Identification of Dipeptidyl Peptidase-4 and α-Amylase Inhibitors from Melicope glabra (Blume) T. G. Hartley (Rutaceae) Using Liquid Chromatography Tandem Mass Spectrometry, In Vitro and In Silico Methods
Abstract
1. Introduction
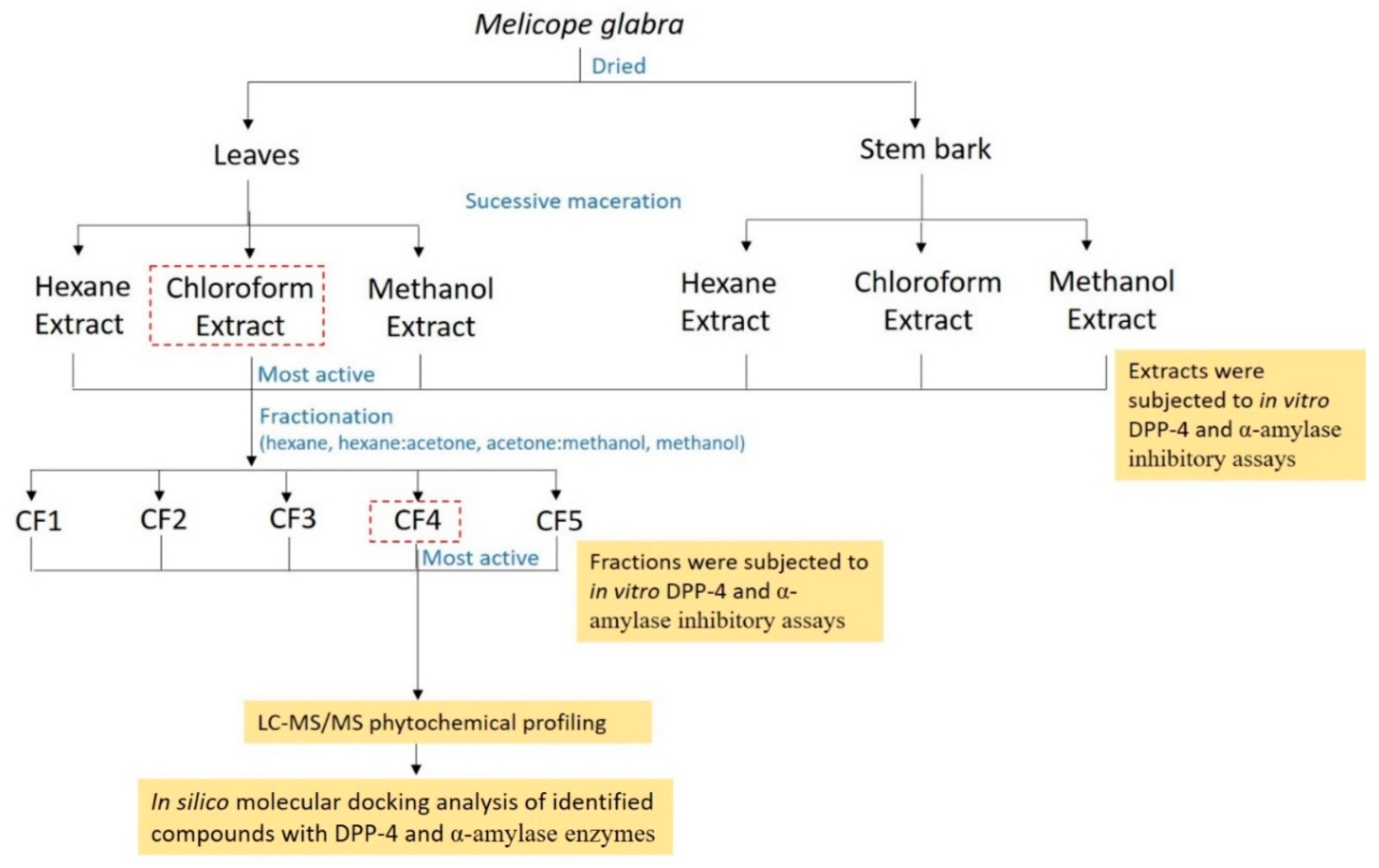
2. Results and Discussion
2.1. Extraction Yield
2.2. Dipeptidyl Peptidase-4 Inhibitory Activity
2.3. α-Amylase Inhibitory Activity
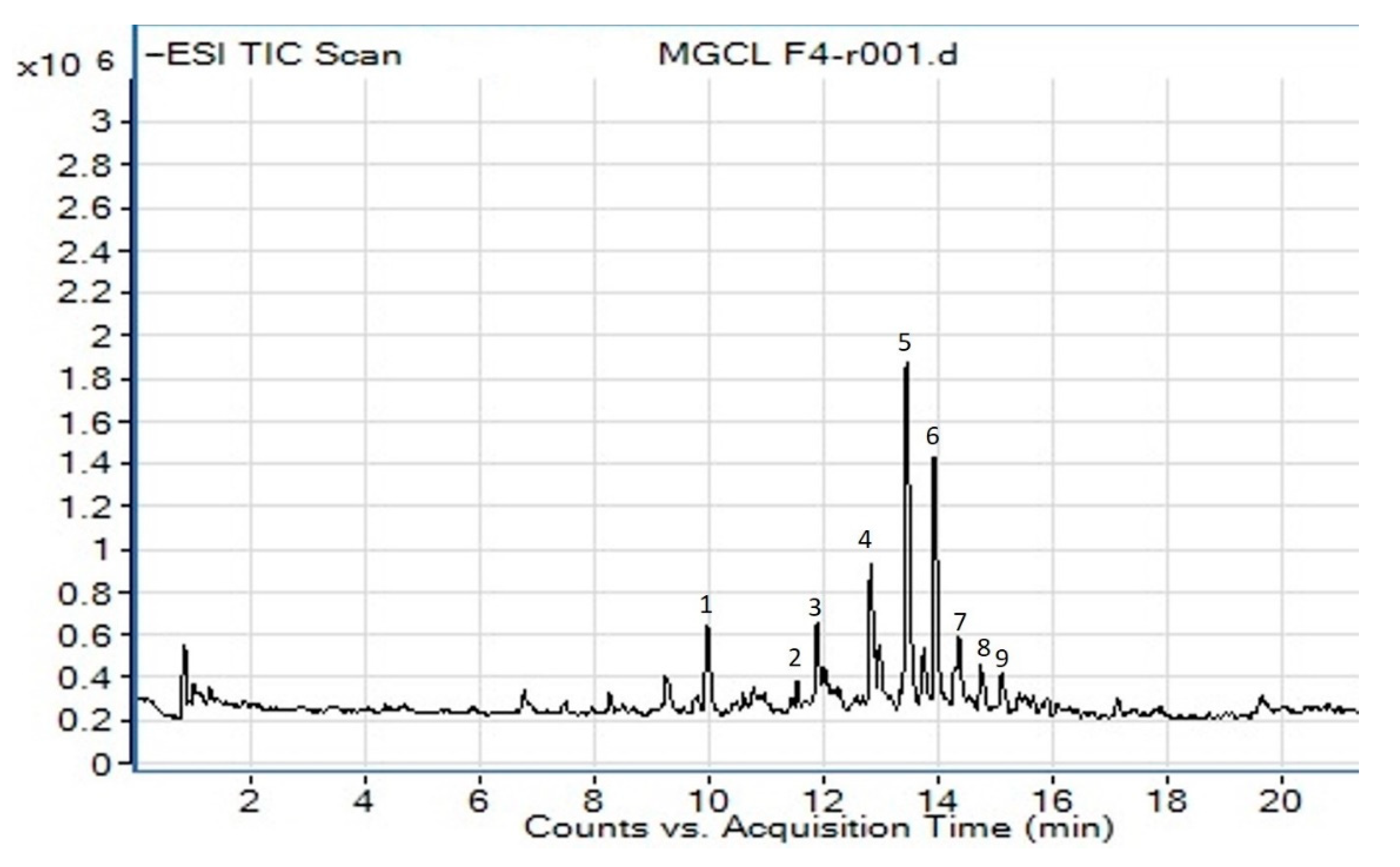
2.4. Phytochemical Profiling of Active Fraction Using LC-MS/MS
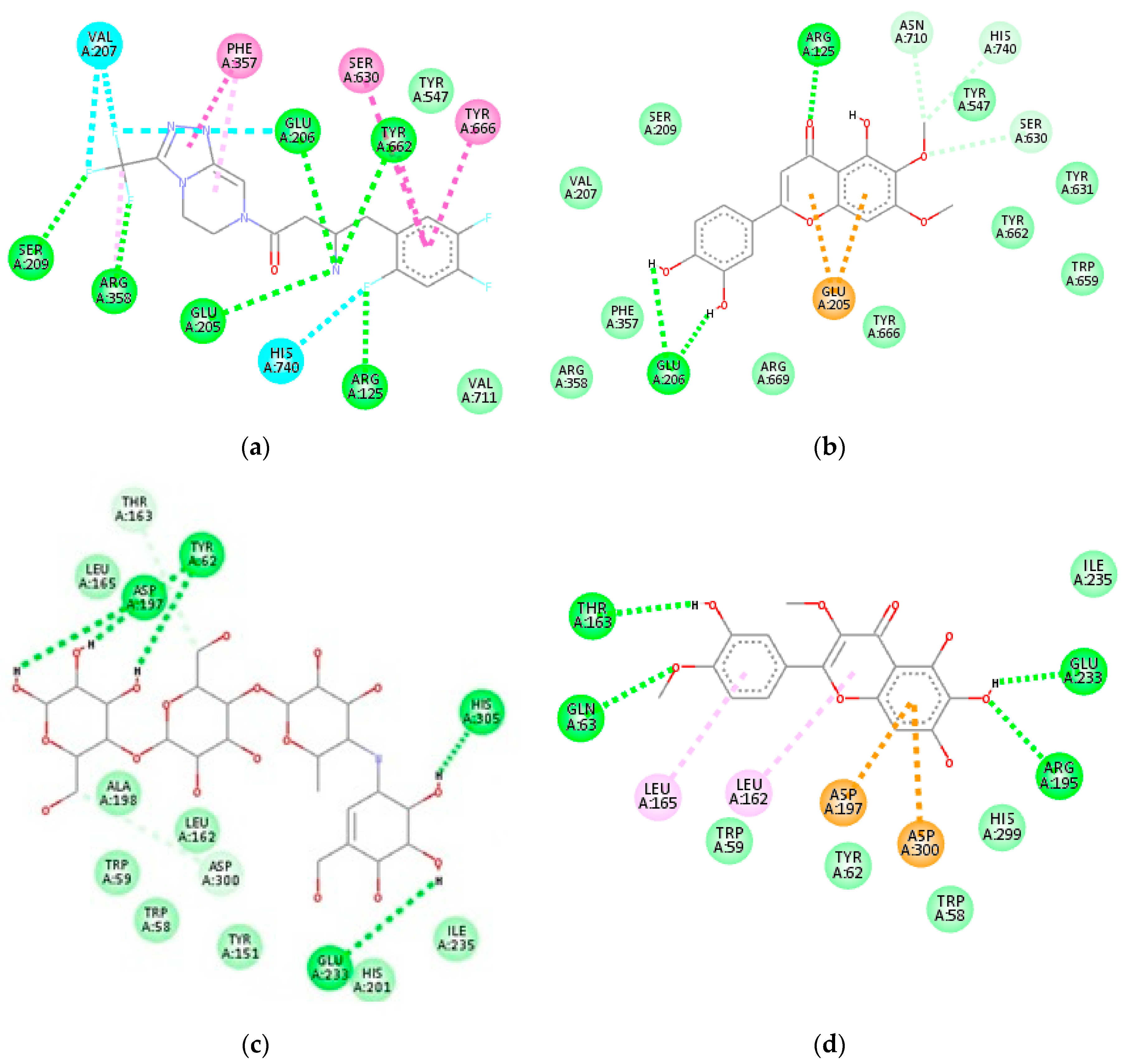
2.5. In Silico Inhibitory Analysis of Identified Compounds
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Plant Materials
3.3. Preparation of Extracts and Fractionation
3.4. Determination of Extraction Yield
3.5. Antidiabetic Assays
3.5.1. DPP-4 Inhibitory Assay
3.5.2. α-Amylase Inhibitory Assay
3.6. LC-MS/MS Analysis
3.7. In Silico Inhibitory Analysis of Identified Compounds
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bindu, J.; Narendhirakannan, R.T. Role of medicinal plants in the management of diabetes mellitus: A review. 3 Biotech 2018, 9, 4–17. [Google Scholar] [CrossRef]
- Costa, J.F.O.; Juiz, P.; São Pedro, A.; David, J.P.D.L.; David, J.M.; Giulietti, A.M.; Franca, F.; dos Santos, R.R.; Soares, M.B.P. Immunomodulatory and antibacterial activities of extracts from Rutaceae species. Rev. Bras. Farmacogn. 2010, 20, 502–505. [Google Scholar] [CrossRef][Green Version]
- Huang, L.; Feng, Z.-L.; Wang, Y.-T.; Lin, L.-G. Anticancer carbazole alkaloids and coumarins from Clausena plants: A review. Chin. J. Nat. Med. 2017, 15, 881–888. [Google Scholar] [CrossRef]
