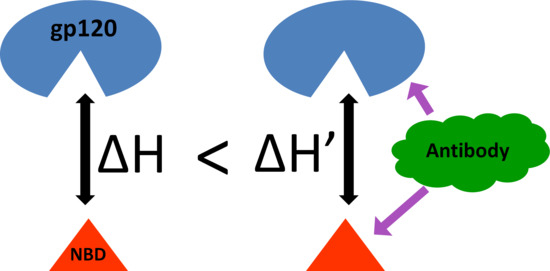
Does Antibody Stabilize the Ligand Binding in GP120 of HIV-1 Envelope Protein? Evidence from MD Simulation
Abstract
:1. Introduction
2. Methods
2.1. System Setup
2.2. MD Simulations
2.3. Clustering of the Trajectories
2.4. Free Energy Calculations
3. Results and Discussion
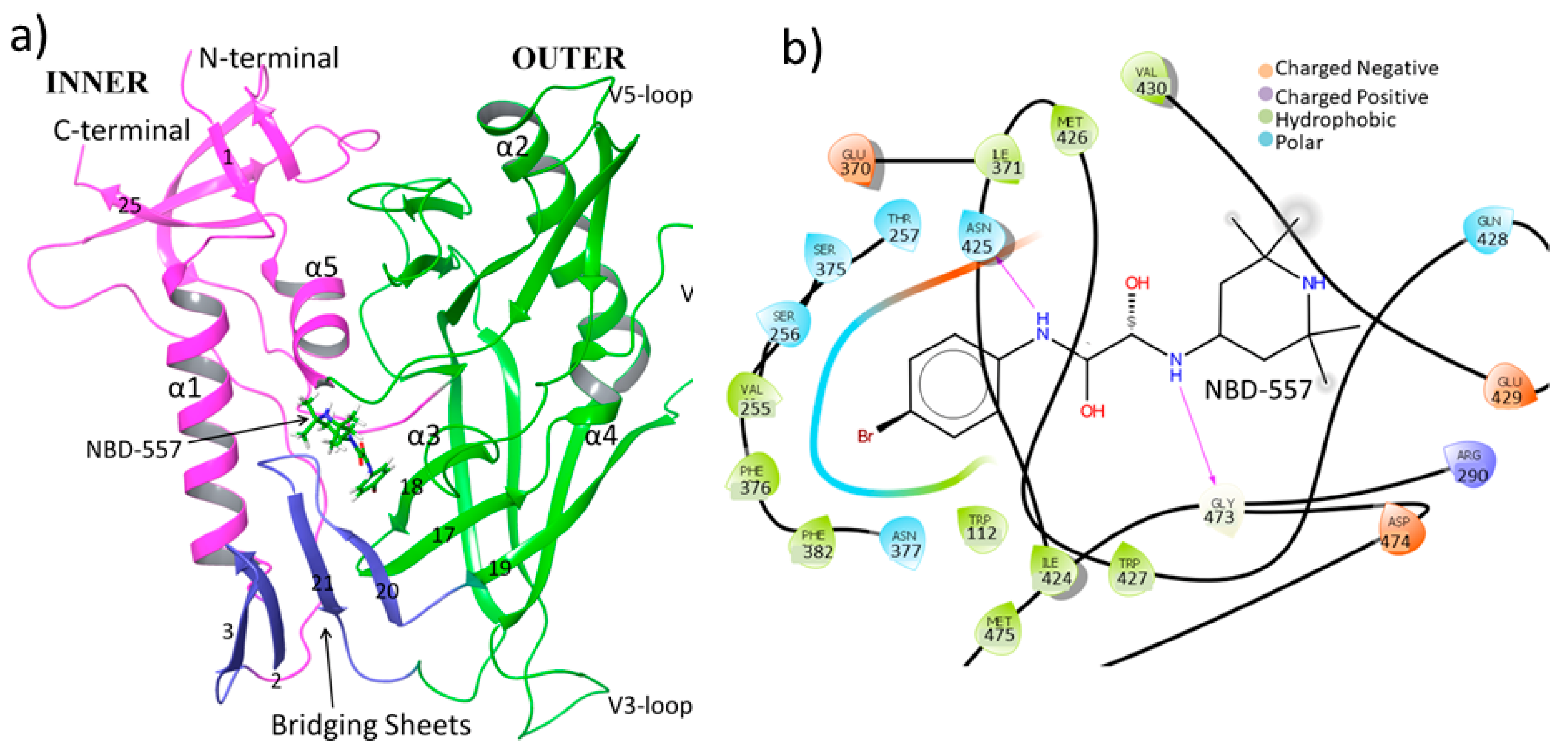
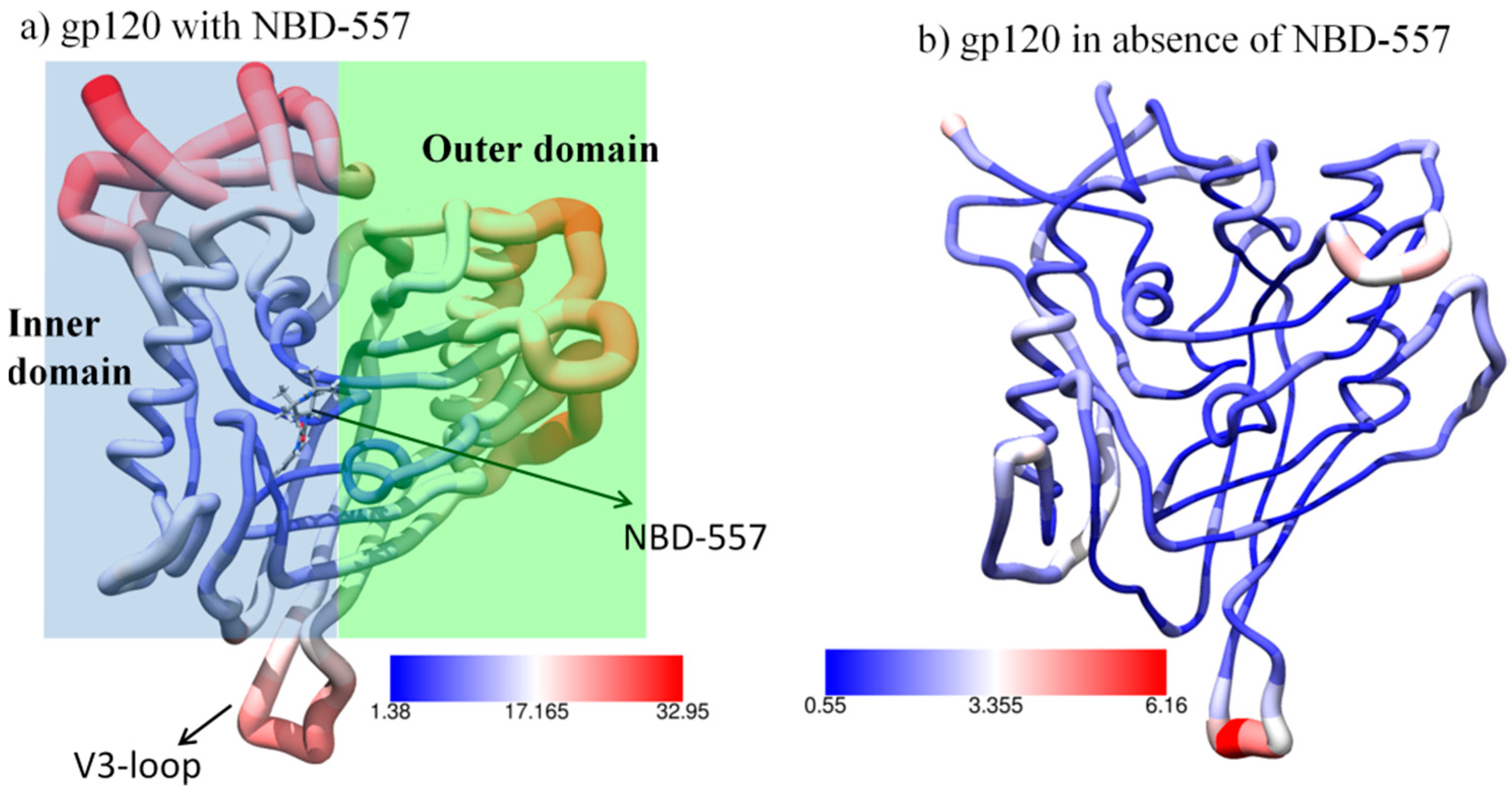
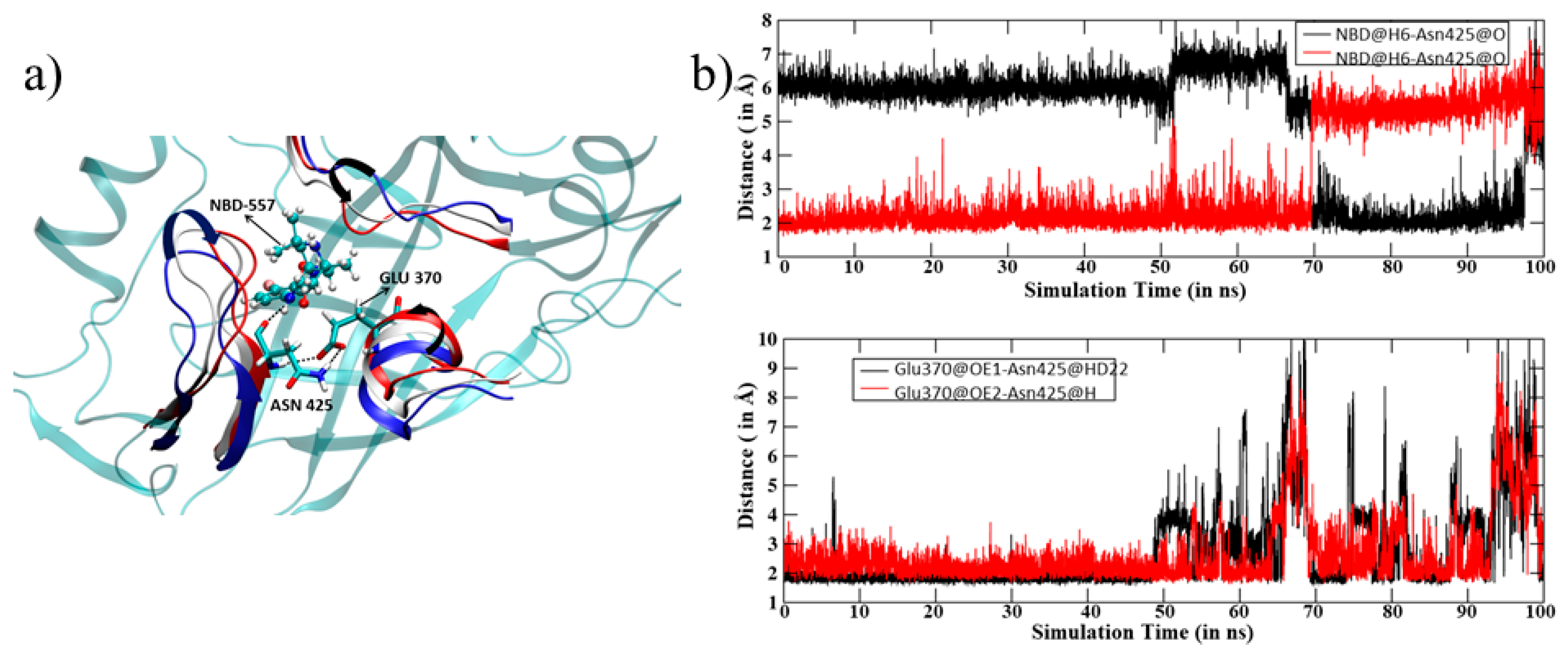
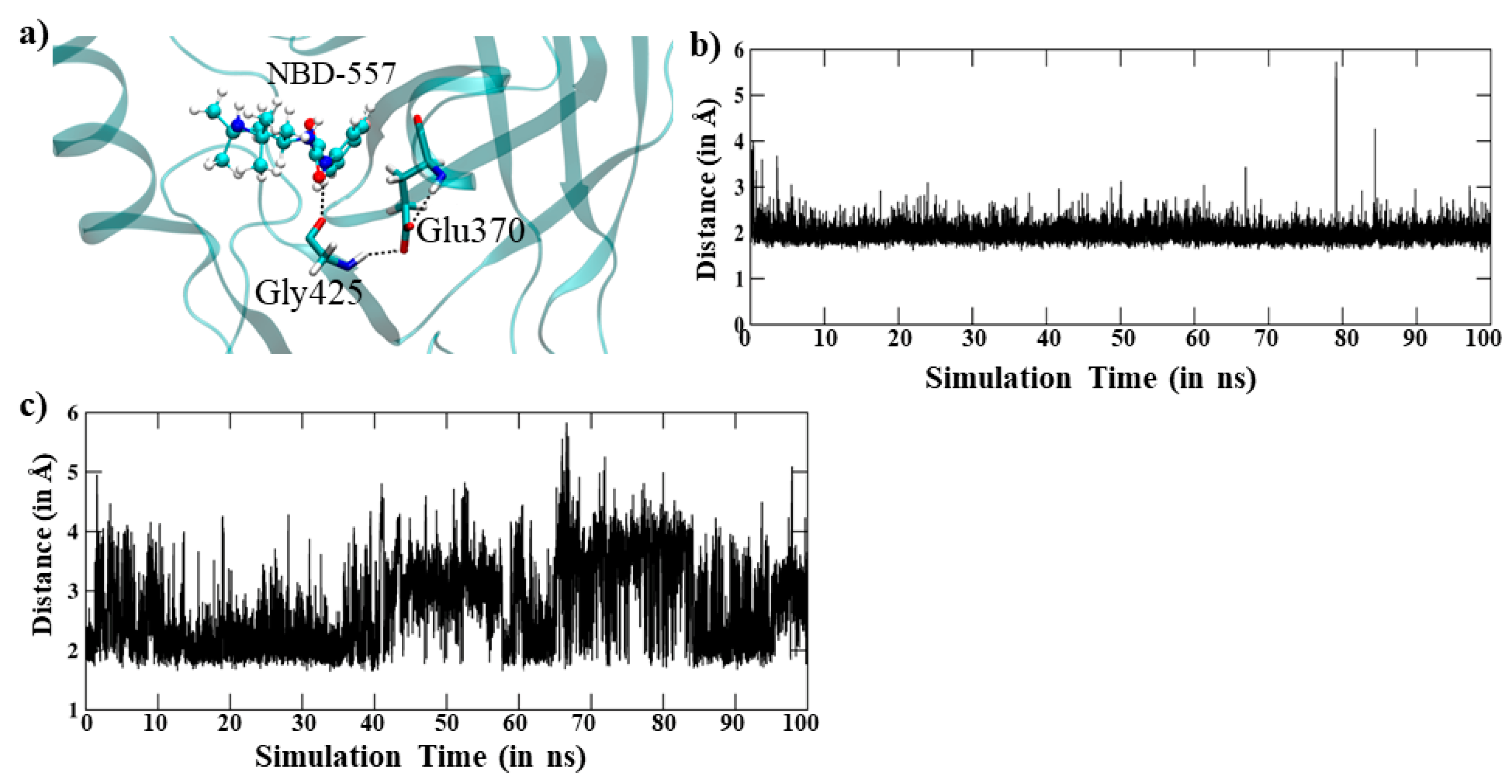
3.1. MD Simulation of gp120 with NBD-557
3.2. Thermo-Chemistry of gp120 Complex with NBD-557
3.3. Dynamics of the In-Silico Mutant Asn425Gly
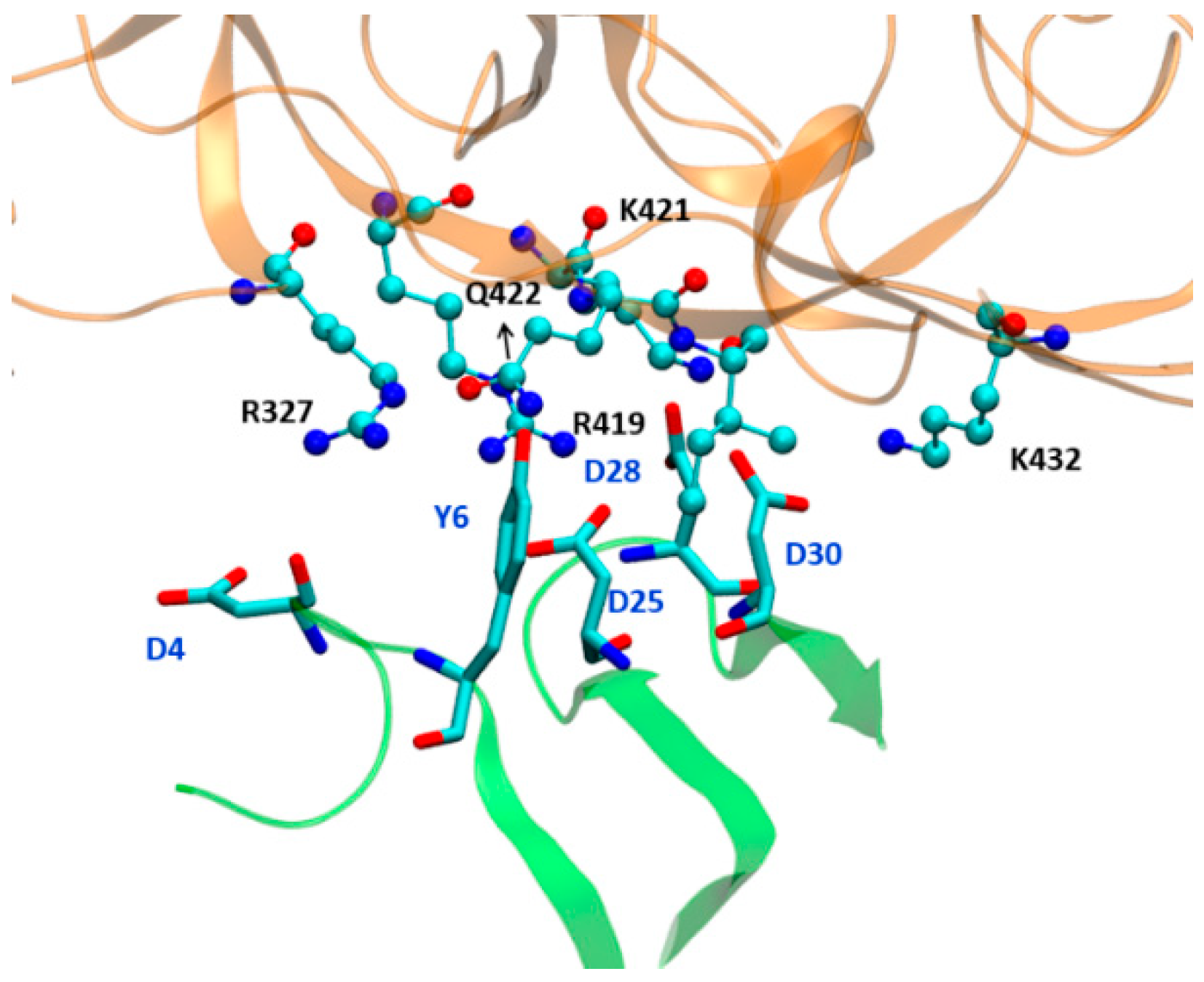
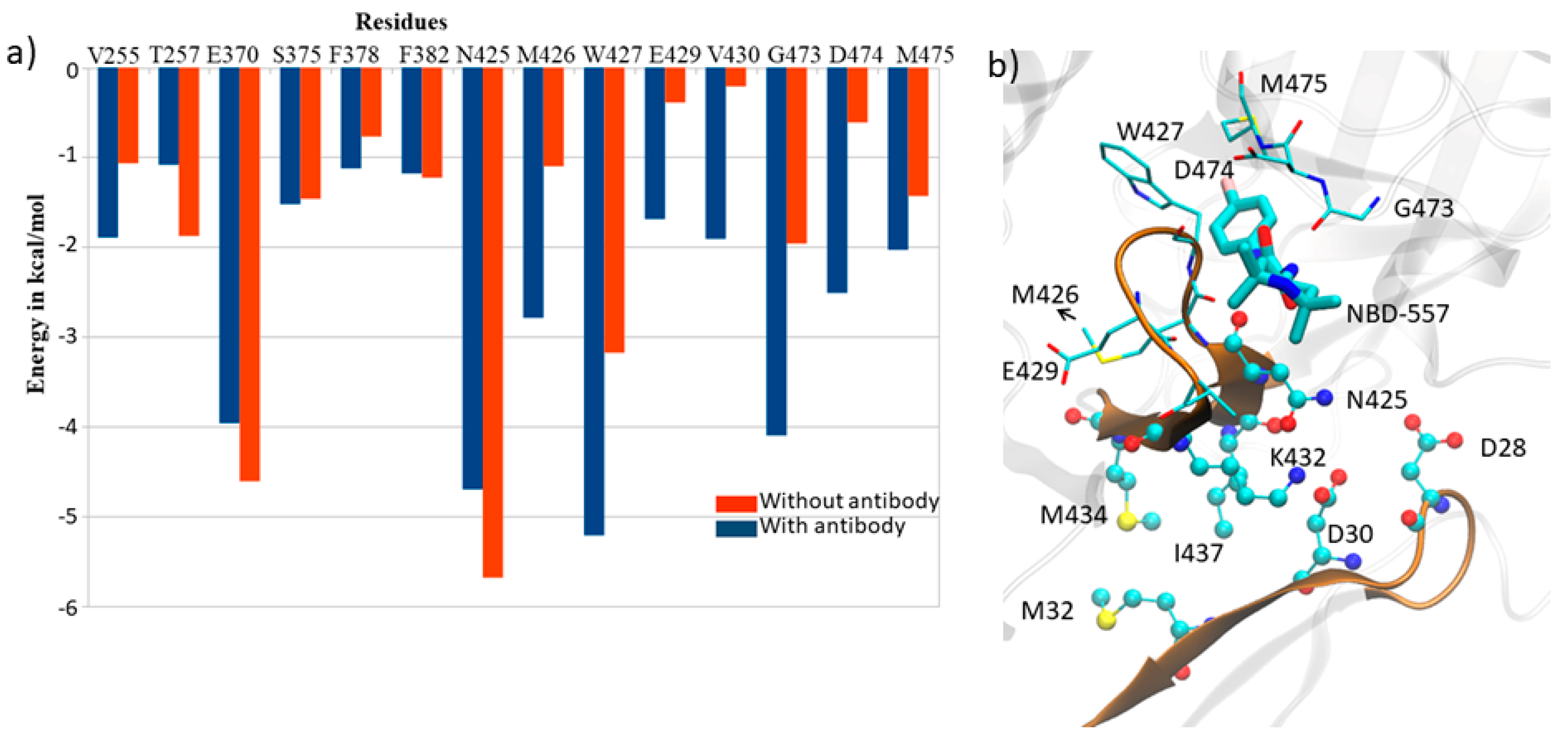
3.4. Effect of Antibody Binding on gp120 Interaction with NBD-557
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Sample Availability
References
- Rife, B.; Salemi, M. On the early dynamics and spread of HIV-1. Trends. Microbiol. 2015, 23, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Marston, H.D. Ending the HIV–AIDS Pandemic—Follow the Science. N. Engl. J. Med. 2015, 373, 2197–2199. [Google Scholar] [CrossRef] [PubMed]
- Barré-Sinoussi, F.; Ross, A.L.; Delfraissy, J.-F. Past, present and future: 30 years of HIV research. Nat. Rev. Microbiol. 2013, 11, 877–883. [Google Scholar]
