomposition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- World Health Organization. OMS Ultraviolet (UV) Radiation and Skin Cancer. 2017. Available online: https://www.who.int/news-room/q-a-detail/radiation-ultraviolet-(uv)-radiation-and-skin-cancer (accessed on 10 March 2020).
- Ando, H. Melanogenesis. In Cosmetic Science and Technology: Theoretical Principles and Applications; Sakamoto, K., Lochhead, R.Y., Maibach, H.I., Yamashita, Y.B.T.-C.S.T., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 729–736. ISBN 9780128020548. [Google Scholar]
- Panzella, L.; Napolitano, A. Natural and bioinspired phenolic compounds as tyrosinase inhibitors for the treatment of skin hyperpigmentation: Recent advances. Cosmetics 2019, 6, 57. [Google Scholar] [CrossRef]
- Yousef, E.; Mitwally, N.; Noufal, N.; Tahir, M.R. Shift work and risk of skin cancer: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 2012. [Google Scholar] [CrossRef]
- Luo, H.; Ma, C. Identification of prognostic genes in uveal melanoma microenvironment. PLoS ONE 2020, 15, e0242263. [Google Scholar] [CrossRef]
- Taylor, N.J.; Gaynanova, I.; Eschrich, S.A.; Welsh, E.A.; Garrett, T.J.; Beecher, C.; Sharma, R.; Koomen, J.M.; Smalley, K.S.M.; Messina, J.L.; et al. Metabolomics of primary cutaneous melanoma and matched adjacent extratumoral microenvironment. PLoS ONE 2020, 15, e0240849. [Google Scholar] [CrossRef]
- Solano, F. On the metal cofactor in the tyrosinase family. Int. J. Mol. Sci. 2018, 19, 633. [Google Scholar] [CrossRef] [PubMed]
- Deri, B.; Kanteev, M.; Goldfeder, M.; Lecina, D.; Guallar, V.; Adir, N.; Fishman, A. The unravelling of the complex pattern of tyrosinase inhibition. Sci. Rep. 2016, 6, 34993. [Google Scholar] [CrossRef]
- Garcia-Jimenez, A.; Teruel-Puche, J.A.; Garcia-Ruiz, P.A.; Saura-Sanmartin, A.; Berna, J.; Garcia-Canovas, F.; Rodriguez-Lopez, J.N. Structural and kinetic considerations on the catalysis of deoxyarbutin by tyrosinase. PLoS ONE 2017, 12, e0187845. [Google Scholar] [CrossRef]
- Sánchez-Ferrer, Á.; Neptuno Rodríguez-López, J.; García-Cánovas, F.; García-Carmona, F. Tyrosinase: A comprehensive review of its mechanism. Biochim. Biophys. Acta (BBA)/Protein Struct. Mol. 1995, 1247, 1–11. [Google Scholar] [CrossRef]
- Rescigno, A.; Sollai, F.; Pisu, B.; Rinaldi, A.; Sanjust, E. Tyrosinase inhibition: General and applied aspects. J. Enzyme Inhib. Med. Chem. 2002, 17, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Şöhretoğlu, D.; Sari, S.; Barut, B.; Özel, A. Tyrosinase inhibition by a rare neolignan: Inhibition kinetics and mechanistic insights through in vitro and in silico studies. Comput. Biol. Chem. 2018, 76, 61–66. [Google Scholar] [CrossRef]
- Ai, N.; Welsh, W.J.; Santhanam, U.; Hu, H.; Lyga, J. Novel Virtual Screening Approach for the Discovery of Human Tyrosinase Inhibitors. PLoS ONE 2014, 9, e112788. [Google Scholar] [CrossRef]
- Park, J.; Jung, H.; Jang, B.; Song, H.-K.; Han, I.-O.; Oh, E.-S. D-tyrosine adds an anti-melanogenic effect to cosmetic peptides. Sci. Rep. 2020, 10, 262. [Google Scholar] [CrossRef]
