1H-NMR Metabolomics Analysis of the Effect of Rubusoside on Serum Metabolites of Golden Hamsters on a High-Fat Diet
Abstract
1. Introduction
2. Results
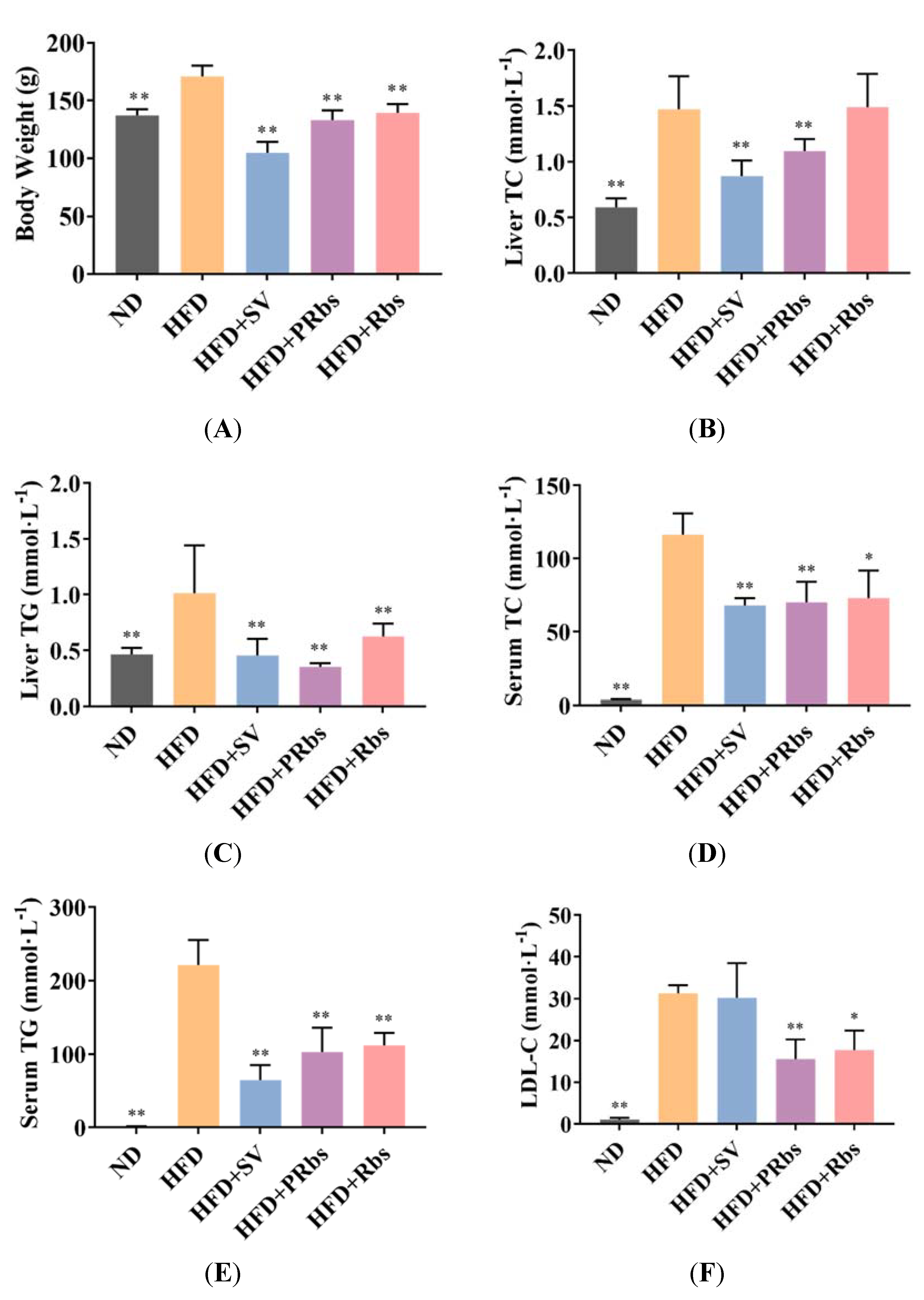
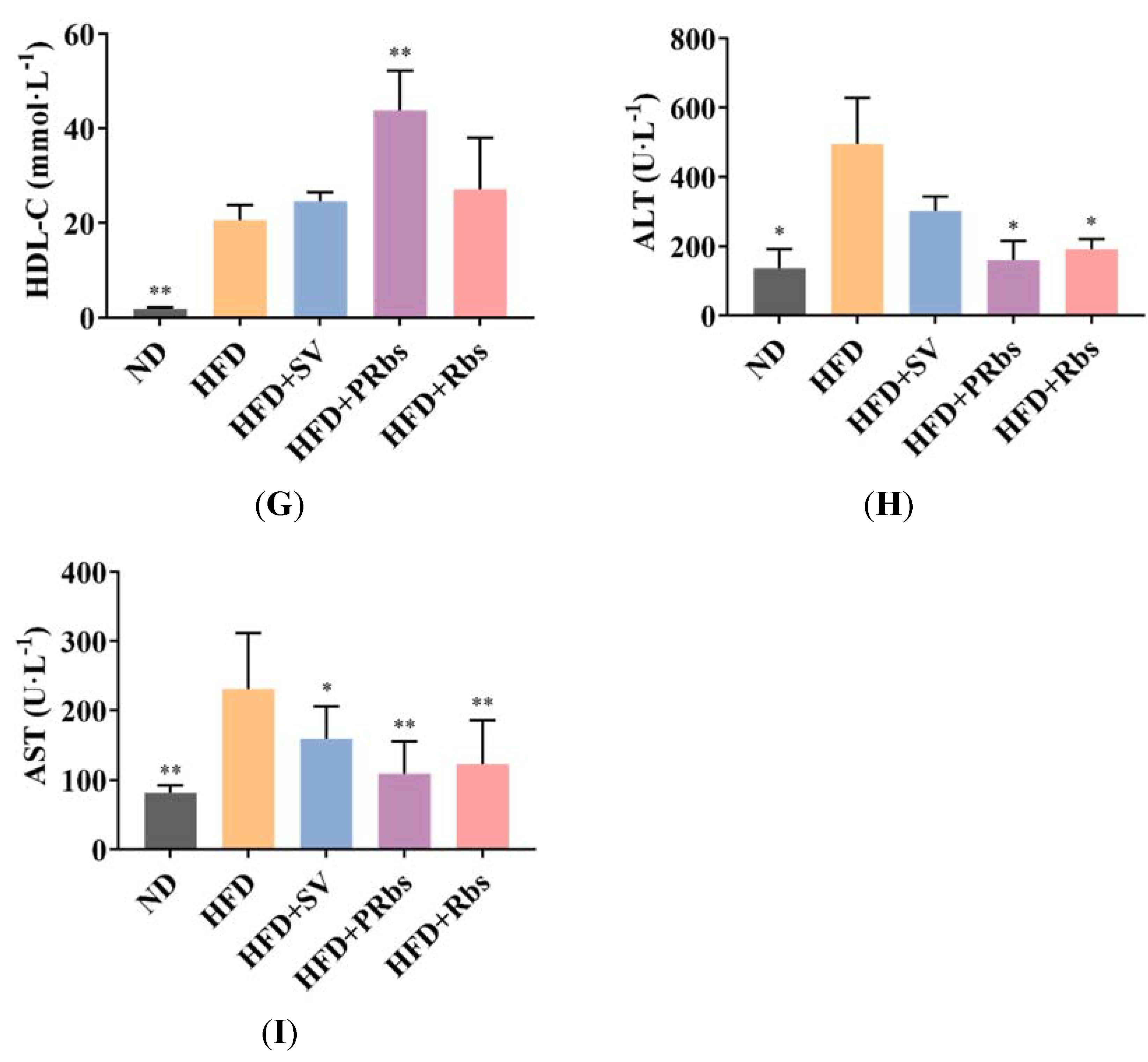
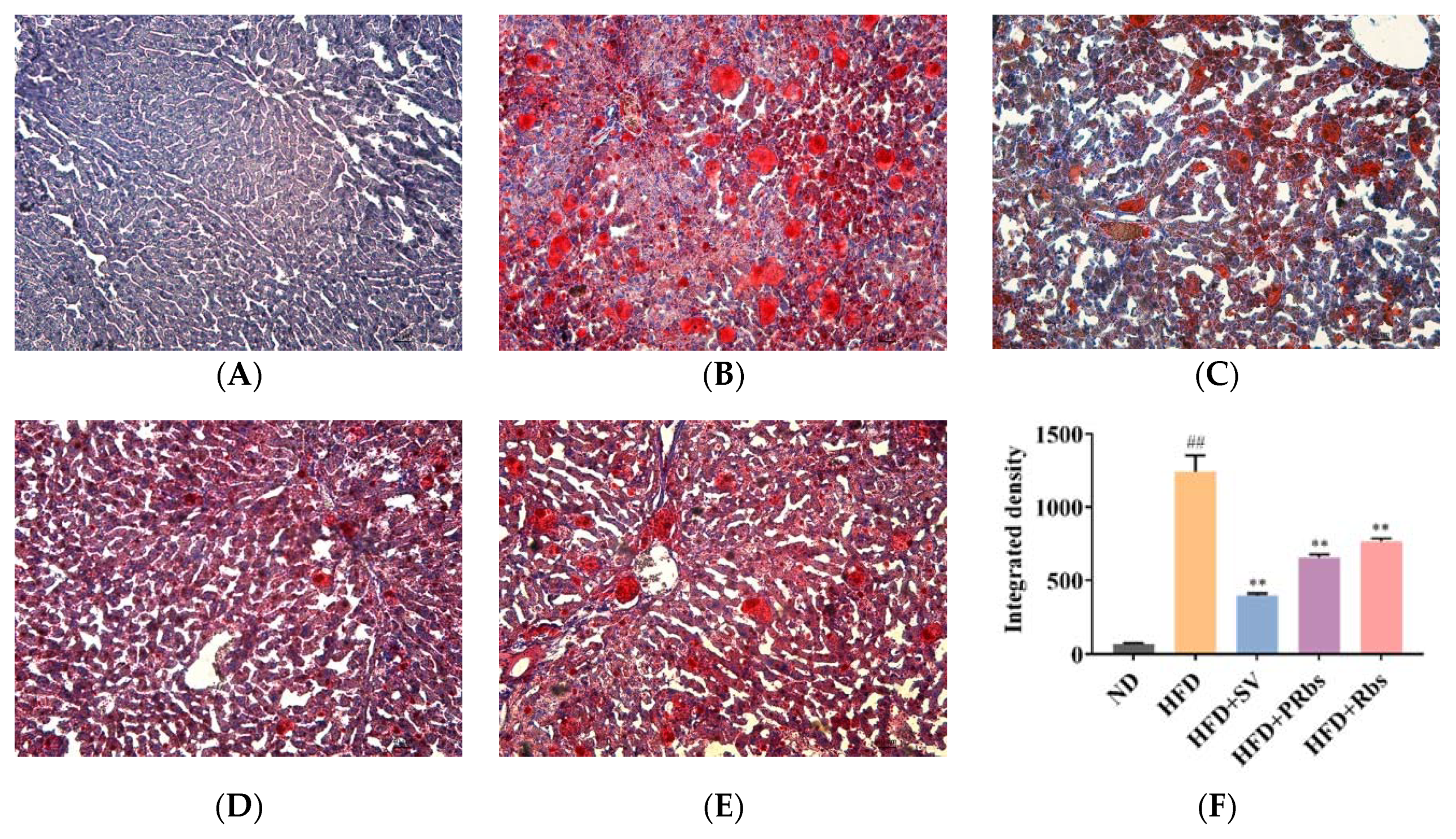
2.1. Animal Experiment Results
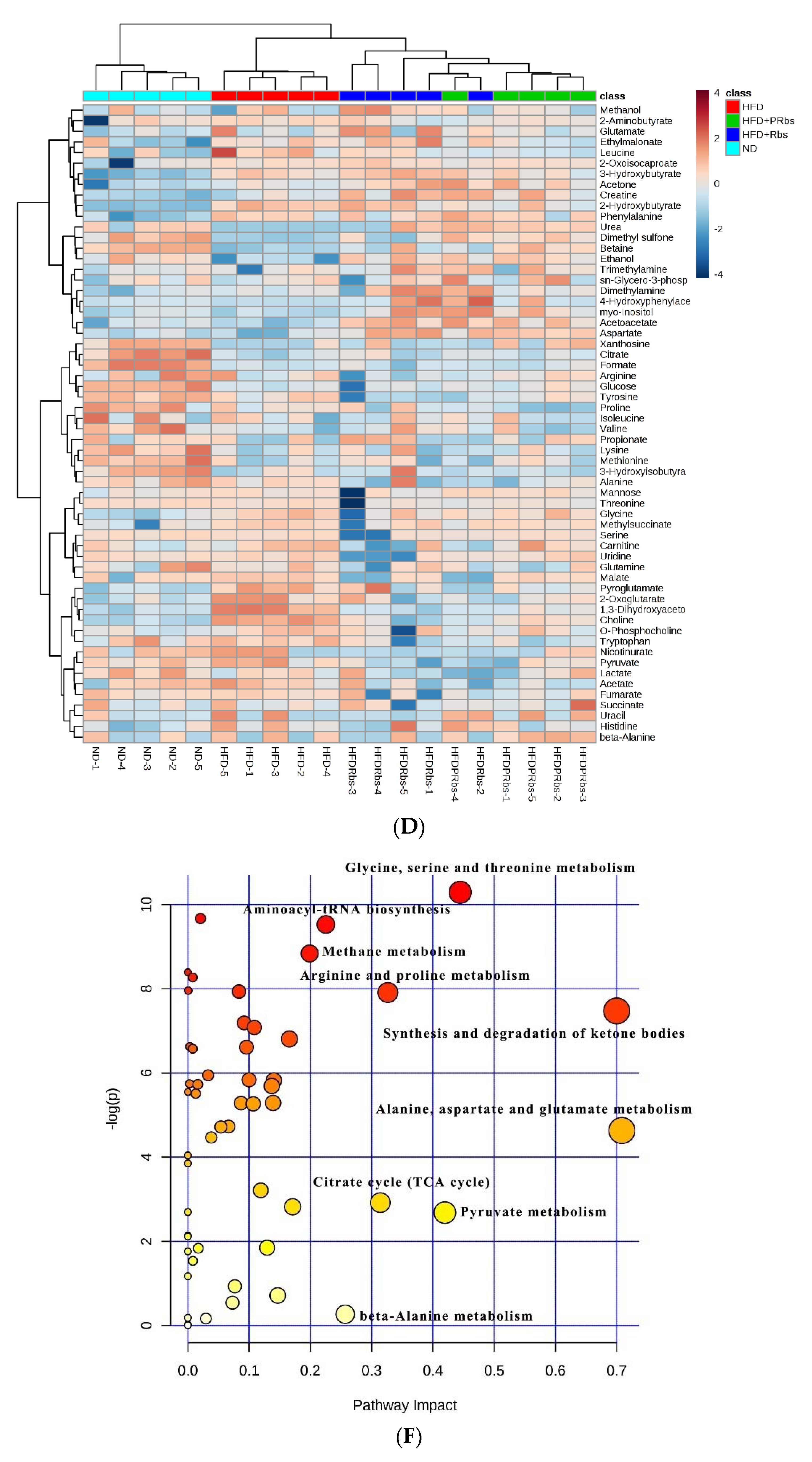
2.2. 1H-NMR Metabolomics Analysis
2.2.1. Analysis of Serum Metabolites
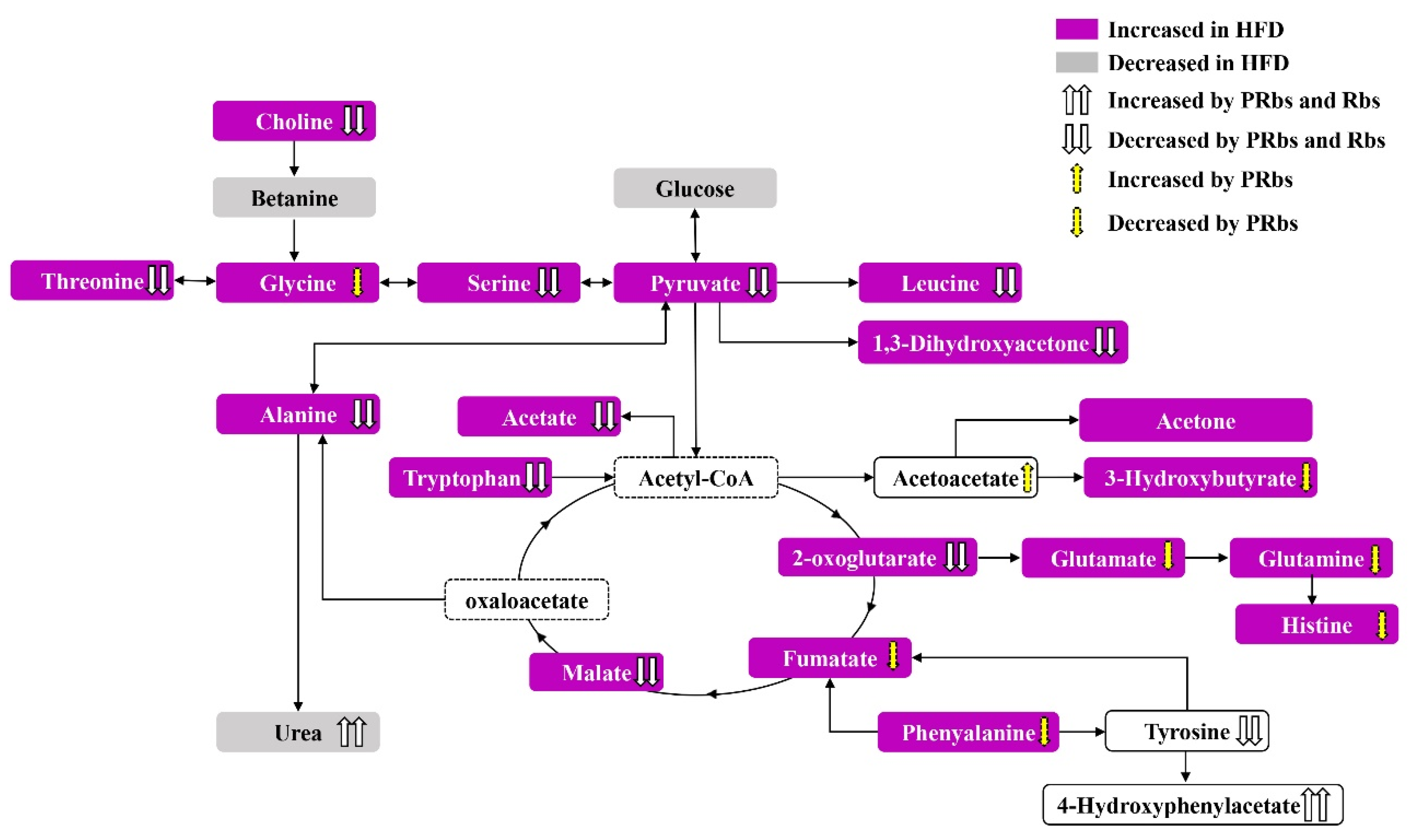
2.2.2. Analysis of Serum Metabolite Pathways
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Animal Experiment
4.3. Serum Sample Processing
4.4. 1H-NMR Spectroscopy
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- China Cardiovascular Disease Reporting Group. Summary of China Cardiovascular Disease Report 2017. Chin. Circ. J. 2018, 33, 1–8. [Google Scholar]
- Zhou, W.H.; Satomi, H.; Ryoji, K.; Osamu, T. A New Sweet Diterpene-Glucoside in Leaves of Rubus suavissimus. Bull. Bot. 1992, 34, 315–318. [Google Scholar]
- Hirono, S.; Chou, W.H.; Kasai, R.; Tanaka, O.; Tada, T. Sweet and bitter diterpene-glucosides from leaves of Rubus suavissimus. Chem. Pharm. Bull. 1990, 38, 1743–1744. [Google Scholar] [CrossRef]
- Ohtani, K.; Aikawa, Y.; Kasai, R.; Chou, W.H.; Yamasaki, K.; Tanaka, O. Minor diterpene glycosides from sweet leaves of Rubus suavissimus. Phytochemistry 1992, 31, 1553–1559. [Google Scholar] [CrossRef]
- Wei-ping, H. Natural sweetener—Industrial trial of rubusoside. Light Ind. Sci. Technol. 1999, 1–5. [Google Scholar]
- Guangxi Zhuang Autonomous Region Food and Drug Administration. Quality Standard for Zhuang Medicine in Guangxi Zhuang Autonomous Region Volume II (2011 Edition); Guangxi Science and Technology Press: Nanning, China, 2011. [Google Scholar]
- Bin, S.; Li, L.; Jipei, L. The Preventive and Curative Effect of Sweetener on the Experimental Rats with Hyperlipidemia. J. Guangxi Med Univ. 2001, 5, 627–629. [Google Scholar]