- Pavani, P.; Raja, N. Anti-Inflammatory Properties of Zanthoxylum Ovalifolium Weight Methanolic Extract on Albino Wistar Rats. Asian J. Pharm. Clin. Res. 2019, 12, 274–277. [Google Scholar] [CrossRef]
- Kassim, N.K.; Rahmani, M.; Ismail, A.; Sukari, M.A.; Gwendoline, C.L.E.; Nadiah, M.N.; Khalijah, A. Antioxidant activity-guided separation of coumarins and lignan from Melicope glabra (Rutaceae). Food Chem. 2013, 139, 87–92. [Google Scholar] [CrossRef]
- Saputri, R.D.; Srie Tjahjandarie, T.; Tanjung, M.; Meliglabrin, A. New Flavonol Derivative from the leaves of Melicope glabra (Blume) T.G. Hartley. Nat. Prod. Sci. 2018, 24, 155. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Tu, Z.; Yuan, T.; Wang, H.; Xie, X.; Fu, Z. Antioxidants and α-glucosidase inhibitors from Ipomoea batatas leaves identified by bioassay-guided approach and structure-activity relationships. Food Chem. 2016, 208, 61–67. [Google Scholar] [CrossRef]
- Vinayagam, R.; Xu, B. Antidiabetic properties of dietary flavonoids: A cellular mechanism review. Nutr. Metab. 2015, 12, 60. [Google Scholar] [CrossRef]
- Gowd, V.; Bao, T.; Wang, L.; Huang, Y.; Chen, S.; Zheng, X.; Cui, S.; Chen, W. Antioxidant and antidiabetic activity of blackberry after gastrointestinal digestion and human gut microbiota fermentation. Food Chem. 2018, 269, 618–627. [Google Scholar] [CrossRef]
- Ming, D.; Bhupathiraju, S.N.; Mu, C.; Dam, R.M.V.; Hu, F.B. Caffeinated and decaffeinated coffee consumption and risk of type 2 diabetes: A systematic review and a dose-response meta-analysis. Diabetes Care 2014, 37, 569–586. [Google Scholar]