- Wagle, A.; Seong, S.H.; Joung, E.-J.; Kim, H.-R.; Jung, H.A.; Choi, J.S. Discovery of a Highly Potent Tyrosinase Inhibitor, Luteolin 5- O -β- d -Glucopyranoside, Isolated from Cirsium japonicum var. maackii (Maxim.) Matsum., Korean Thistle: Kinetics and Computational Molecular Docking Simulation. ACS Omega 2018, 3, 17236–17245. [Google Scholar] [CrossRef]
- Lima, C.R.; Silva, J.R.A.; De Tássia Carvalho Cardoso, É.; Silva, E.O.; Lameira, J.; Do Nascimento, J.L.M.; Do Socorro Barros Brasil, D.; Alves, C.N. Combined kinetic studies and computational analysis on kojic acid analogs as tyrosinase inhibitors. Molecules 2014, 19, 9591–9605. [Google Scholar] [CrossRef]
- Asadzadeh, A.; Fassihi, A.; Yaghmaei, P.; Pourfarzam, M. Docking studies of some novel Kojic acid Derivatives as possible tyrosinase inhibitors. Biomed. Pharmacol. J. 2015, 8, 535–545. [Google Scholar] [CrossRef]
- Karakaya, G.; Türe, A.; Ercan, A.; Öncül, S.; Aytemir, M.D. Synthesis, computational molecular docking analysis and effectiveness on tyrosinase inhibition of kojic acid derivatives. Bioorg. Chem. 2019, 88, 102950. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, M.; Eslamifar, M.; Khezri, K. Kojic acid applications in cosmetic and pharmaceutical preparations. Biomed. Pharmacother. 2019, 110, 582–593. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Li, C.; Zhang, W.J.; Shi, Y.; Wen, Z.J.; Chen, Q.X.; Wang, Q. Kinetic and computational molecular docking simulation study of novel kojic acid derivatives as anti-tyrosinase and antioxidant agents. J. Enzyme Inhib. Med. Chem. 2019, 34, 990–998. [Google Scholar] [CrossRef]
- Ashooriha, M.; Khoshneviszadeh, M.; Khoshneviszadeh, M.; Moradi, S.E.; Rafiei, A.; Kardan, M.; Emami, S. 1,2,3-Triazole-based kojic acid analogs as potent tyrosinase inhibitors: Design, synthesis and biological evaluation. Bioorg. Chem. 2019, 82, 414–422. [Google Scholar] [CrossRef]
- Xie, W.; Zhang, H.; He, J.; Zhang, J.; Yu, Q.; Luo, C.; Li, S. Synthesis and biological evaluation of novel hydroxybenzaldehyde-based kojic acid analogues as inhibitors of mushroom tyrosinase. Bioorg. Med. Chem. Lett. 2017, 27, 530–532. [Google Scholar] [CrossRef]
- Kataev, E.A.; Ramana Reddy, M.; Niranjan Reddy, G.; Reddy, V.H.; Suresh Reddy, C.; Subba Reddy, B.V. Supramolecular catalysis by β-cyclodextrin for the synthesis of kojic acid derivatives in water. New J. Chem. 2016, 40, 1693–1697. [Google Scholar] [CrossRef]
- Sarrafi, Y.; Mehrasbi, E.; Mashalchi, S.Z. MCM-41-SO3H: An efficient, reusable, heterogeneous catalyst for the one-pot, three-component synthesis of pyrano[3,2-b]pyrans. Res. Chem. Intermed. 2015, 47, 1729–1741. [Google Scholar] [CrossRef]
- Chang, T.-S. An Updated Review of Tyrosinase Inhibitors. Int. J. Mol. Sci. 2009, 10, 2440–2475. [Google Scholar] [CrossRef]
- ChemAxon Marvin. Available online: https://chemaxon.com/products/marvin (accessed on 10 March 2020).
- Stewart, J.J.P. Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters. J. Mol. Model. 2013, 19, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J.P. Mopac2009. Available online: http://openmopac.net/background.html (accessed on 10 March 2020).