- Blumenthal, R.; Durell, S.; Viard, M. HIV Entry and Envelope Glycoprotein-mediated Fusion. J. Biol. Chem. 2012, 287, 40841–40849. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.L.; Korte, T.; Blumenthal, R. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 1998, 273, 404–409. [Google Scholar] [CrossRef] [Green Version]
- Chien, M.P.; Jiang, S.; Chang, D.K. The function of coreceptor as a basis for the kinetic dissection of HIV type 1 envelope protein-mediated cell fusion. FASEB J. 2008, 22, 1179–1192. [Google Scholar] [CrossRef]
- Sougrat, R.; Bartesaghi, A.; Lifson, J.D.; Bennett, A.E.; Bess, J.W.; Zabransky, D.J.; Subramaniam, S. Electron tomography of the contact between T cells and SIV/HIV-1: Implications for viral entry. PLoS Pathog. 2007, 3, e63. [Google Scholar] [CrossRef] [Green Version]
- Dobrowsky, T.M.; Daniels, B.R.; Siliciano, R.F.; Sun, S.X.; Wirtz, D. Organization of cellular receptors into a nanoscale junction during HIV-1 adhesion. PLoS Comput. Biol. 2010, 6, e1000855. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Barroso, I.; Durell, S.; Sakaguchi, K.; Appella, E.; Blumenthal, R. Dilation of the human immune deficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 1998, 140, 315–323. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.C.; Tang, M.; Zhang, M.Y.; Majeed, S.; Montabana, E.; Stanfield, R.L.; Dimitrov, D.S.; Korber, B.; Sodroski, J.; Wilson, I.A.; et al. Structure of a V3-containing HIV-1 gp120 core. Science 2005, 310, 1025–1028. [Google Scholar] [CrossRef] [Green Version]
- Kwong, P.D.; Wyatt, R.; James Robinson, J.; Sweet, R.W.; Joseph Sodroski, J.; Hendrickson, W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998, 395, 648–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Ma, L.; Jiang, S.; Liu, S.; He, Y.; Strick, N.; Neamati, N.; Debnath, A.K. Identification of N-phenyl-N′-(2,2,6,6tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology 2005, 339, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, Z.; Wallace, O.B.; Deshpande, M.; Fang, H.; Yang, Z.; Zadjura, L.M.; Tweedie, D.L.; Huang, S.; Zhao, F.; et al. Discovery of 4-benzoyl-1-[(4methoxy-1 H-pyrrolo [2,3-b] pyridin-3-yl) oxoacetyl]-2-(R)-methylpiperazine (BMS-378806): A novel HIV-1 attachment inhibitor that interferes with CD4-gp120 interactions. J. Med. Chem. 2003, 46, 4236–4239. [Google Scholar] [CrossRef]
