Fatty-Acid Uptake in Prostate Cancer Cells Using Dynamic Microfluidic Raman Technology
Abstract
1. Introduction
2. Results
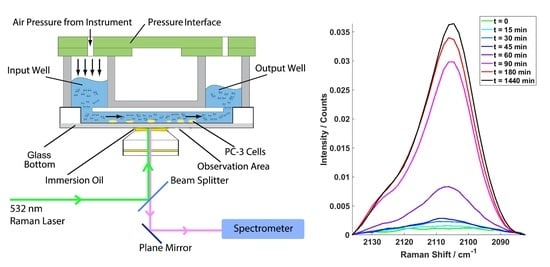
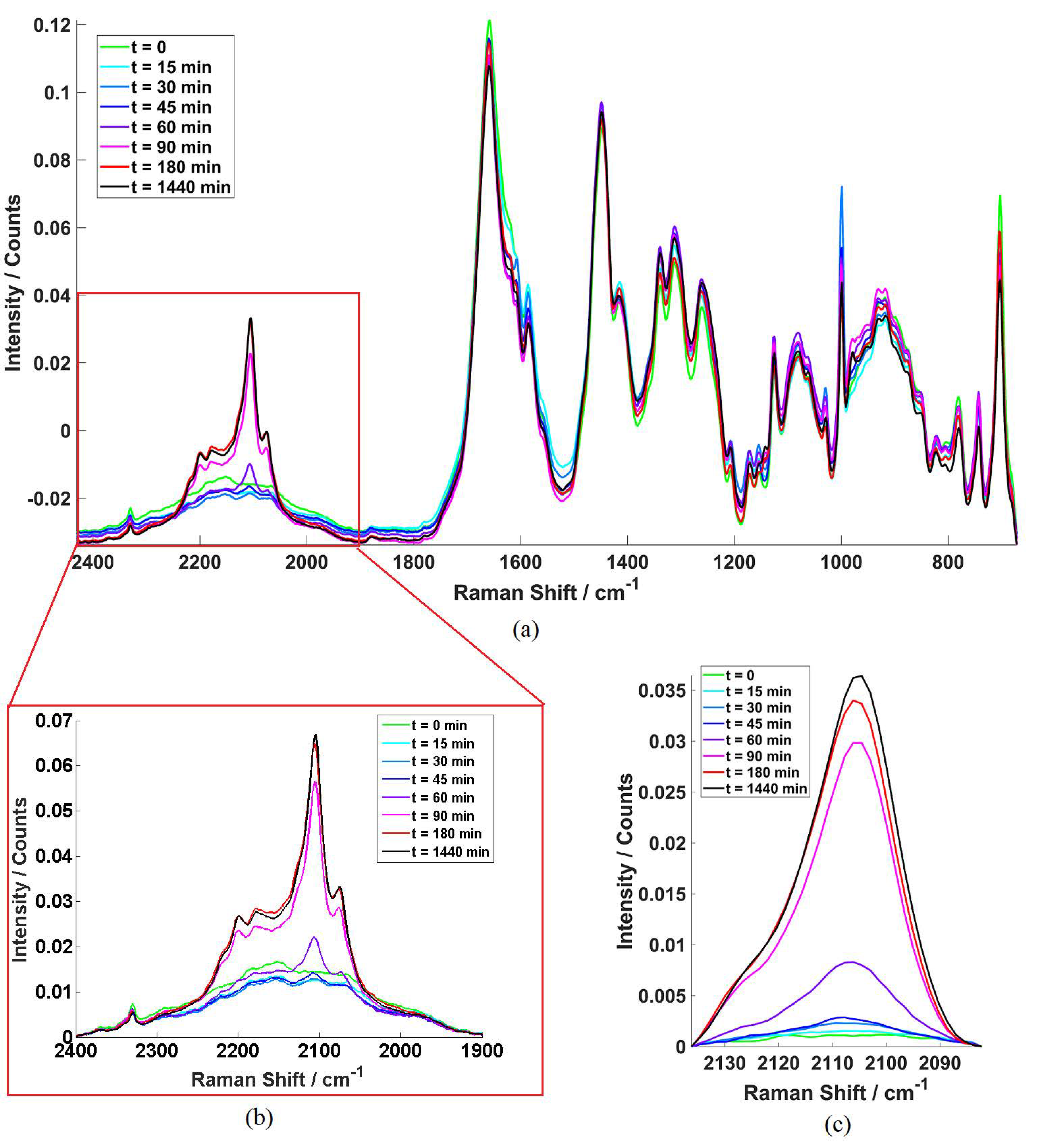
2.1. Example Cell Spectra
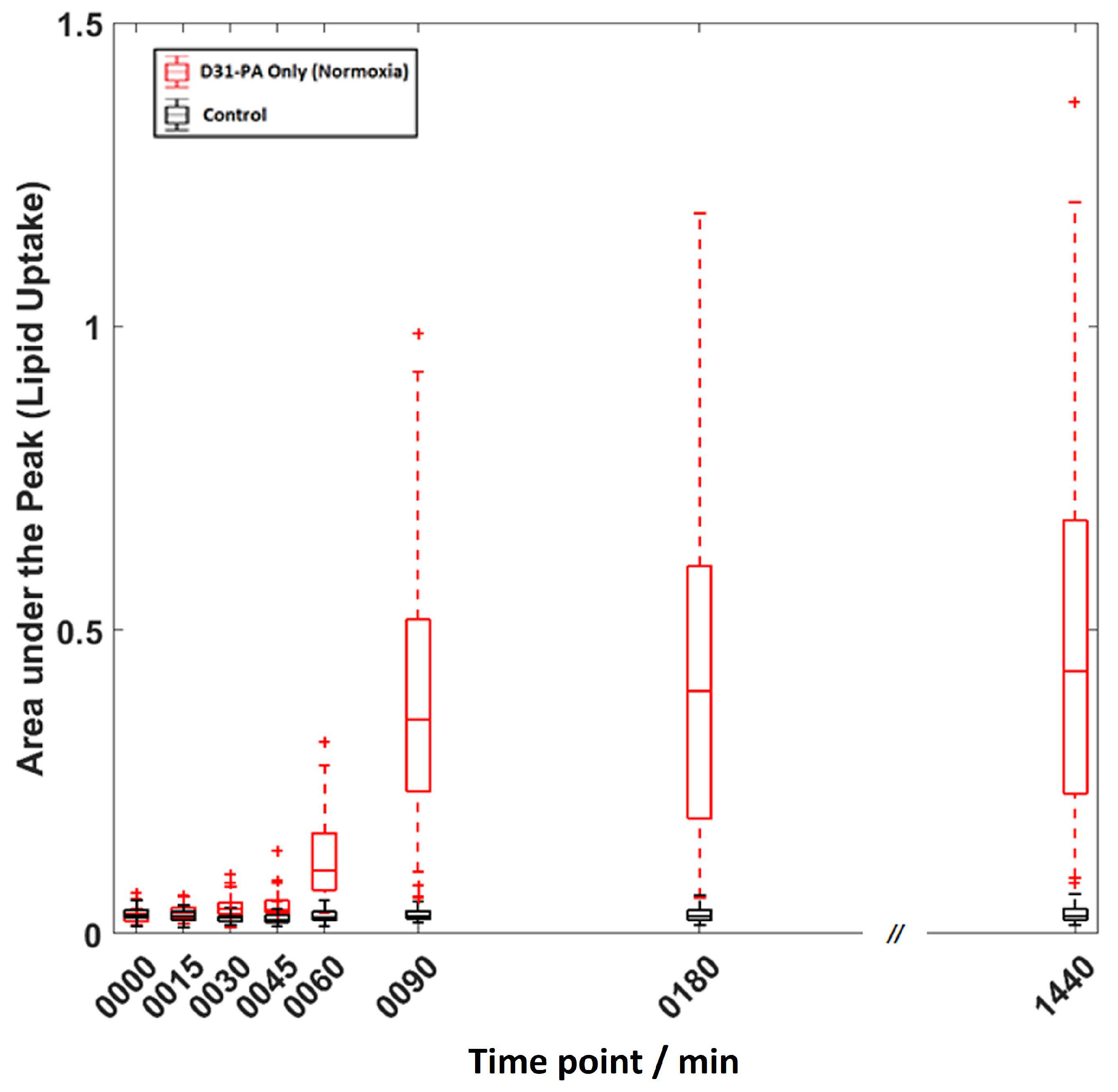
2.2. Competative Fatty Acid Uptake.
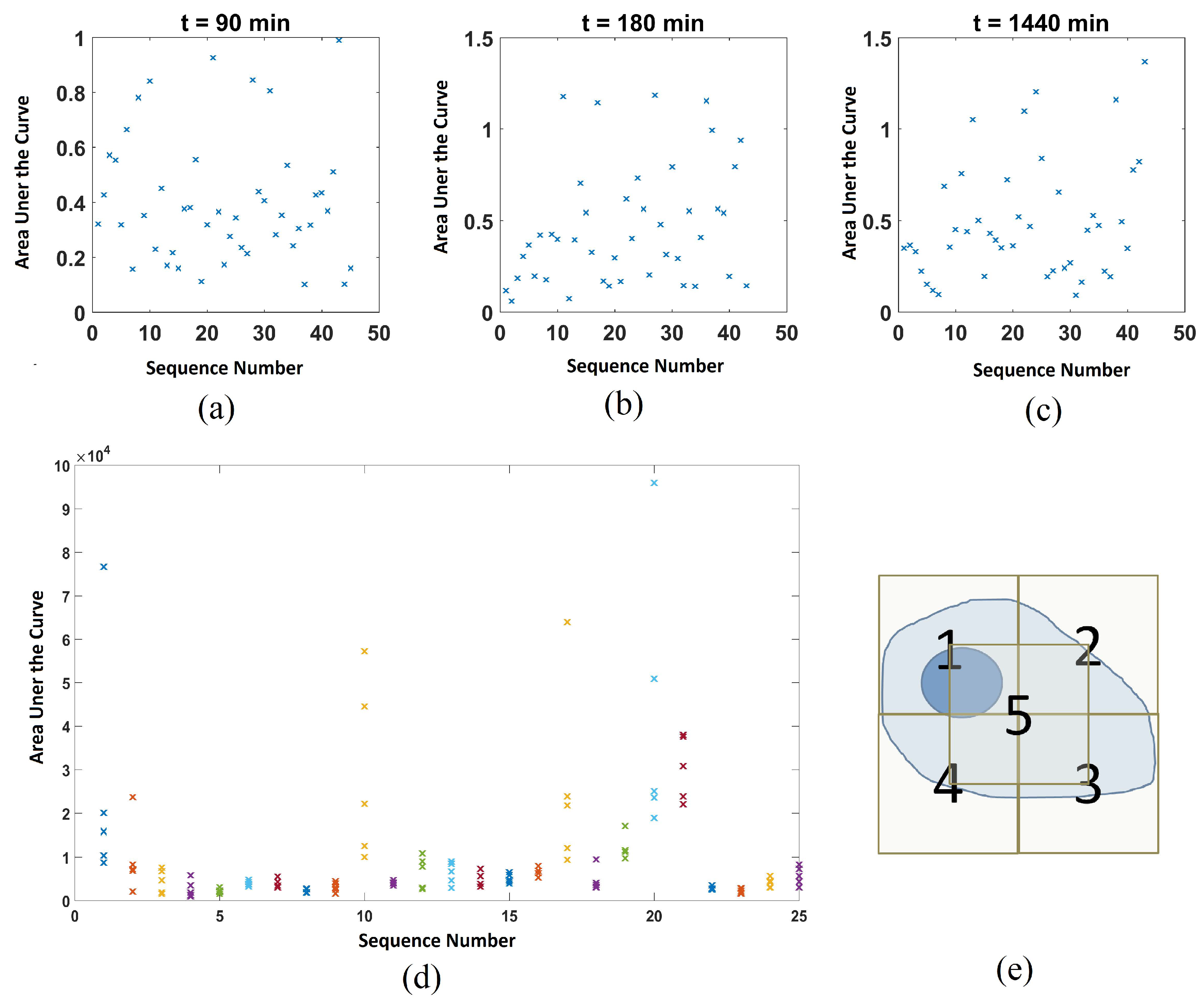
2.3. Distribution of Data Points
3. Discussion
4. Materials and Methods
4.1. Instrumentation
4.1.1. Microfluidic Device
4.1.2. Raman Microspectroscopy
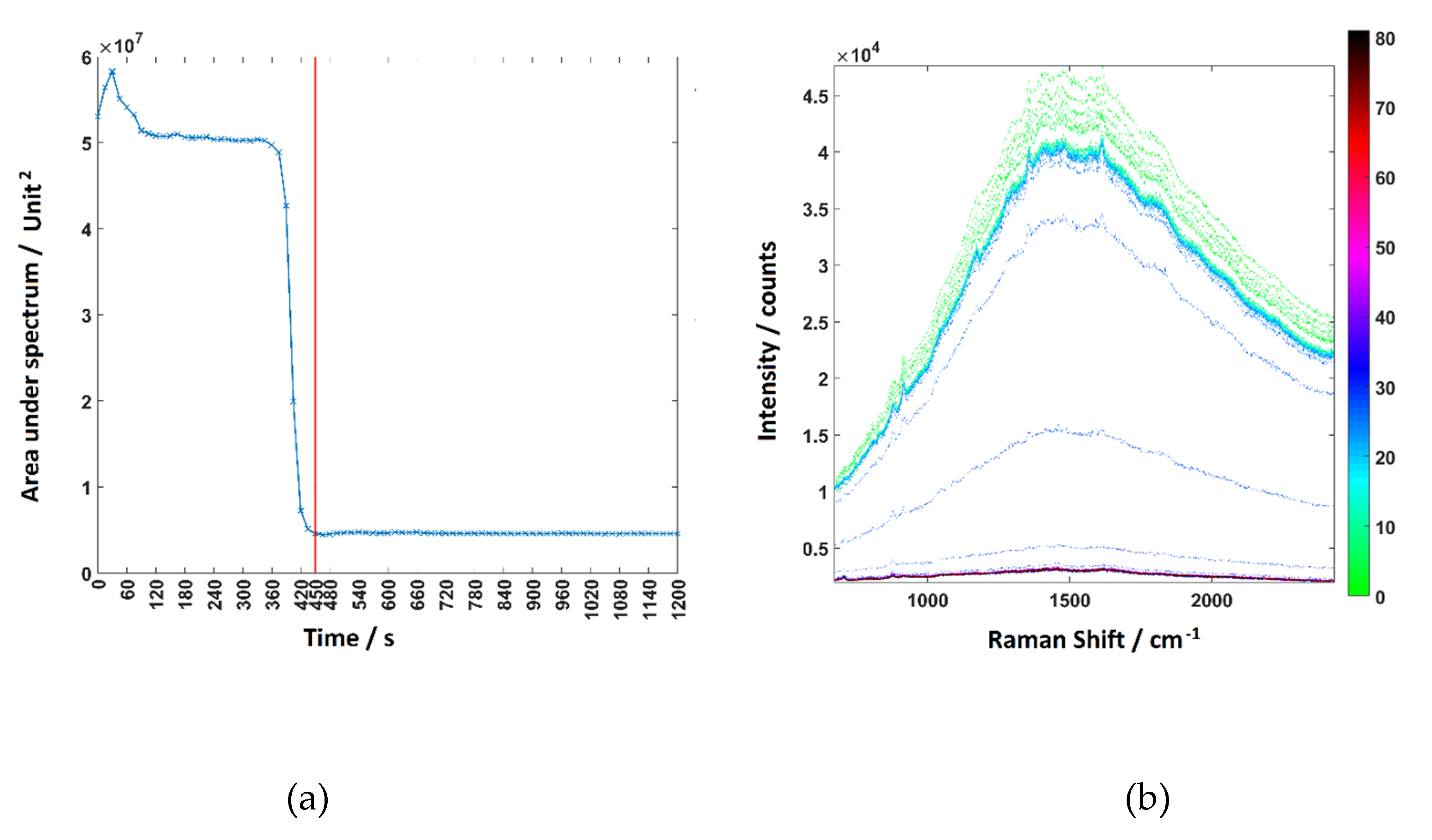
4.1.3. System Repeatability
- (i)
- Repeat measurements on the same bead
- (ii)
- Repeat measurements on different beads
4.2. Cell Culture
4.3. Fatty Acid Treatments
4.4. Data Processing and Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
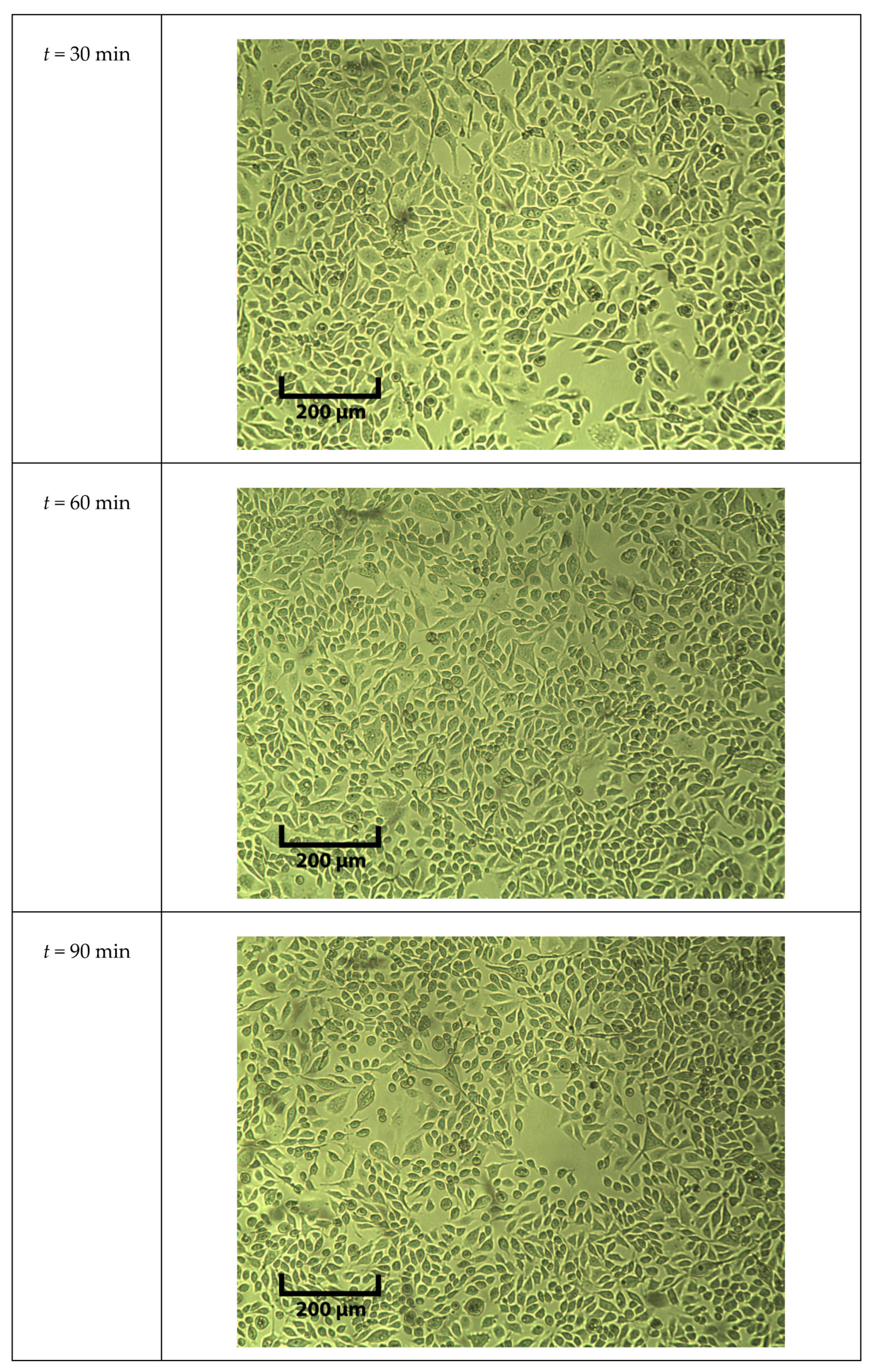
Appendix A.1. Example Cell Image Taken under the Microscope

Appendix A.2. Additional Data on the 5-Point Measurements
Appendix A.3. Statistical Significance for the Uptake Experiments
| RSD/% | ||||||||
|---|---|---|---|---|---|---|---|---|
| 0 min | 15 min | 30 min | 45 min | 60 min | 90 min | 180 min | 1440 min | |
| D31-PA | 32.1 | 32.5 | 40.1 | 46.1 | 54.1 | 56.4 | 69.9 | 65.2 |
| vs. DHA | 27.2 | 35.2 | 29.4 | 31.4 | 63.3 | 51.1 | 56.9 | 84.9 |
| vs. EPA | 40.3 | 33.3 | 36.2 | 29.2 | 33.8 | 41.6 | 54.0 | 49.3 |
| vs. AA | 32.1 | 34.8 | 61.9 | 61.2 | 86.1 | 78.1 | 63.2 | 51.2 |
| D31-PA (Hypoxic) | 32.7 | 39.1 | 62.3 | 64.1 | 63.0 | 76.5 | 69.9 | 90.9 |
| Ctrl | 35.1 | 34.8 | 28.5 | 33.7 | 35.3 | 37.8 | 35.3 | 44.0 |
| RSD/% | ||||||||
|---|---|---|---|---|---|---|---|---|