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J.P.J.; Lombardo, F. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Muegge, I. Selection criteria for drug-like compounds. Med. Res. Rev. 2003, 23, 302–321. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Stierand, K.; Rarey, M. Drawing the PDB: Protein-ligand complexes in two dimensions. ACS Med. Chem. Lett. 2010, 1, 540–545. [Google Scholar] [CrossRef]
- Goldberg, D.E. Genetic algorithms in search, optimization, and machine learning. Choice Rev. Online 1989, 27, 27–0936-27–0936. [Google Scholar]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins Struct. Funct. Bioinforma. 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Marelius, J.; Kolmodin, K.; Feierberg, I.; Åqvist, J. Q: A molecular dynamics program for free energy calculations and empirical valence bond simulations in biomolecular systems. J. Mol. Graph. Model. 1998, 16, 213–225. [Google Scholar] [CrossRef]
- Duarte, F.; Bauer, P.; Barrozo, A.; Amrein, B.A.; Purg, M.; Åqvist, J.; Kamerlin, S.C.L. Force Field Independent Metal Parameters Using a Nonbonded Dummy Model. J. Phys. Chem. B 2014, 118, 4351–4362. [Google Scholar] [CrossRef] [PubMed]
- Liao, Q.; Kamerlin, S.C.L.; Strodel, B. Development and Application of a Nonbonded Cu2+ Model That Includes the Jahn-Teller Effect. J. Phys. Chem. Lett. 2015, 6, 2657–2662. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef] [PubMed]
- Valipour, E.; Arikan, B. Characterization of tyrosinase enzyme from native Bacillus megaterium SP. STRAIN M36. J. Microbiol. Biotechnol. Food Sci. 2016, 05, 465–469. [Google Scholar]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Lee, F.S.; Warshel, A. A local reaction field method for fast evaluation of long-range electrostatic interactions in molecular simulations. J. Chem. Phys. 1992, 97, 3100–3107. [Google Scholar] [CrossRef]
- Grant, B.J.; Rodrigues, A.P.C.C.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S.D.D. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef]
- Gutiérrez-De-Terán, H.; Åqvist, J. Linear interaction energy: Method and applications in drug design. Methods Mol. Biol. 2012, 819, 305–323. [Google Scholar]