- Akhtar, N.; Jafri, L.; Green, B.D.; Kalsoom, S.; Mirza, B. A Multi-Mode Bioactive Agent Isolated from Ficus microcarpa L. Fill. With Therapeutic Potential for Type 2 Diabetes Mellitus. Front. Pharmacol. 2018, 9, 1376. [Google Scholar] [CrossRef]
- Kalhotra, P.; Chittepu, V.; Osorio-Revilla, G.; Gallardo-Velazquez, T. Discovery of Galangin as a Potential DPP-4 Inhibitor That Improves Insulin-Stimulated Skeletal Muscle Glucose Uptake: A Combinational Therapy for Diabetes. Int. J. Mol. Sci. 2019, 20, 1228. [Google Scholar] [CrossRef]
- Dehdari, L.; Dehdari, T. The determinants of anti-diabetic medication adherence based on the experiences of patients with type 2 diabetes. Arch. Public Health 2019, 77, 21. [Google Scholar] [CrossRef] [PubMed]
- Akula, P.; Sree, A.; Santosh, B.; Sandeep, K.; Raviteja, K.; Keerthi, T. Evaluation of anti-microbial activity of leaf and bark extracts of Murraya koeniggi (curry leaves). J. Pharmacogn. Phytochem. 2016, 5, 101–105. [Google Scholar]
- Overgaard, H.; Sirisopa, P.; Mikolo, B.; Malterud, K.; Wangensteen, H.; Zou, Y.-F.; Paulsen, B.S.; Massamba, D.; Duchon, S.; Corbel, V.; et al. Insecticidal Activities of Bark, Leaf and Seed Extracts of Zanthoxylum heitzii against the African Malaria Vector Anopheles gambiae. Molecules 2014, 19, 21276–21290. [Google Scholar] [CrossRef] [PubMed]