- Tian, C.P.; Qu, W.J.; Zhang, W.; Wang, H.; Li, M.J. Effect of Rubusoside on Mice Gluconeogenesis and Metabolism of Blood Lipid. Guangxi J. Tradit. Chin. Med. 2001, 24, 59–61. [Google Scholar]
- Koh, G.Y.; Mccutcheon, K.; Zhang, F.; Liu, D.; Cartwright, C.A.; Martin, R.; Yang, P.; Liu, Z. Improvement of Obesity Phenotype by Chinese Sweet Leaf Tea (Rubus suavissimus) Components in High-Fat Diet-Induced Obese Rats. J. Agric. Food Chem. 2011, 59, 98–104. [Google Scholar] [CrossRef]
- Ezure, T.; Amano, S. Rubus suavissimus S. Lee extract increases early adipogenesis in 3T3-L1 preadipocytes. J. Nat. Med. 2011, 65, 247–253. [Google Scholar] [CrossRef]
- Tian, C.P.; Qu, W.J.; Sun, B.; Li, M.J.; Wang, H.; Hung, X.Q. Effect of Rubusoside Extracts on Hyperglycemic Rats Induced by Streptozotocin. Acta Nutr. Sin. 2003, 25, 29–33. [Google Scholar]
- Deng, S.L. Study on Nutritional Components and Development and Utilization Value of Sweet Tea Leaves in Guangxi. For. Prod. Spec. China 2000, 54, 18–19. [Google Scholar]
- Zheng, H.; Wu, J.; Huang, H.; Meng, C.; Li, W.; Wei, T.; Su, Z. Metabolomics analysis of the protective effect of rubusoside on palmitic acid-induced lipotoxicity in INS-1 cells using UPLC-Q/TOF MS. J. Mol. Omics 2019, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.L.; He, K.X. A Comparative Study between of Rubus Suarissimus S. Lee Saponin and Xylitolon Insoluble Exopolysaccharides Produced by Streptococcus Mutans. Guangxi Med. J. 2010, 32, 265–266. [Google Scholar]
- Kim, J.; Nguyen, T.T.H.; Jin, J.; Septiana, I.; Son, G.M.; Lee, G.H.; Jung, Y.J.; Qureshi, D.; Mok, I.K.; Pal, K. Anti-cariogenic Characteristics of Rubusoside. Biotechnol. Bioprocess Eng. 2019, 24, 282–287. [Google Scholar] [CrossRef]