- Herschhorn, A.; Gu, C.; Espy, N.; Richard, J.; Finzi, A.; Sodroski, J.G. A broad HIV-1 inhibitor blocks envelope glycoprotein transitions critical for entry. Nat. Chem. Biol. 2014, 10, 845–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courter, J.R.; Madani, N.; Sodroski, J.; Schön, A.; Freire, E.; Kwong, P.D.; Hendrickson, W.A.; Chaiken, I.M.; LaLonde, J.M.; Smith, A.B., 3rd. Structure-based design, synthesis and validation of CD4-mimetic small molecule inhibitors of HIV-1 entry: Conversion of a viral entry agonist to an antagonist. Acc. Chem. Res. 2014, 47, 1228–1237. [Google Scholar] [CrossRef]
- Li, W.; Lu, L.; Li, W.; Jiang, S. Small-molecule HIV-1 entry inhibitors targeting gp120 and gp41: A patent review (2010–2015). Exp. Opin. Therap. Patent 2017. [Google Scholar] [CrossRef] [PubMed]
- Kuritzkes, D.R. HIV-1 entry inhibitors: An overview. Curr. Opin. 2009, 4, 82–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Huang, B.; Zhan, P.; De Clercq, E.; Liu, X. Discovery of small molecular inhibitors targeting HIV-1 gp120-CD4 interaction drived from BMS-378806. Eur. J. Med. Chem. 2014, 86, 481–490. [Google Scholar] [CrossRef]
- Schön, A.; Madani, N.; Klein, J.C.; Hubicki, A.; Ng, D.; Yang, X.; Smith, A.B.; Sodroski, S.; Freire, E. Thermodynamics of Binding of a Low-Molecular-Weight CD4 Mimetic to HIV-1 gp120. Biochemistry 2006, 45, 10973–10980. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.D.; Lalonde, J.M.; Yang, Y.; Elban, M.A.; Sugawara, A.; Courter, J.R.; Jones, D.M.; Smith, A.B.; Debnath, A.K.; Kwong, P.D. Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site. PLoS ONE 2014, 9, e85940. [Google Scholar] [CrossRef]
- Olsson, M.H.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. Amber 14; University of California: San Francisco, CA, USA, 2015. [Google Scholar]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A well behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Kollmann, P.A. Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J. Am. Chem. Soc. 1993, 115, 9620–9631. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Izaguirre, J.A.; Catarello, D.P.; Wozniak, J.M.; Skeel, R.D. Langevin stabilization of molecular dynamics. J. Chem. Phys. 2001, 114, 2090–2098. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the Cartesian equation of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond molecular dynamics simulations with AMBER on GPU. 2. Explicit solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Dubey, K.D.; Tiwari, R.K.; Ojha, R.P. Recent Advances in Protein–Ligand Interactions: MD Simulations and Binding Free Energy. Curr. Comput. Aided Drug Des. 2013, 9, 518–531. [Google Scholar] [CrossRef]
- Gohlke, H.; Case, D.A. Converging free energy estimates: MM-PB (GB) SA studies on the protein-protein complex RasRaf. J. Comput. Chem. 2003, 25, 238–250. [Google Scholar] [CrossRef]
- Fogolari, F.; Brigo, A.; Molinari, H. Protocols for MM/PBSA molecular dynamics simulations of proteins. Biophys. J. 2003, 85, 159–166. [Google Scholar] [CrossRef] [Green Version]
- Grochowaski, P.; Trylska, J. Continuum molecular electrostatics, salt effects, and counterion binding—A review of the Poisson Boltzmann theory and its modifications. Biopolymers 2007, 89, 93–113. [Google Scholar] [CrossRef]
- Tsui, V.; Case, D.A. Theory and application of generalized born solvation model in macromolecular simulations. Biopolymers 2001, 56, 271–291. [Google Scholar] [CrossRef]
- Dubey, K.D.; Chaubey, A.K.; Ojha, R.P. Stability of trimeric DENV envelope protein at low and neutral pH: An insight from MD study. Biochim. Biophys. Acta 2013, 1834, 53–64. [Google Scholar] [CrossRef]
- Dubey, K.D.; Chaubey, A.K.; Ojha, R.P. Role of pH on dimeric interactions for DENVenvelope protein: An insight from molecular dynamics study. Biochim. Biophys. Acta 2011, 1814, 1796–1801. [Google Scholar] [CrossRef]