- Feduraev, P.; Chupakhina, G.; Maslennikov, P.; Tacenko, N.; Skrypnik, L. Variation in Phenolic Compounds Content and Antioxidant Activity of Different Plant Organs from Rumex crispus L. and Rumex obtusifolius L. at Different Growth Stages. Antioxidants 2019, 8, 237. [Google Scholar] [CrossRef]
- Kim, B.R.; Kim, H.; Choi, I.; Kim, J.B.; Jin, C.; Han, A.R. DPP-IV Inhibitory Potentials of Flavonol Glycosides Isolated from the Seeds of Lens culinaris: In Vitro and Molecular Docking Analyses. Molecules 2018, 23, 1998. [Google Scholar] [CrossRef]
- Holst, J.J. The incretin system in healthy humans: The role of GIP and GLP-1. Metab. Clin. Exp. 2019, 96, 46–55. [Google Scholar] [CrossRef]
- Geng, Y.; Lu, Z.M.; Huang, W.; Xu, H.Y.; Shi, J.S.; Xu, Z.H. Bioassay-Guided Isolation of DPP-4 Inhibitory Fractions from Extracts of Submerged Cultured of Inonotus obliquus. Molecules 2013, 18, 1150–1161. [Google Scholar] [CrossRef]
- Bower, A.M.; Real Hernandez, L.M.; Berhow, M.A.; de Mejia, E.G. Bioactive Compounds from Culinary Herbs Inhibit a Molecular Target for Type 2 Diabetes Management, Dipeptidyl Peptidase IV. J. Agric. Food Chem. 2014, 62, 6147–6158. [Google Scholar] [CrossRef]
- Alam, F.; Islam, M.A.; Kamal, M.; Gan, S. Updates on Managing Type 2 Diabetes Mellitus with Natural Products: Towards Antidiabetic Drug Development. Curr. Med. 2016, 23, 5395–5431. [Google Scholar] [CrossRef]
- Tan, D.C.; Idris, K.I.; Kassim, N.K.; Lim, P.C.; Safinar Ismail, I.; Hamid, M.; Ng, R.C. Comparative study of the antidiabetic potential of Paederia foetida twig extracts and compounds from two different locations in Malaysia. Pharm. Biol. 2019, 57, 345–354. [Google Scholar] [CrossRef]