- Hansson, T.; Marelius, J.; Åqvist, J. Ligand binding affinity prediction by linear interaction energy methods. J. Comput. Aided. Mol. Des. 1998, 12, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Rifai, E.A.; van Dijk, M.; Geerke, D.P. Recent Developments in Linear Interaction Energy Based Binding Free Energy Calculations. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Cong, Y.; Li, M.; Bao, J.; Qi, Y.; Zhang, J.Z.H. Molecular Mechanism of Selective Binding of NMS-P118 to PARP-1 and PARP-2: A Computational Perspective. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef]
- De Alencar, N.A.N.D.; Sousa, P.R.M.; Silva, J.R.A.; Lameira, J.; Alves, C.N.; Martí, S.; Moliner, V. Computational analysis of human OGA structure in complex with PUGNAc and NAG-thiazoline derivatives. J. Chem. Inf. Model. 2012, 52, 2775–2783. [Google Scholar] [CrossRef]
- Do Nascimento, L.D.; de Moraes, A.A.B.; da Costa, K.S.; Galúcio, J.M.P.; Taube, P.S.; Costa, C.M.L.; Cruz, J.N.; Andrade, E.H.d.A.; de Faria, L.J.G. Bioactive natural compounds and antioxidant activity of essential oils from spice plants: New findings and potential applications. Biomolecules 2020, 10, 988. [Google Scholar] [CrossRef]
- Rayan, A.; Raiyn, J.; Falah, M. Nature is the best source of anticancer drugs: Indexing natural products for their anticancer bioactivity. PLoS ONE 2017, 12, e0187925. [Google Scholar] [CrossRef] [PubMed]
- Basmadjian, C.; Zhao, Q.; Bentouhami, E.; Djehal, A.; Nebigil, C.G.; Johnson, R.A.; Serova, M.; de Gramont, A.; Faivre, S.; Raymond, E.; et al. Cancer wars: Natural products strike back. Front. Chem. 2014, 2, 20. [Google Scholar] [CrossRef]
- de Oliveira, M.D.; Araújo, J.d.O.; Galúcio, J.M.P.; Santana, K.; Lima, A.H. Targeting shikimate pathway: In silico analysis of phosphoenolpyruvate derivatives as inhibitors of EPSP synthase and DAHP synthase. J. Mol. Graph. Model. 2020, 101, 107735. [Google Scholar] [CrossRef]
- Da Costa, K.S.; Galúcio, J.M.; Da Costa, C.H.S.; Santana, A.R.; Dos Santos Carvalho, V.; Do Nascimento, L.D.; Lima E Lima, A.H.; Neves Cruz, J.; Alves, C.N.; Lameira, J. Exploring the Potentiality of Natural Products from Essential Oils as Inhibitors of Odorant-Binding Proteins: A Structure- And Ligand-Based Virtual Screening Approach to Find Novel Mosquito Repellents. ACS Omega 2019, 4, 22475–22486. [Google Scholar] [CrossRef] [PubMed]