- Dubey, K.D.; Tiwari, G.; Ojha, R.P. Targeting domain-III hinging of dengue envelope (DENV-2) protein by MD simulations, docking and free energy calculations. J. Mol. Model. 2017, 23, 102. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Virtual molecular dynamics. J. Mol. Graph. Model. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Haung, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]



| Energy Contribution | Values (in kcal Mol) | Standard Error |
|---|---|---|
| ∆EVDW | −36.94 | 2.19 |
| ∆EEEL | −5.22 | 1.93 |
| ∆ENPOLAR | 21.33 | 1.71 |
| ∆EDISPER | −4.32 | 0.17 |
| ∆Ggas | −42.16 | 3.04 |
| ∆G solv | 17 | 1.67 |
| ∆HTOTAL | −25.16 | 3.24 |
| ∆HEXPref | −24.50 |
| Energy Contribution | Values (In kcal Mol) | Standard Error |
|---|---|---|
| ∆EVDW | −46.19 | 3.23 |
| ∆EEEL | −14.88 | 3.3 |
| ∆ENPOLAR | 29.65 | 2.52 |
| ∆EDISPER | −5.28 | 0.32 |
| ∆Ggas | −61.07 | 5.25 |
| ∆Gsolv | 24.36 | 2.32 |
| ∆HTOTAL | −36.71 | 3.66 |
| Residues (Antibody) | Residues (gp120) | Energy (kcal/mol) |
|---|---|---|
| TYR6 | ARG327 | −3.928 |
| TYR6 | GLN422 | −2.059 |
| ASP25 | ARG419 | −18.346 |
| ASP25 | LYS421 | −2.437 |
| ASP28 | PRO369 | −1.679 |
| ASP28 | ARG419 | −7.616 |
| ASP28 | LYS421 | −10.152 |
| ASP30 | LYS421 | −11.001 |
| ASP30 | ILE 423 | −2.028 |
| ASP30 | LYS432 | −7.517 |
| MET32 | MET434 | −1.656 |
| Energy Contribution | Values (in kcal Mol) | Standard Error |