- Lim, J.; Zhang, X.; Ferruzzi, M.G.; Hamaker, B.R. Starch digested product analysis by HPAEC reveals structural specificity of flavonoids in the inhibition of mammalian α-Amylase and α-glucosidases. Food Chem. 2019, 288, 413–421. [Google Scholar] [CrossRef]
- Li, L.; Chen, Y.; Feng, X.; Yin, J.; Li, S.; Sun, Y.; Zhang, L. Identification of Metabolites of Eupatorin in Vivo and in Vitro Based on UHPLC-Q-TOF-MS/MS. Molecules 2019, 24, 2658. [Google Scholar] [CrossRef]
- Malviya, N.; Malviya, S. Bioassay guided fractionation-an emerging technique influence the isolation, identification and characterization of lead phytomolecules. Int. J. Hosp. Pharm. 2017, 2, 5. [Google Scholar] [CrossRef][Green Version]
- Abdullah, N.; Kasim, K. In-Vitro Antidiabetic Activity of Clinacanthus nutans extracts. Int. J. Pharmacogn. Phytochem. Res. 2017, 9, 846–852. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]

| Plant Part | Extracts | Extraction Yield |
|---|---|---|
| (%) | ||
| Leaves | Hexane | 2.03 ± 0.31 c |
| Chloroform | 2.79 ± 0.19 d | |
| Methanol | 7.30 ± 1.10 e | |
| Stem bark | Hexane | 0.3 ± 0.16 a |
| Chloroform | 0.7 ± 0.03 b | |
| Methanol | 2.13 ± 0.25 c |
| Plant Part | Extracts | IC50 (μg/mL) | |
|---|---|---|---|
| DPP-4 | α-Amylase | ||
| Leaves | Hexane | 1623.60 ± 121.61 g | 4230.12 ± 324.76 h |
| Chloroform | 169.40 ± 9.30 c | 303.64 ± 10.10 c | |
| Methanol | 1086.48 ± 142.69 f | 2488.13 ± 231.54 g | |
| Stem bark | Hexane | 8408.36 ± 102.23 i | 5447.01 ± 243.16 i |
| Chloroform | 332.31 ± 10.07 d | 975.80 ± 17.10 e | |
| Methanol | 4992.33 ± 0.84 h | 3946.12 ± 143.21 h | |