| 0 min | 15 min | 30 min | 45 min | 60 min | 90 min | 180 min | 1440 min | |
| D31-PA | 3.08 | 2.59 | 2.73 | 2.61 | 2.47 | 4.08 | 5.15 | 4.22 |
| vs. DHA | 2.91 | 2.48 | 3.08 | 3.61 | 3.16 | 2.56 | 3.43 | 4.38 |
| vs. EPA | 2.09 | 1.94 | 3.55 | 3.11 | 3.33 | 2.94 | 3.23 | 2.30 |
| vs. AA | 3.21 | 4.16 | 2.57 | 2.51 | 3.08 | 3.30 | 2.87 | 2.99 |
| D31-PA (Hypoxic) | 4.03 | 3.10 | 2.46 | 3.32 | 3.85 | 6.02 | 6.34 | 11.5 |
| Ctrl | 2.91 | 3.39 | 3.51 | 2.46 | 2.67 | 3.21 | 3.41 | 3.47 |
| p Value | Control | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 min | 15 min | 30 min | 45 min | 60 min | 90 min | 180 min | 1440 min | |
| vs. D31-PA (Normoxic) | 0.998 (98) | 0.012 (93) * | <0.001 (92) ** | <0.001 (94) ** | <0.001 (95) ** | <0.001 (95) ** | <0.001 (94) ** | <0.001 (91) ** |
| vs. DHA | 0.543 (98) | <0.001 (88) ** | <0.001 (97) ** | <0.001 (95) ** | <0.001 (90) ** | <0.001 (92) ** | <0.001 (89) ** | <0.001 (96) ** |
| vs. EPA | 0.013 (99) | 0.272 (93) | <0.001 (97) ** | 0.025 (90) * | <0.001 (94) ** | <0.001 (96) ** | <0.001 (100) ** | <0.001 (96) ** |
| vs. AA | 0.769 (96) * | <0.001 (99) ** | <0.001 (99) ** | <0.001 (98) ** | <0.001 (94) ** | <0.001 (104) ** | <0.001 (93) ** | <0.001 (98) ** |
| vs. D31-PA (Hypoxic) | 0.942 (99) | <0.001 (96) ** | <0.001 (96) ** | <0.001 (96) ** | <0.001 (97) ** | <0.001 (99) ** | <0.001 (100) ** | <0.001 (98) ** |
Appendix A.4. Biological Optimisation of the System: Flow Rate and Background Test
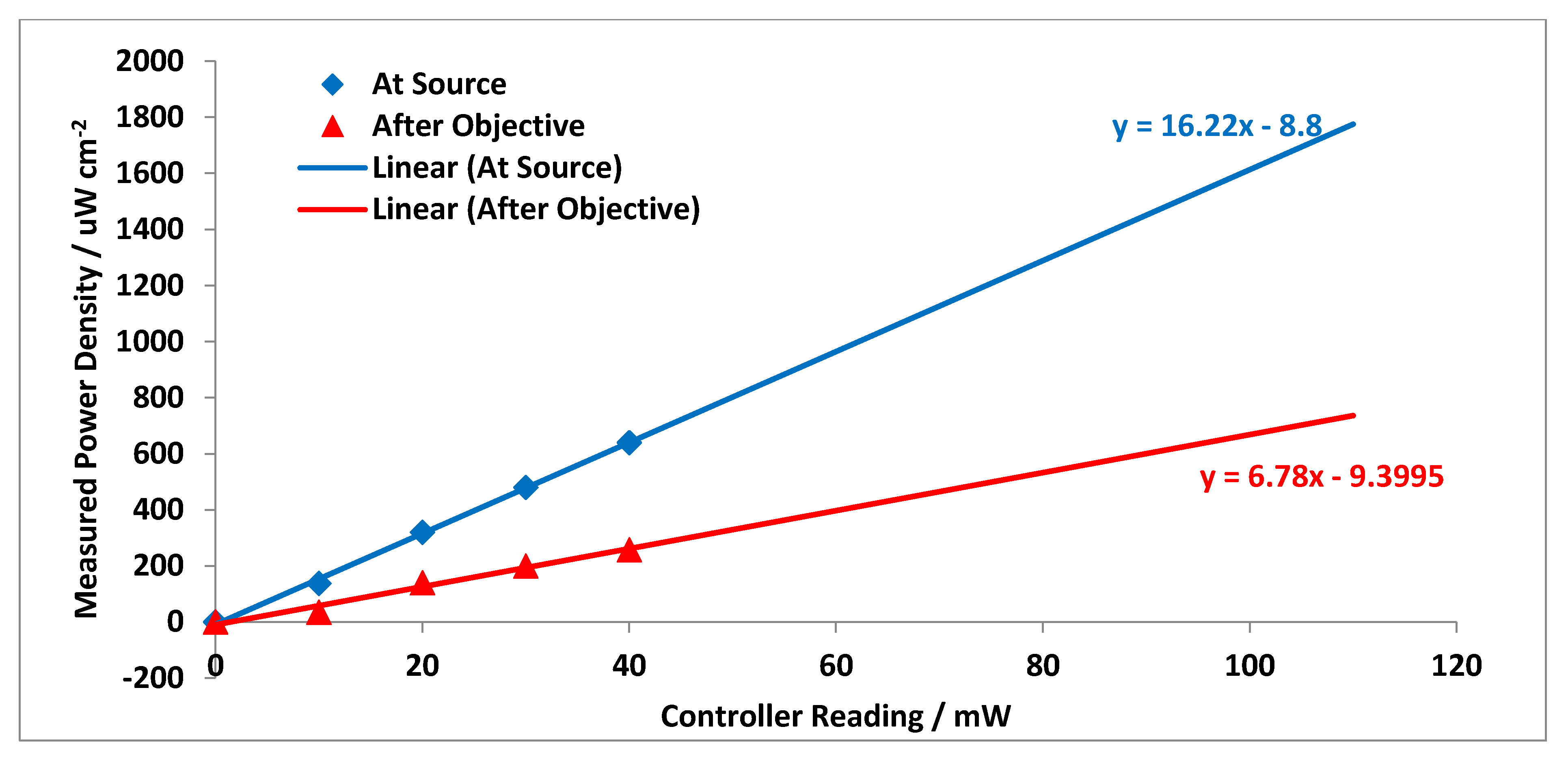
Appendix A.5. Relative Raman Laser Power Ratio Measurement Results
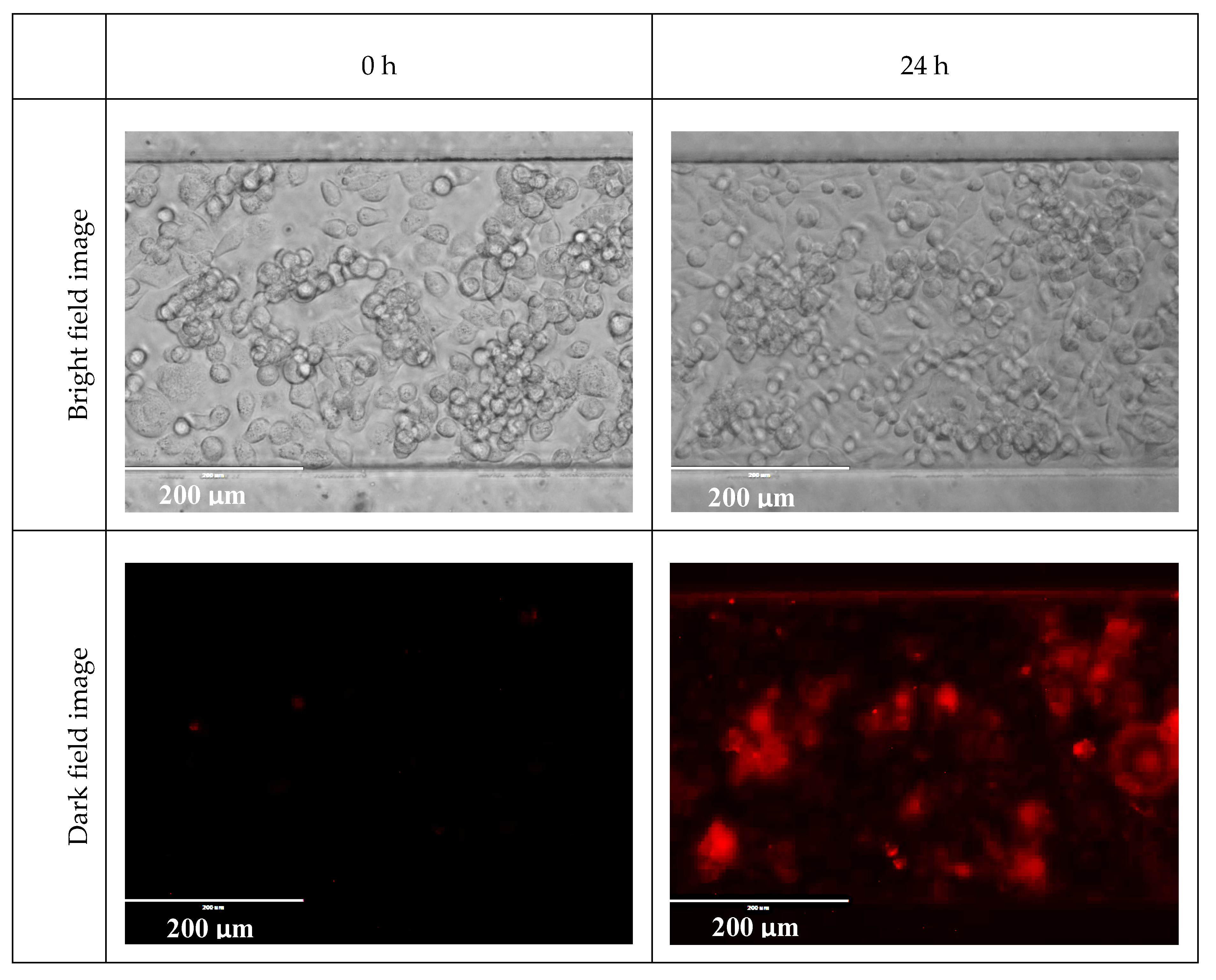