|---|---|---|
| ∆EVDW | −43.94 | 2.16 |
| ∆EEEL | −14.12 | 2.19 |
| ∆ENPOLAR | 29.40 | 1.46 |
| ∆EDISPER | −4.90 | 0.15 |
| ∆Ggas | −58.07 | 2.75 |
| ∆G solv | 24.49 | 1.40 |
| ∆HTOTAL | −33.57 | 2.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, S.; Pandey, V.; Kumar Tiwari, R.; Ojha, R.P.; Dubey, K.D. Does Antibody Stabilize the Ligand Binding in GP120 of HIV-1 Envelope Protein? Evidence from MD Simulation. Molecules 2021, 26, 239. https://doi.org/10.3390/molecules26010239
Yadav S, Pandey V, Kumar Tiwari R, Ojha RP, Dubey KD. Does Antibody Stabilize the Ligand Binding in GP120 of HIV-1 Envelope Protein? Evidence from MD Simulation. Molecules. 2021; 26(1):239. https://doi.org/10.3390/molecules26010239
Chicago/Turabian StyleYadav, Shalini, Vishnudatt Pandey, Rakesh Kumar Tiwari, Rajendra Prasad Ojha, and Kshatresh Dutta Dubey. 2021. "Does Antibody Stabilize the Ligand Binding in GP120 of HIV-1 Envelope Protein? Evidence from MD Simulation" Molecules 26, no. 1: 239. https://doi.org/10.3390/molecules26010239
APA StyleYadav, S., Pandey, V., Kumar Tiwari, R., Ojha, R. P., & Dubey, K. D. (2021). Does Antibody Stabilize the Ligand Binding in GP120 of HIV-1 Envelope Protein? Evidence from MD Simulation. Molecules, 26(1), 239. https://doi.org/10.3390/molecules26010239