- Rampogu, S.; Parate, S.; Parameswaran, S.; Park, C.; Baek, A.; Son, M.; Park, Y.; Park, S.J.; Lee, K.W. Natural compounds as potential Hsp90 inhibitors for breast cancer-Pharmacophore guided molecular modelling studies. Comput. Biol. Chem. 2019, 83, 107113. [Google Scholar] [CrossRef]
- Galúcio, J.M.; Monteiro, E.F.; de Jesus, D.A.; Costa, C.H.; Siqueira, R.C.; dos Santos, G.B.; Lameira, J.; da Costa, K.S. In silico identification of natural products with anticancer activity using a chemo-structural database of Brazilian biodiversity. Comput. Biol. Chem. 2019, 83, 107102. [Google Scholar] [CrossRef] [PubMed]
- Şenkardeş, S.; Han, M.İ.; Kulabaş, N.; Abbak, M.; Çevik, Ö.; Küçükgüzel, İ.; Küçükgüzel, Ş.G. Synthesis, molecular docking and evaluation of novel sulfonyl hydrazones as anticancer agents and COX-2 inhibitors. Mol. Divers. 2020, 24, 673–689. [Google Scholar] [CrossRef]
- da Costa, K.S.; Galúcio, J.M.; de Jesus, D.A.; Gomes, G.C.; Lima e Lima, A.H.; Taube, P.S.; dos Santos, A.M.; Lameira, J. Targeting Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1: A Structure-based Virtual Screening Approach to Find Novel Inhibitors. Curr. Comput. Aided. Drug Des. 2019, 15, 605–617. [Google Scholar] [CrossRef]
- Jiang, X.; Li, S.; Zhang, H.; Wang, L.-L. Discovery of potentially biased agonists of mu-opioid receptor (MOR) through molecular docking, pharmacophore modeling, and MD simulation. Comput. Biol. Chem. 2021, 90, 107405. [Google Scholar] [CrossRef] [PubMed]
- Mercado-Camargo, J.; Cervantes-Ceballos, L.; Vivas-Reyes, R.; Pedretti, A.; Serrano-García, M.L.; Gómez-Estrada, H. Homology Modeling of Leishmanolysin (gp63) from Leishmania panamensis and Molecular Docking of Flavonoids. ACS Omega 2020, 5, 14741–14749. [Google Scholar] [CrossRef]
- da Costa, K.S.; Galúcio, J.M.P.; Leonardo, E.S.; Cardoso, G.; Leal, É.; Conde, G.; Lameira, J. Structural and evolutionary analyses of Leishmania Alba proteins. Mol. Biochem. Parasitol. 2017, 217, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Paudel, P.; Wagle, A.; Seong, S.H.; Park, H.J.; Jung, H.A.; Choi, J.S. A new tyrosinase inhibitor from the red alga symphyocladia latiuscula (harvey) yamada (rhodomelaceae). Mar. Drugs 2019, 17, 295. [Google Scholar] [CrossRef] [PubMed]