Appendix A.6. Cell Viability in Room Temperature DPBS

| Sample 1 | Sample 2 | Sample 3 | Average |
|---|---|---|---|
| 90.9% | 88% | 94.1% | 91.0% |
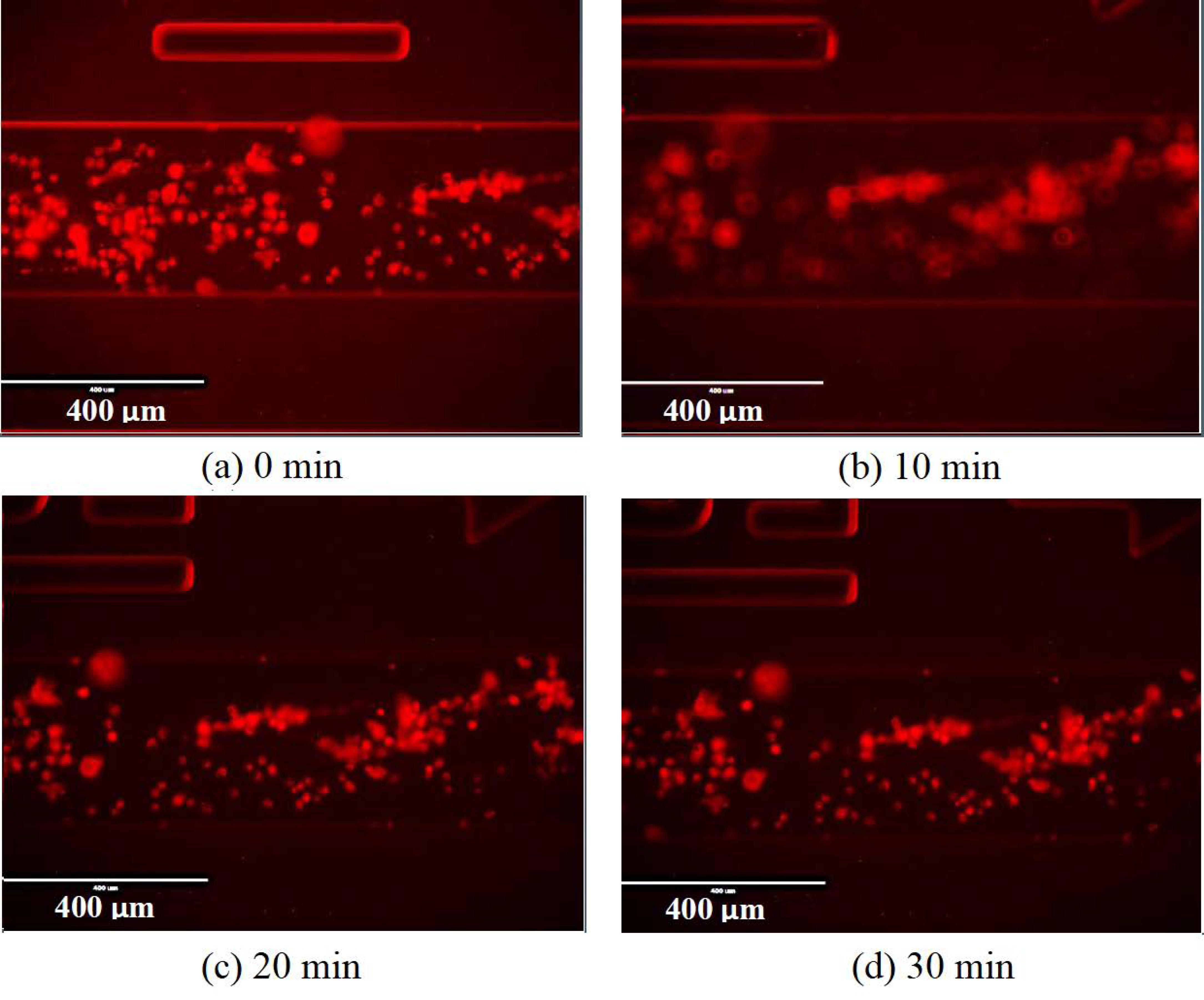
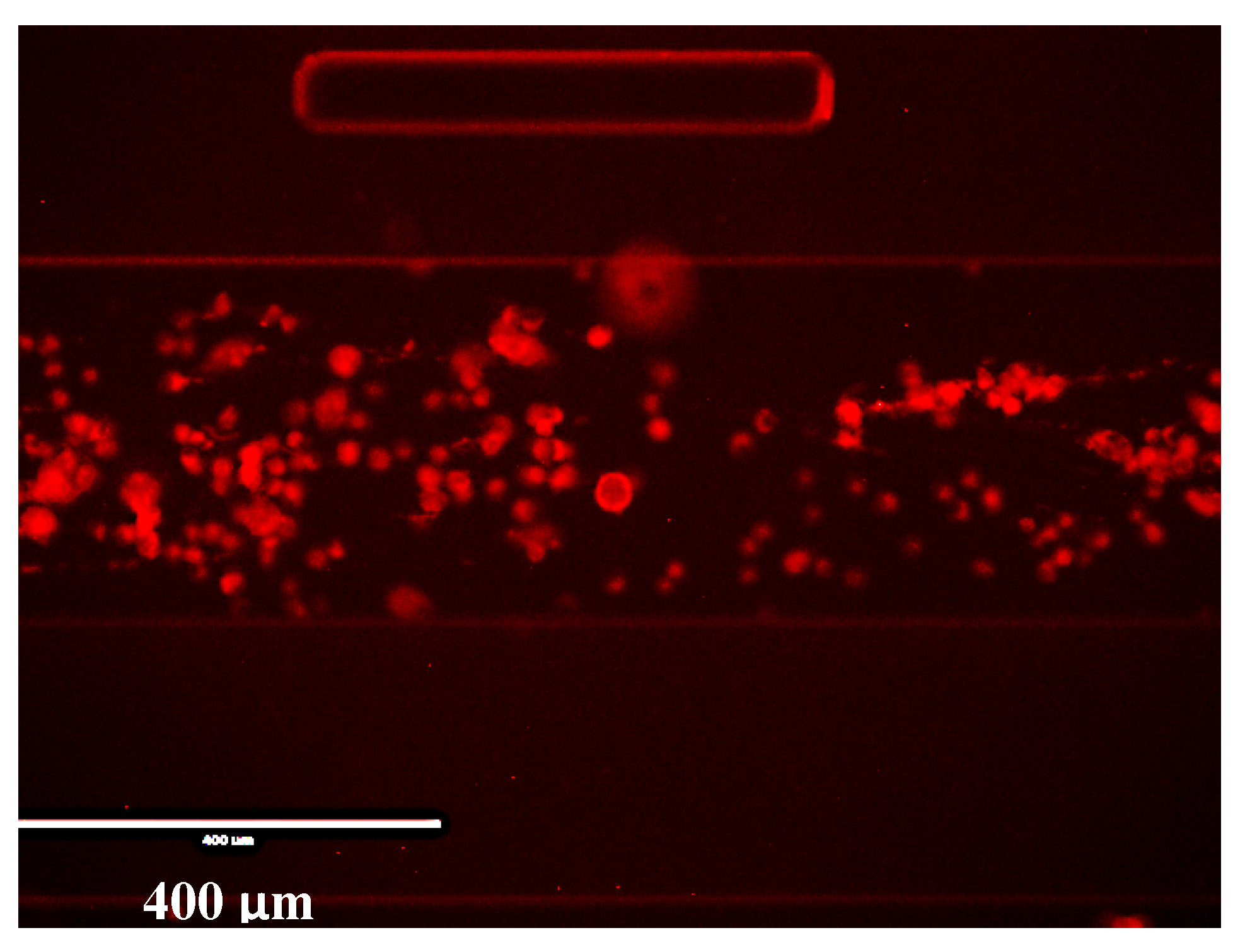
Appendix A.7. Monitoring the Change of States of Cells Using Hypoxic Reagent
References
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G.; Thompson, C.B. Q&A: Clues from cell metabolism. Nature 2010, 465, 3–5. [Google Scholar]
- Benjamin, D.I.; Cravatt, B.F.; Nomura, D.K. Global Profiling Strategies for Mapping Dysregulated Metabolic Pathways in Cancer. Cell Metab. 2012, 16, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Cheng, C.; Tan, Z.; Li, N.; Tang, M.; Yang, L.; Cao, Y. Emerging roles of lipid metabolism in cancer metastasis. Mol. Cancer 2017, 16, 76. [Google Scholar] [CrossRef]
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef]
- Röhrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef]
- Dubikovskaya, E.; Chudnovskiy, R.; Karateev, G.; Park, H.M.; Stahl, A. Measurement of long-chain fatty acid uptake into adipocytes. Methods Enzym. 2014, 538, 107–134. [Google Scholar]
- Maier, O.; Oberle, V.; Hoekstra, D. Fluorescent lipid probes: Some properties and applications (a review). Chem. Phys. Lipids 2002, 116, 3–18. [Google Scholar] [CrossRef]
- Blanksby, S.J.; Mitchell, T.W. Advances in Mass Spectrometry for Lipidomics. Annu. Rev. Anal. Chem. 2010, 3, 433–465. [Google Scholar] [CrossRef] [PubMed]
- Denbigh, J.L.; Lockyer, N.P. ToF-SIMS as a tool for profiling lipids in cancer and other diseases. Mater. Sci. Technol. 2014, 31, 137–147. [Google Scholar] [CrossRef]