- George Thompson, A.M.; Iancu, C.V.; Nguyen, T.T.H.; Kim, D.; Choe, J.Y. Inhibition of human GLUT1 and GLUT5 by plant carbohydrate products; insights into transport specificity. Sci. Rep. 2015, 5, 12804. [Google Scholar] [CrossRef]
- Gougeon, L.; da Costa, G.; Guyon, F.; Richard, T. 1H-NMR metabolomics applied to Bordeaux red wines. Food Chem. 2019, 301, 125257. [Google Scholar] [CrossRef]
- Liu, R.; Xu, H.; Zhang, X.; Wang, X.; Yuan, Z.; Sui, Z.; Wang, D.; Bi, K.; Li, Q. Metabolomics Strategy Using High Resolution Mass Spectrometry Reveals Novel Biomarkers and Pain-Relief Effect of Traditional Chinese Medicine Prescription Wu-Zhu-Yu Decoction Acting on Headache Modelling Rats. Molecules 2017, 22, 2110. [Google Scholar] [CrossRef]
- Wang, H.; Liu, A.; Zhao, W.; Zhao, H.; Gong, L.; Chen, E.; Cui, N.; Ji, X.; Wang, S.; Jiang, H. Metabolomics Research Reveals the Mechanism of Action of Astragalus Polysaccharide in Rats with Digestive System Disorders. Molecules 2018, 23, 3333. [Google Scholar] [CrossRef]
- Guo, F.; Zi, T.; Liu, L.; Feng, R.; Sun, C. A 1H-NMR based metabolomics study of the intervention effect of mangiferin on hyperlipidemia hamsters induced by a high-fat diet. Food Funct. 2017, 8, 2455–2464. [Google Scholar] [CrossRef]
- Fan, L.L.; Wei, W.; He, L.L.; Cao, F.; Miao, J.H. Quantification of Rubusoside in Preparations of Rubus suavissimus by RSLC-DAD. Lishizhen Med. Mater. Med. Res. 2012, 23, 129–131. [Google Scholar]
- Fan, L.L.; Qu, X.S.; Yi, T.; Peng, Y.; Jiang, M.J.; Miao, J.H.; Xiao, P.G. Metabolomics of the protective effect of Ampelopsis grossedentata and its major active compound dihydromyricetin on the liver of high-fat diet hamster. Evid. Based Complement. Altern. Med. 2020, 2020, 3472578. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Ji, D.; Li, L.; Su, L.; Gu, W.; Gu, L.; Wang, Q.; Lu, T.; Mao, C. Mechanism of Curcuma wenyujin Rhizoma on Acute Blood Stasis in Rats Based on a UPLC-Q/TOF-MS Metabolomics and Network Approach. Molecules 2019, 24, 82. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.H.; Wang, C.G.; Luo, W.Z.; Wang, J.; Li, B.; Lv, J.P.; Dong, R.J.; Ge, D.Y.; Han, Y.; Yang, Y.J. Metabolomic Study to Determine the Mechanism Underlying the Effects of Sagittaria sagittifolia Polysaccharide on Isoniazid- and Rifampicin-Induced Hepatotoxicity in Mice. Molecules 2018, 23, 3087. [Google Scholar] [CrossRef]
- Rothhammer, V.; Mascanfroni, I.D.; Bunse, L.; Takenaka, M.C.; Kenison, J.E.; Mayo, L.; Chao, C.C.; Patel, B.; Yan, R.; Blain, M. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016, 22, 586. [Google Scholar] [CrossRef]
- Akhtar, M.T.; Mushtaq, M.Y.; Verpoorte, R.; Richardson, M.K.; Choi, Y.H. Metabolic effects of cannabinoids in zebrafish (Danio rerio) embryos determined by 1H-NMR metabolomics. Metabolomics 2016, 12, 44. [Google Scholar] [CrossRef]
- Schimke, R.T. Studies on factors affecting the levels of urea cycle enzymes in rat liver. J. Biol. Chem. 1963, 238, 1012–1018. [Google Scholar]
- Whitington, P.F.; Alonso, E.M.; Boyle, J.T.; Molleston, J.P.; Rosenthal, P.; Emond, J.C.; Millis, J.M. Liver transplantation for the treatment of urea cycle disorders. J. Inherit. Metab. Dis. 1998, 21, 112–118. [Google Scholar] [CrossRef]
- Hao, J.J.; Hu, H.W.; Liu, J.; Wang, X.; Liu, X.; Wang, J.B.; Niu, M.; Zhao, Y.L.; Xiao, X.L. Integrated Metabolomics and Network Pharmacology Study on Immunoregulation Mechanisms of Panax ginseng through Macrophages. Evid. Based Complement. Altern. Med. 2019, 2019, 3630260. [Google Scholar] [CrossRef]
- Randle, P.J. Carbohydrate metabolism and lipid storage and breakdown in diabetes. Diabetologia 1966, 2, 237–247. [Google Scholar] [CrossRef]