- Brasil, E.M.; Canavieira, L.M.; Cardoso, É.T.C.; Silva, E.O.; Lameira, J.; Nascimento, J.L.M.; Eifler-Lima, V.L.; Macchi, B.M.; Sriram, D.; Bernhardt, P.V.; et al. Inhibition of tyrosinase by 4H-chromene analogs: Synthesis, kinetic studies, and computational analysis. Chem. Biol. Drug Des. 2017, 90, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Valasatava, Y.; Rosato, A.; Furnham, N.; Thornton, J.M.; Andreini, C. To what extent do structural changes in catalytic metal sites affect enzyme function? J. Inorg. Biochem. 2018, 179, 40–53. [Google Scholar] [CrossRef]
- Carvalho, A.T.P.; Barrozo, A.; Doron, D.; Kilshtain, A.V.; Major, D.T.; Kamerlin, S.C.L. Challenges in computational studies of enzyme structure, function and dynamics. J. Mol. Graph. Model. 2014, 54, 62–79. [Google Scholar] [CrossRef]
- Burian, M.; Amenitsch, H. Dummy-atom modelling of stacked and helical nanostructures from solution scattering data. IUCrJ 2018, 5, 390–401. [Google Scholar] [CrossRef]
- Wang, C.; Vernon, R.; Lange, O.; Tyka, M.; Baker, D. Prediction of structures of zinc-binding proteins through explicit modeling of metal coordination geometry. Protein Sci. 2010, 19, 494–506. [Google Scholar] [CrossRef]
- Da Costa, K.S.; Leal, E.; Dos Santos, A.M.; Lima E Lima, A.H.; Alves, C.N.; Lameira, J. Structural analysis of viral infectivity factor of HIV type 1 and its interaction with A3G, EloC and EloB. PLoS ONE 2014, 9, e89116. [Google Scholar] [CrossRef]
- Liao, Q.; Pabis, A.; Strodel, B.; Kamerlin, S.C.L. Extending the Nonbonded Cationic Dummy Model to Account for Ion-Induced Dipole Interactions. J. Phys. Chem. Lett. 2017, 8, 5408–5414. [Google Scholar] [CrossRef]
- Oelschlaeger, P.; Schmid, R.D.; Pleiss, J. Insight into the mechanism of the IMP-1 metallo-β-lactamase by molecular dynamics simulations. Protein Eng. Des. Sel. 2003, 16, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Tomar, V.; Mazumder, M.; Chandra, R.; Yang, J.; Sakharkar, M.K. Small molecule drug design. In Encyclopedia of Bioinformatics and Computational Biology: ABC of Bioinformatics; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C.B.T.-E., Eds.; Academic Press: Oxford, UK, 2018; Volume 1–3, pp. 741–760. ISBN 9780128114322. [Google Scholar]