- Gazi, E.; Harvey, T.J.; Brown, M.D.; Lockyer, N.P.; Gardner, P.; Clarke, N.W. A FTIR microspectroscopic study of the uptake and metabolism of isotopically labelled fatty acids by metastatic prostate cancer. Vib. Spectrosc. 2009, 50, 99–105. [Google Scholar] [CrossRef]
- Gazi, E.; Gardner, P.; Lockyer, N.P.; Hart, C.A.; Brown, M.D.; Clarke, N.W. Direct evidence of lipid translocation between adipocytes and prostate cancer cells with imaging FTIR microspectroscopy. J. Lipid Res. 2007, 48, 1846–1856. [Google Scholar] [CrossRef]
- Majzner, K.; Tott, S.; Roussille, L.; Deckert, V.; Chlopicki, S.; Baranska, M. Uptake of fatty acids by a single endothelial cell investigated by Raman spectroscopy supported by AFM. Analyst 2018, 143, 970–980. [Google Scholar] [CrossRef]
- Jamieson, L.E.; Greaves, J.; McLellan, J.A.; Munro, K.R.; Tomkinson, N.C.O.; Chamberlain, L.H.; Faulds, K.; Graham, D. Tracking intracellular uptake and localisation of alkyne tagged fatty acids using Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 197, 30–36. [Google Scholar] [CrossRef]
- Wei, L.; Hu, F.; Shen, Y.; Chen, Z.; Yu, Y.; Lin, C.-C.; Wang, M.C.; Min, W. Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat. Methods 2014, 11, 410–412. [Google Scholar] [CrossRef]
- Hong, S.; Chen, T.; Zhu, Y.; Li, A.; Huang, Y.; Chen, X. Live-cell stimulated Raman scattering imaging of alkyne-tagged biomolecules. Angew. Chem. Int. Ed. 2014, 53, 5827–5831. [Google Scholar] [CrossRef]
- Clemens, G.; Flower, K.R.; Gardner, P.; Henderson, A.P.; Knowles, J.P.; Marder, T.B.; Whiting, A.; Przyborski, S. Design and biological evaluation of synthetic retinoids: Probing length vs. stability vs. activity. Mol. Biosyst. 2013, 9, 3124–3134. [Google Scholar] [CrossRef]