- Gallego-Ortega, D.; de Molina, A.R.; Ramos, M.A.; Valdes-Mora, F.; Barderas, M.G.; Sarmentero-Estrada, J.; Lacal, J.C. Differential role of human choline kinase α and β enzymes in lipid metabolism: Implications in cancer onset and treatment. PLos ONE 2009, 4, e7819. [Google Scholar] [CrossRef]
- Almanza-Aguilera, E.; Urpi-Sarda, M.; Llorach, R.; Vázquez-Fresno, R.; Garcia-Aloy, M.; Carmona, F.; Sanchez, A.; Madrid-Gambin, F.; Estruch, R.; Corella, D. Microbial metabolites are associated with a high adherence to a Mediterranean dietary pattern using a 1H–NMR-based untargeted metabolomics approach. J. Nutr. Biochem. 2017, 48, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.L.; Li, J.C.; Tang, R.Y.; Zhang, G.D.; Zeng, H.W.; Wood, R.J.; Liu, Z.H. High Fat Diet Alters Gut Microbiota and the Expression of Paneth Cell-Antimicrobial Peptides Preceding Changes of Circulating Inflammatory Cytokines. Mediat. Inflamm. 2017, 2017, 9474896. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Xie, M.; Wan, P.; Chen, D.; Dai, Z.; Ye, H.; Hu, B.; Zeng, X.; Liu, Z.J.J.O.A.; Chemistry, F. Fuzhuan brick tea polysaccharides attenuate metabolic syndrome in high-fat diet induced mice in association with modulation in the gut microbiota. J. Agric. Food Chem. 2018, 66, 2783–2795. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.J.; Li, S.Q.; Zhang, Y.Y.; Wang, S.L.; Hou, S.L. Expression changes of lipid droplets during alcohol induced liver injury in mice. World Chin. J. Dig. 2016, 24, 3683–3688. [Google Scholar] [CrossRef]
- Tang, C.; Zhang, K.; Liang, X.; Zhao, Q.; Zhang, J. Application of a NMR-based untargeted quantitative metabonomic approach to screen for illicit salbutamol administration in cattle. Anal. Bioanal. Chem. 2016, 408, 4777–4783. [Google Scholar] [CrossRef] [PubMed]