- Park, Y.-D.D.; Lyou, Y.-J.J.; Hahn, H.-S.S.; Hahn, M.-J.J.; Yang, J.-M.M. Complex Inhibition of Tyrosinase by Thiol-Composed Cu 2+ Chelators: A Clue for Designing Whitening Agents. J. Biomol. Struct. Dyn. 2006, 24, 131–138. [Google Scholar] [CrossRef]
- Aqvist, J.; Luzhkov, V.B.; Brandsdal, B.O. Ligand binding affinities from MD simulations. Acc. Chem. Res. 2002, 35, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Noskov, S.Y.; Lim, C. Free Energy Decomposition of Protein-Protein Interactions. Biophys. J. 2001, 81, 737–750. [Google Scholar] [CrossRef]
- Li, M.; Cong, Y.; Li, Y.; Zhong, S.; Wang, R.; Li, H.; Duan, L. Insight Into the Binding Mechanism of p53/pDIQ-MDMX/MDM2 With the Interaction Entropy Method. Front. Chem. 2019, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, E.C.M.; da Costa, K.S.; Lameira, J.; Alves, C.N.; Lima, A.H. Investigation of the target-site resistance of EPSP synthase mutants P106T and T102I/P106S against glyphosate. RSC Adv. 2020, 10, 44352–44360. [Google Scholar] [CrossRef]
- da Costa, C.H.S.; Bichara, T.W.; Gomes, G.C.; dos Santos, A.M.; da Costa, K.S.; e Lima, A.H.L.; Alves, C.N.; Lameira, J. Unraveling the conformational dynamics of glycerol 3-phosphate dehydrogenase, a nicotinamide adenine dinucleotide-dependent enzyme of Leishmania mexicana. J. Biomol. Struct. Dyn. 2020, 1–12. [Google Scholar] [CrossRef]
- Costa, C.H.S.; Oliveira, A.R.S.; dos Santos, A.M.; da Costa, K.S.; e Lima, A.H.L.; Alves, C.N.; Lameira, J. Computational study of conformational changes in human 3-hydroxy-3-methylglutaryl coenzyme reductase induced by substrate binding. J. Biomol. Struct. Dyn. 2019, 37, 4374–4383. [Google Scholar] [CrossRef]
- Neves Cruz, J.; da Costa, K.S.; de Carvalho, T.A.A.; de Alencar, N.A.N. Measuring the structural impact of mutations on cytochrome P450 21A2, the major steroid 21-hydroxylase related to congenital adrenal hyperplasia. J. Biomol. Struct. Dyn. 2020, 38, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
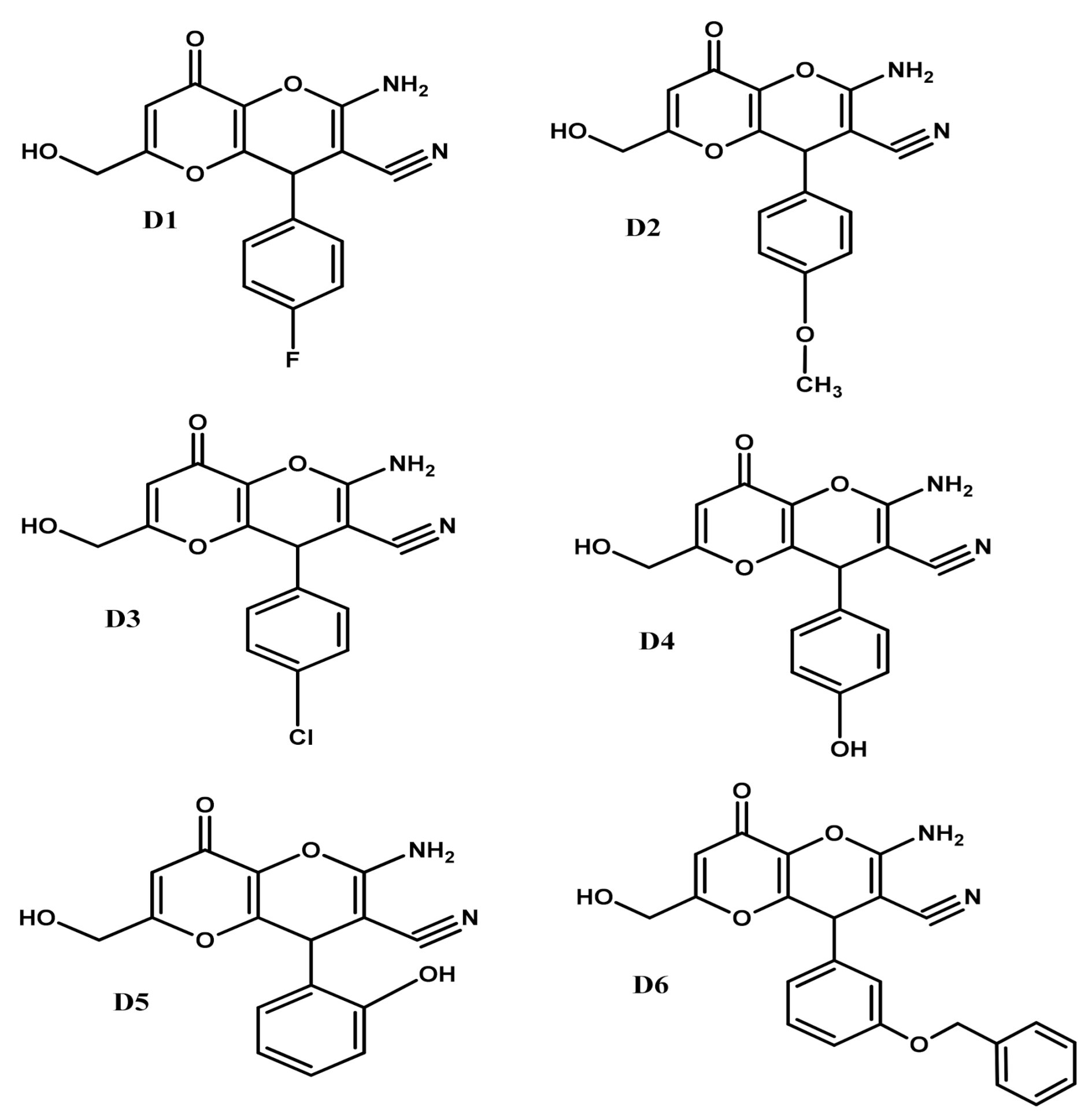
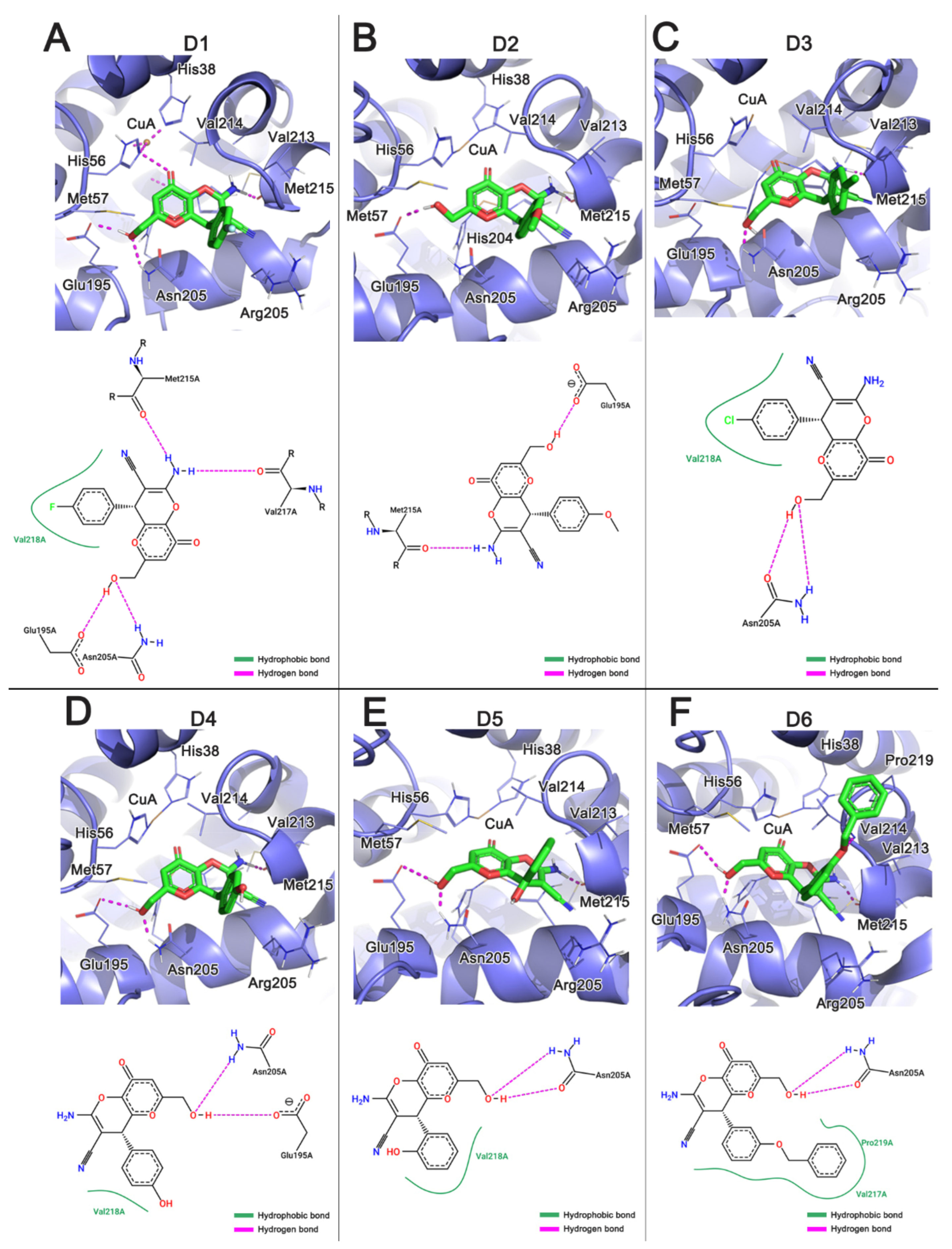
| Type | Compounds | GoldScore |
|---|---|---|
| Cofactors | L-DOPA | 48.31 |
| L-tyrosine | 48.38 | |
| Inhibitors | Kojic acid (KA) | 34.98 |
| D1 | 43.81 | |
| D2 | 42.24 | |
| D3 | 44.45 | |
| D4 | 43.48 | |
| D5 | 43.52 | |
| D6 | 53.20 | |
| D7 | 50.26 | |
| D8 | 41.91 | |
| D9 | 42.84 | |
| D10 | 53.29 | |
| D11 | 52.73 | |
| D12 | 46.08 | |