- Strehle, K.R.; Cialla, D.; Rösch, P.; Henkel, T.; Köhler, M.; Popp, J. A reproducible surface-enhanced Raman spectroscopy approach. Online SERS measurements in a segmented microfluidic system. Anal. Chem. 2007, 79, 1542–1547. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.A.; Brown, L.O.; Gaskill, D.F.; Naivar, M.; Graves, S.W.; Doorn, S.K.; Nolan, J.P. A flow cytometer for the measurement of raman spectra. Cytom. Part A 2008, 73, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Dochow, S.; Krafft, C.; Neugebauer, U.; Bocklitz, T.; Henkel, T.; Mayer, G.; Albert, J.; Popp, J. Tumour cell identification by means of Raman spectroscopy in combination with optical traps and microfluidic environments. Lab Chip 2011, 11, 1484–1490. [Google Scholar] [CrossRef] [PubMed]
- Perozziello, G.; Candeloro, P.; De Grazia, A.; Esposito, F.; Allione, M.; Coluccio, M.L.; Tallerico, R.; Valpapuram, I.; Tirinato, L.; Das, G.; et al. Microfluidic device for continuous single cells analysis via Raman spectroscopy enhanced by integrated plasmonic nanodimers. Opt. Express 2016, 24, A180. [Google Scholar] [CrossRef]
- Casabella, S.; Scully, P.; Goddard, N.; Gardner, P. Automated analysis of single cells using Laser Tweezers Raman Spectroscopy. Analyst 2016, 141, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Cancer Research UK Prostate Cancer Incidence Statistics. Available online: http://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/prostate-cancer/incidence#heading-Two (accessed on 19 January 2016).