| Fractions | |||
| CF1 | 1711.06 ± 70.32 g | 8663.12 ± 110.75 j | |
| CF2 | 619.31 ± 9.21 e | 253.30 ± 19.21 b | |
| CF3 | 313.18 ± 20.92 d | 531.44 ± 11.38 d | |
| CF4 | 128.35 ± 12.77 b | 170.19 ± 20.66 a | |
| CF5 | 711.42 ± 10.26 e | 1817.83 ± 209.42 f | |
| Sitagliptin | 0.01 ± 0.01 a | - | |
| Acarbose | - | 188.60 ± 14.31 a | |
| Compound | RT (min) | Identification | Molecular Formula | Observed Neutral Mass (Da) | Mass Error (mDa) | Precursor ion [M−H] (m/z) | Major Fragments (m/z) |
|---|---|---|---|---|---|---|---|
| Flavonoid | |||||||
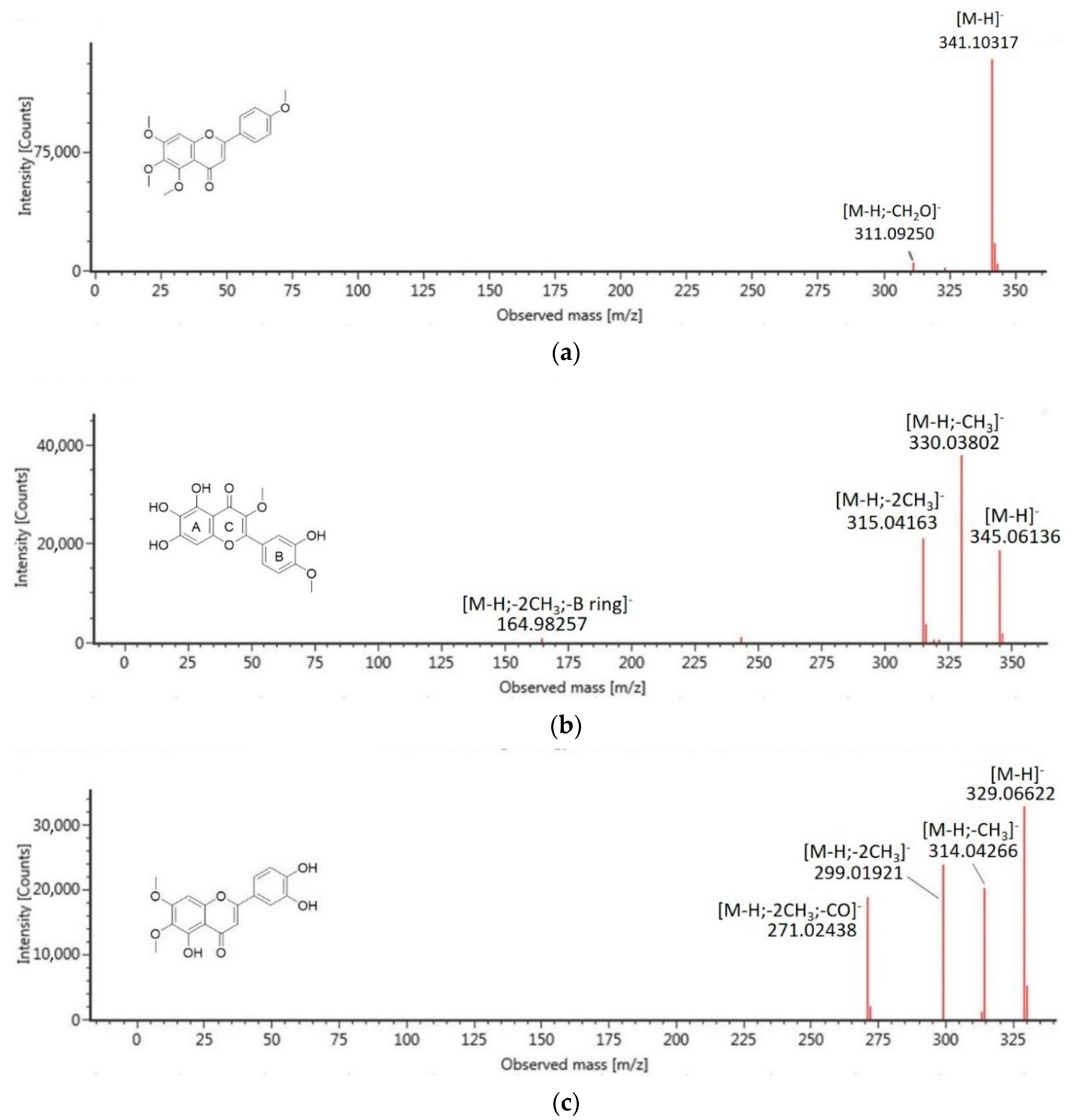
| 5 | 13.82 | 4′,5,6,7-tetramethoxy-flavone | C19H18O6 | 342.1103 | 0.8 | 341.1032 | 311.0925 |
| 6 | 14.07 | Isorhamnetin | C16H12O7 | 316.0583 | −0.8 | 315.0503 | 300.0270, 272.0280 |
| 7 | 14.22 | quercetagetin-3,4′-dimethyl ether | C17H14O8 | 346.0689 | 0.4 | 345.0617 | 330.0380, 315.0416, 164.9826 |
| 8 | 14.91 | 5,3′,4′-trihydroxy-6,7-dimethoxy-flavone | C17H14O7 | 330.0740 | 0.4 | 329.0662 | 314.0427, 299.0192, 271.0244 |
| Cinnamic acid derivative | |||||||
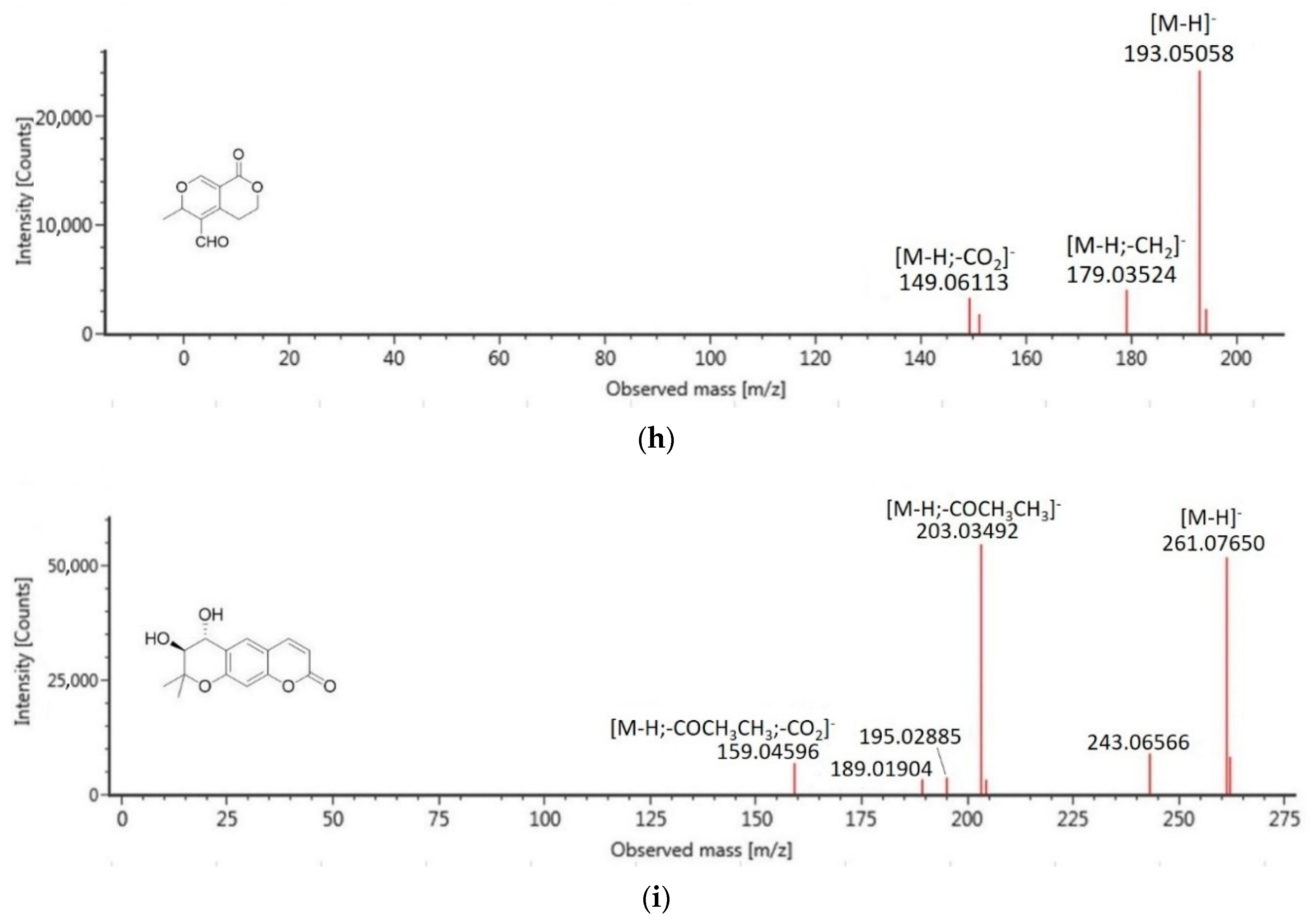
| 3 | 11.91 | methyl 3,4,5-trimethoxycinnamate | C13H16O5 | 252.0998 | −0.4 | 251.0923 | 193.0506, 179.0352 |
| Glucoside | |||||||
| 4 | 13.04 | Renifolin | C18H24O7 | 352.1522 | 0.4 | 351.1452 | 199.0990 |
| Lactone | |||||||
| 2 | 11.63 | swermirin | C10H10O4 | 194.0579 | −0.4 | 193.0506 | 179.0352, 149.0611 |
| 9 | 15.15 | 2-methoxy-5-acetoxy-fruranogermacr-1(10)-en-6-one | C18H24O5 | 320.1624 | 0.7 | 319.1554 | 305.1400, 179.0356 |
| Coumarin | |||||||