- Yue, D.; Zhang, Y.; Cheng, L.; Ma, J.; Xi, Y.; Yang, L.; Su, C.; Shao, B.; Huang, A.; Xiang, R. Hepatitis B virus X protein (HBx)-induced abnormalities of nucleic acid metabolism revealed by 1H-NMR-based metabonomics. Sci. Rep. 2016, 6, 24430. [Google Scholar]
- Kim, M.J.; Lee, M.Y.; Shon, J.C.; Kwon, Y.S.; Liu, K.H.; Lee, C.H.; Ku, K.M. Untargeted and targeted metabolomics analyses of blackberries—Understanding postharvest red drupelet disorder. Food Chem. 2019, 300, 125169. [Google Scholar] [CrossRef]
- Chong, J.; Yamamoto, M.; Xia, J. MetaboAnalystR 2.0: From Raw Spectra to Biological Insights. Metabolites 2019, 9, 57. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds used in the study are available from the authors. |

| Label | HFD/ND | HFD+PRbs/HFD | HFD+Rbs/HFD |
|---|---|---|---|
| 1,3-Dihydroxyacetone | 32.06 ** | 0.06 ** | 0.03 ** |
| 2-Aminobutyrate | 2.09 * | 0.57 ** | 0.63 |
| 2-Hydroxybutyrate | 4.26 ** | 0.60 * | 0.80 |
| 2-Oxoglutarate | 3.68 ** | 0.39 ** | 0.38 * |
| 2-Oxoisocaproate | 2.39 ** | 0.76 * | 0.91 |
| 3-Hydroxybutyrate | 4.69 ** | 0.73 * | 1.09 |
| 3-Hydroxyisobutyrate | 0.93 | 0.71 * | 1.03 |
| 4-Hydroxyphenylacetate | ≈0 a | >0 a | >0 a |
| Acetate | 1.53 * | 0.50 ** | 0.56 * |
| Acetoacetate | 0.91 | 4.21 * | 4.94 |
| Acetone | 2.21 ** | 1.44 | 1.46 |
| Alanine | 1.34 * | 0.59 ** | 0.48 ** |
| Arginine | 1.13 | 0.74 | 0.53 |
| Aspartate | 1.42 | 1.09 | 1.31 |
| Betaine | 0.50 ** | 1.75 | 1.45 |
| Carnitine | 1.68 | 0.80 | 0.47 * |
| Choline | 4.10 ** | 0.45 ** | 0.34 ** |
| Citrate | 0.48 ** | 0.92 | 0.99 |
| Creatine | 1.97 ** | 1.18 | 1.55 |
| Dimethyl sulfone | 0.54 ** | 1.73 ** | 1.23 |
| Dimethylamine | 2.10 ** | 1.10 | 2.49 |
| Ethanol | 0.04 | 27.51 ** | 35.81 |
| Ethylmalonate | 1.79 * | 0.67 ** | 0.95 |
| Formate | 0.16 ** | 0.89 | 0.81 |
| Fumarate | 1.46 | 0.58 ** | 0.48 |
| Glucose | 0.46 ** | 0.98 | 0.75 |