- Norrish, A.E.; Skeaff, C.M.; Arribas, G.L.; Sharpe, S.J.; Jackson, R.T. Prostate cancer risk and consumption of fish oils: A dietary biomarker-based case-control study. Br. J. Cancer 1999, 81, 1238–1242. [Google Scholar] [CrossRef]
- Dennis, L.K.; Snetselaar, L.G.; Smith, B.J.; Stewart, R.E.; Robbins, M.E.C. Problems with the assessment of dietary fat in prostate cancer studies. Am. J. Epidemiol. 2004, 160, 436–444. [Google Scholar] [CrossRef]
- Giovannucci, E.; Rimm, E.B.; Colditz, G.; Stampfer, M.J.; Ascherio, A.; Chute, C.; Willett, W. A Prospective Study of Dietary Fat and Risk of Prostate Cancer. JNCI J. Natl. Cancer Inst. 1993, 85, 1571–1579. [Google Scholar] [CrossRef]
- Brown, M.D.; Hart, C.A.; Gazi, E.; Bagley, S.; Clarke, N.W. Promotion of prostatic metastatic migration towards human bone marrow stoma by Omega 6 and its inhibition by Omega 3 PUFAs. Br. J. Cancer 2006, 94, 842–853. [Google Scholar] [CrossRef]
- Šebek, J.; Pele, L.; Potma, E.O.; Benny Gerber, R. Raman spectra of long chain hydrocarbons: Anharmonic calculations, experiment and implications for imaging of biomembranes. Phys. Chem. Chem. Phys. 2011, 13, 12724–12733. [Google Scholar] [CrossRef]
- Wenger, R.; Kurtcuoglu, V.; Scholz, C.; Marti, H.; Hoogewijs, D. Frequently asked questions in hypoxia research. Hypoxia 2015, 3, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Milosevic, M.; Warde, P.; Meńard, C.; Chung, P.; Toi, A.; Ishkanian, A.; McLean, M.; Pintilie, M.; Sykes, J.; Gospodarowicz, M.; et al. Tumor hypoxia predicts biochemical failure following radiotherapy for clinically localized prostate cancer. Clin. Cancer Res. 2012, 18, 2108–2114. [Google Scholar] [CrossRef] [PubMed]