| 1 | 10.20 | trans-decursidinol | C14H14O5 | 262.0841 | −0.2 | 261.0765 | 203.0349, 159.0460 |
| Compounds | DPP-4 | α-Amylase | ||||
|---|---|---|---|---|---|---|
| Binding Affinity (kcal/mol) | H-Bond | Hydrophobic | Binding Affinity (kcal/mol) | H-Bond | Hydrophobic | |
| 1 | −7.7 | Glu206, Arg125, Ser630, Glu205 | Glu205, Tyr666 | −8.2 | Glu233, Asp300 | Ala198, Leu162, His201 |
| 2 | −5.4 | Lys122, Asp739 | His740, Arg125 | −6.4 | His299, Asp197, Tyr62, Asp300 | Ala198, Trp58 |
| 3 | −5.6 | Arg125, Tyr547, Arg669, Val 207, Tyr662 | Tyr666, Glu205 | −6.0 | His299, Tyr62, Thr163 | Trp59 |
| 4 | −7.8 | Glu206, Glu205, Asn710, Arg125, Ser630 | Tyr547, Phe357 | −8.2 | Asp300, His299, Asp197, Glu233, His305 | Trp59, Leu165 |
| 5 | −7.7 | Arg125, Ser630, His740, Asn710, Tyr547, Val207 | Glu205, Phe357 | −8.4 | His299, Gln63, Asp300, Glu233 | Tyr62, Trp59 |
| 6 | −7.8 | Glu206, Arg358, Tyr547 | Phe357, Tyr666 | −8.0 | Lys200, Tyr151, Asp197 | Ile235, Ala198, His201, Leu162, Lys200 |
| 7 | −7.9 | Ser209, Arg125, Tyr631 | Glu205, Glu206, Tyr666, Phe662 | −8.6 | Thr163, Glu233, Gln63, Arg195 | Leu162, Leu165, Asp197, Asp300 |
| 8 | −8.1 | Glu206, Arg125, Asn710, His740, Ser630 | Glu205 | −8.1 | Glu233, Asp197, Asp300 | Trp59, Leu165 |
| 9 | −6.3 | His126, Arg125 | Glu205, Tyr666, Phe357 | −7.5 | Gln63, Asp197 | Leu162, His299, Tyr62, His305, Trp59, Trp58 |
| sitagliptin | −8.6 | Ser209, Arg125, Arg358, Glu205, Glu206, Tyr 662 | Phe357, Ser630, Tyr666, Val207, His740 | - | - | - |