| Glutamate | 1.86 * | 0.68 * | 0.83 |
| Glutamine | 1.39 * | 0.69 ** | 0.71 |
| Glycine | 2.29 ** | 0.71 ** | 0.63 |
| Histidine | 1.84 * | 0.69 * | 0.79 |
| Isoleucine | 1.39 | 0.72 * | 0.79 |
| Lactate | 1.29 * | 0.74 | 0.74 * |
| Leucine | 2.17* | 0.53 * | 0.63 * |
| Lysine | 1.00 | 0.76 | 0.89 |
| Malate | 2.35 ** | 0.42 ** | 0.23 ** |
| Mannose | 2.15 ** | 0.77 | 0.61 |
| Methanol | 1.38 | 0.86 | 1.23 |
| Methionine | 1.18 | 0.79 * | 0.82 |
| Methylsuccinate | 2.91 ** | 0.67 * | 0.68 |
| Nicotinurate | 3.09 | 0.13 * | 0.00 * |
| O-Phosphocholine | 1.88 | 0.73 | 0.67 |
| Phenylalanine | 1.90 ** | 0.75 * | 0.80 |
| Proline | 1.00 | 0.46 ** | 0.81 |
| Propionate | 0.57 | 1.11 | 2.56 |
| Pyroglutamate | 3.42 ** | 0.43 ** | 0.59 * |
| Pyruvate | 1.95 * | 0.40 ** | 0.29 ** |
| Serine | 1.27 * | 0.63 ** | 0.39 * |
| Succinate | 1.26 | 1.25 | 0.68 |
| Threonine | 1.40 | 0.48 ** | 0.45 * |
| Trimethylamine | 1.57 | 1.20 | 1.89 |
| Tryptophan | 1.50 ** | 0.57 ** | 0.56 ** |
| Tyrosine | 1.10 | 0.62 ** | 0.48 ** |
| Uracil | 10.44 | 0.59 | 0.24 |
| Urea | 0.17 * | 13.37 * | 14.49 ** |
| Uridine | 1.65 * | 0.71 | 0.23 ** |
| Valine | 1.18 | 0.78 ** | 0.91 |
| Xanthosine | 0.07 ** | 7.91 | 2.34 |
| myo-Inositol | 1.30 * | 1.54 | 2.05 |
| sn-Glycero-3-phosphocholine | 1.68 * | 0.92 | 0.72 |
| beta-Alanine | 1.87 | 1.33 | 0.45 |
| Pathway Name | Total Compounds | Hits | Raw p | -Log (p) | Holm Adjust | FDR | Impact |
|---|---|---|---|---|---|---|---|
| Alanine, aspartate and glutamate metabolism | 24 | 8 | 0.01 | 4.63 | 0.25 | 0.02 | 0.71 |
| Synthesis and degradation of ketone bodies | 6 | 3 | 0.00 | 7.48 | 0.03 | 0.00 | 0.70 |
| Glycine, serine and threonine metabolism | 48 | 9 | 0.00 | 10.29 | 0.00 | 0.00 | 0.44 |
| Arginine and proline metabolism | 77 | 9 | 0.00 | 7.91 | 0.02 | 0.00 | 0.33 |
| Aminoacyl-tRNA biosynthesis | 75 | 18 | 0.00 | 9.53 | 0.00 | 0.00 | 0.23 |