- Furlong, S.T.; Thibault, K.S.; Morbelli, L.M.; Quinn, J.J.; Rogers, R.A. Uptake and Compartmentalization of Fluorescent Lipid Analogs in Larval Schistosoma-Mansoni. J. Lipid Res. 1995, 36, 1–12. [Google Scholar]
- Mojumdar, E.H.; Groen, D.; Gooris, G.S.; Barlow, D.J.; Lawrence, M.J.; Deme, B.; Bouwstra, J.A. Localization of cholesterol and fatty acid in a model lipid membrane: A neutron diffraction approach. Biophys. J. 2013, 105, 911–918. [Google Scholar] [CrossRef] [PubMed]
- Hayes, V.; Johnston, I.; Arepally, G.M.; McKenzie, S.E.; Cines, D.B.; Rauova, L.; Poncz, M. Endothelial antigen assembly leads to thrombotic complications in heparin-induced thrombocytopenia. J. Clin. Invest. 2017, 127, 1090–1098. [Google Scholar] [CrossRef]
- Diebel, L.N.; Diebel, M.E.; Martin, J.V.; Liberati, D.M. Acute hyperglycemia exacerbates trauma-induced endothelial and glycocalyx injury: An in vitro model. J. Trauma Acute Care Surg. 2018, 85, 960–967. [Google Scholar] [CrossRef]
- Abumrad, N.; Coburn, C.; Ibrahimi, A. Membrane proteins implicated in long-chain fatty acid uptake by mammalian cells: CD36, FATP and FABPm. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1999, 1441, 4–13. [Google Scholar] [CrossRef]
- Williams, D.H.; Fleming, I. Spectroscopic Methods in Organic Chemistry, 5th ed.; McGraw-Hill: New York, NY, USA, 1995; ISBN 9780077091477. [Google Scholar]
- Shimanouchi, T. Tables of Molecular Vibrational Frequencies; National Bureau of Standards: Gaithersburg, MD, USA, 1972; Volume 1.