| D13 | 43.51 | |
| D14 | 44.81 |
| Ligands | D1 | D2 | D3 | D4 | D5 | D6 | KA | |
|---|---|---|---|---|---|---|---|---|
| Average distance (Å) | Cu-A | 4.46 (± 0.6) | 3.27 (± 0.6) | 3.94 (± 0.7) | 3.96 (± 1.0) | 2.77 (± 0.9) | 2.14 (± 0.2) | 5.78 (± 0.7) |
| Cu-B | 6.12 (± 0.4) | 5.69 (± 1.0) | 6.27 (± 0.7) | 5.95 (± 0.6) | 5.00 (± 0.3) | 5.66 (± 0.4) | 4.65 (± 0.4) |
| Ligands | |||||
|---|---|---|---|---|---|
| D1 | −36.93 | −20.15 | −50.81 | −44.58 | −5.09 |
| D2 | −90.91 | −46.27 | −40.81 | −22.41 | −18.07 |
| D3 | −39.06 | −21.76 | −48.00 | −44.10 | −4.42 |
| D4 | −37.12 | −18.68 | −78.25 | −57.11 | −10.31 |
| D5 | −98.00 | −54.04 | −38.86 | −18.86 | −18.13 |
| D6 | −92.73 | −49.36 | −47.53 | −29.11 | −17.65 |
| KA | −34.82 | −26.22 | −22.09 | −10.17 | −5.00 |
| L-DOPA | −32.74 | −13.07 | −61.95 | −44.54 | −12.84 |
| L- tyrosine | −26.62 | −13.47 | −67.43 | −41.58 | −9.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardoso, R.; Valente, R.; Souza da Costa, C.H.; da S. Gonçalves Vianez, J.L., Jr.; Santana da Costa, K.; de Molfetta, F.A.; Nahum Alves, C. Analysis of Kojic Acid Derivatives as Competitive Inhibitors of Tyrosinase: A Molecular Modeling Approach. Molecules 2021, 26, 2875. https://doi.org/10.3390/molecules26102875
Cardoso R, Valente R, Souza da Costa CH, da S. Gonçalves Vianez JL Jr., Santana da Costa K, de Molfetta FA, Nahum Alves C. Analysis of Kojic Acid Derivatives as Competitive Inhibitors of Tyrosinase: A Molecular Modeling Approach. Molecules. 2021; 26(10):2875. https://doi.org/10.3390/molecules26102875
Chicago/Turabian StyleCardoso, Richelly, Renan Valente, Clauber Henrique Souza da Costa, João Lidio da S. Gonçalves Vianez, Jr., Kauê Santana da Costa, Fábio Alberto de Molfetta, and Cláudio Nahum Alves. 2021. "Analysis of Kojic Acid Derivatives as Competitive Inhibitors of Tyrosinase: A Molecular Modeling Approach" Molecules 26, no. 10: 2875. https://doi.org/10.3390/molecules26102875
APA StyleCardoso, R., Valente, R., Souza da Costa, C. H., da S. Gonçalves Vianez, J. L., Jr., Santana da Costa, K., de Molfetta, F. A., & Nahum Alves, C. (2021). Analysis of Kojic Acid Derivatives as Competitive Inhibitors of Tyrosinase: A Molecular Modeling Approach. Molecules, 26(10), 2875. https://doi.org/10.3390/molecules26102875