| Methane metabolism | 34 | 7 | 0.00 | 8.84 | 0.01 | 0.00 | 0.20 |
| Butanoate metabolism | 40 | 7 | 0.00 | 6.81 | 0.05 | 0.00 | 0.17 |
| Histidine metabolism | 44 | 4 | 0.00 | 5.82 | 0.11 | 0.01 | 0.14 |
| d-Glutamine and d-glutamate metabolism | 11 | 3 | 0.01 | 5.29 | 0.16 | 0.01 | 0.14 |
| Inositol phosphate metabolism | 39 | 1 | 0.00 | 5.69 | 0.12 | 0.01 | 0.14 |
| Phenylalanine metabolism | 45 | 6 | 0.04 | 3.21 | 0.84 | 0.06 | 0.12 |
| Tryptophan metabolism | 79 | 1 | 0.00 | 7.08 | 0.04 | 0.00 | 0.11 |
| Tyrosine metabolism | 76 | 6 | 0.01 | 5.27 | 0.16 | 0.01 | 0.11 |
| Lysine biosynthesis | 32 | 3 | 0.00 | 5.84 | 0.11 | 0.01 | 0.10 |
| Glycolysis or Gluconeogenesis | 31 | 5 | 0.00 | 6.61 | 0.06 | 0.00 | 0.10 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Jiang, M.; Li, Y.; Su, J.; Li, L.; Qu, X.; Fan, L. 1H-NMR Metabolomics Analysis of the Effect of Rubusoside on Serum Metabolites of Golden Hamsters on a High-Fat Diet. Molecules 2020, 25, 1274. https://doi.org/10.3390/molecules25061274
Li L, Jiang M, Li Y, Su J, Li L, Qu X, Fan L. 1H-NMR Metabolomics Analysis of the Effect of Rubusoside on Serum Metabolites of Golden Hamsters on a High-Fat Diet. Molecules. 2020; 25(6):1274. https://doi.org/10.3390/molecules25061274
Chicago/Turabian StyleLi, Li, Manjing Jiang, Yaohua Li, Jian Su, Li Li, Xiaosheng Qu, and Lanlan Fan. 2020. "1H-NMR Metabolomics Analysis of the Effect of Rubusoside on Serum Metabolites of Golden Hamsters on a High-Fat Diet" Molecules 25, no. 6: 1274. https://doi.org/10.3390/molecules25061274
APA StyleLi, L., Jiang, M., Li, Y., Su, J., Li, L., Qu, X., & Fan, L. (2020). 1H-NMR Metabolomics Analysis of the Effect of Rubusoside on Serum Metabolites of Golden Hamsters on a High-Fat Diet. Molecules, 25(6), 1274. https://doi.org/10.3390/molecules25061274