| acarbose | - | - | - | −6.9 | His305, Glu233, Tyr62, Asp197, Asp300, Thr163 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quek, A.; Kassim, N.K.; Ismail, A.; Latif, M.A.M.; Shaari, K.; Tan, D.C.; Lim, P.C. Identification of Dipeptidyl Peptidase-4 and α-Amylase Inhibitors from Melicope glabra (Blume) T. G. Hartley (Rutaceae) Using Liquid Chromatography Tandem Mass Spectrometry, In Vitro and In Silico Methods. Molecules 2021, 26, 1. https://doi.org/10.3390/molecules26010001
Quek A, Kassim NK, Ismail A, Latif MAM, Shaari K, Tan DC, Lim PC. Identification of Dipeptidyl Peptidase-4 and α-Amylase Inhibitors from Melicope glabra (Blume) T. G. Hartley (Rutaceae) Using Liquid Chromatography Tandem Mass Spectrometry, In Vitro and In Silico Methods. Molecules. 2021; 26(1):1. https://doi.org/10.3390/molecules26010001
Chicago/Turabian StyleQuek, Alexandra, Nur Kartinee Kassim, Amin Ismail, Muhammad Alif Mohammad Latif, Khozirah Shaari, Dai Chuan Tan, and Pei Cee Lim. 2021. "Identification of Dipeptidyl Peptidase-4 and α-Amylase Inhibitors from Melicope glabra (Blume) T. G. Hartley (Rutaceae) Using Liquid Chromatography Tandem Mass Spectrometry, In Vitro and In Silico Methods" Molecules 26, no. 1: 1. https://doi.org/10.3390/molecules26010001
APA StyleQuek, A., Kassim, N. K., Ismail, A., Latif, M. A. M., Shaari, K., Tan, D. C., & Lim, P. C. (2021). Identification of Dipeptidyl Peptidase-4 and α-Amylase Inhibitors from Melicope glabra (Blume) T. G. Hartley (Rutaceae) Using Liquid Chromatography Tandem Mass Spectrometry, In Vitro and In Silico Methods. Molecules, 26(1), 1. https://doi.org/10.3390/molecules26010001