- Brown, M.D.; Hart, C.; Gazi, E.; Gardner, P.; Lockyer, N.; Clarke, N. Influence of omega-6 PUFA arachidonic acid and bone marrow adipocytes on metastatic spread from prostate cancer. Br. J. Cancer 2010, 102, 403–413. [Google Scholar] [CrossRef]
- Rezende, L.P.; Galheigo, M.R.U.; Landim, B.C.; Cruz, A.R.; Botelho, F.V.; Zanon, R.G.; Góes, R.M.; Ribeiro, D.L. Effect of glucose and palmitate environment on proliferation and migration of PC3-prostate cancer cells. Cell Biol. Int. 2019, 43, 373–383. [Google Scholar] [CrossRef]
- Kim, S.; Yang, X.; Yin, A.; Zha, J.; Beharry, Z.; Bai, A.; Bielawska, A.; Bartlett, M.G.; Yin, H.; Cai, H. Dietary palmitate cooperates with Src kinase to promote prostate tumor progression. Prostate 2019, 79, 896–908. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, S.; Ji, Y.; Chen, H.; Zhang, H.; Chen, W.; Gu, Z.; Chen, Y.Q. Dietary intake of n-3 PUFAs modifies the absorption, distribution and bioavailability of fatty acids in the mouse gastrointestinal tract. Lipids Health Dis. 2017, 16, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Holman, R.T. Essential Fatty Acids. Nutr. Rev. 1958, 16, 33–35. [Google Scholar] [CrossRef] [PubMed]
- Bensaad, K.; Favaro, E.; Lewis, C.A.; Peck, B.; Lord, S.; Collins, J.M.; Pinnick, K.E.; Wigfield, S.; Buffa, F.M.; Li, J.L.; et al. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014, 9, 349–365. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Yotnda, P. Induction and Testing of Hypoxia in Cell Culture. J. Vis. Exp. 2011, 54, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Harvey, T.J.; Faria, E.C.; Henderson, A.; Gazi, E.; Ward, A.D.; Clarke, N.W.; Brown, M.D.; Snook, R.D.; Gardner, P. Spectral discrimination of live prostate and bladder cancer cell lines using Raman optical tweezers. J. Biomed. Opt. 2008, 13, 64004. [Google Scholar] [CrossRef][Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |



| p Value | D31-PA Only (Normoxic State) | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 min | 15 min | 30 min | 45 min | 60 min | 90 min | 180 min | 1440 min | |
| vs. DHA | 0.999 (98) | 0.999 (89) | 0.254 (91) | 0.023 (95) * | 0.999 (89) | <0.001 (87) ** | <0.001 (81) ** | 0.144 (91) |
| vs. EPA | 1.000 (99) | 0.043 (94) * | <0.001 (91) ** | <0.001 (90) ** | <0.001 (93) ** | <0.001 (91) ** | <0.001 (92) ** | <0.001 (91) ** |
| vs. AA | 0.987 (96) | 0.933 (100) | 0.690 (93) | 0.964 (98) | <0.001 (93) ** | <0.001 (99) ** | <0.001 (85) ** | <0.001 (93) ** |
| vs. D31-PA (Hypoxic) | 0.918 (99) | 1.000 (97) | 1.000 (90) | 1.000 (96) | 0.901 (96) | 0.016 (94) * | 0.871 (92) | 0.010 (93) * |
| vs. control | 0.998 (98) | 0.012 (93) * | <0.001 (92) ** | <0.001 (94) ** | <0.001 (95) ** | <0.001 (95) ** | <0.001 (93) ** | <0.001 (91) ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, N.-T.; D. Snook, R.; Brown, M.D.; Haines, B.A.; Ridley, A.; Gardner, P.; Denbigh, J.L. Fatty-Acid Uptake in Prostate Cancer Cells Using Dynamic Microfluidic Raman Technology. Molecules 2020, 25, 1652. https://doi.org/10.3390/molecules25071652
Tang N-T, D. Snook R, Brown MD, Haines BA, Ridley A, Gardner P, Denbigh JL. Fatty-Acid Uptake in Prostate Cancer Cells Using Dynamic Microfluidic Raman Technology. Molecules. 2020; 25(7):1652. https://doi.org/10.3390/molecules25071652
Chicago/Turabian StyleTang, Nga-Tsing, Richard D. Snook, Mick D. Brown, Bryan A. Haines, Andrew Ridley, Peter Gardner, and Joanna L. Denbigh. 2020. "Fatty-Acid Uptake in Prostate Cancer Cells Using Dynamic Microfluidic Raman Technology" Molecules 25, no. 7: 1652. https://doi.org/10.3390/molecules25071652
APA StyleTang, N.-T., D. Snook, R., Brown, M. D., Haines, B. A., Ridley, A., Gardner, P., & Denbigh, J. L. (2020). Fatty-Acid Uptake in Prostate Cancer Cells Using Dynamic Microfluidic Raman Technology. Molecules, 25(7), 1652. https://doi.org/10.3390/molecules25071652