Abstract
This study aimed to generate a comparative data on biological response of yttrium oxide nanoparticles (Y2O3 NPs) with the antioxidant CeO2 NPs and pro-oxidant ZnO NPs. Sizes of Y2O3 NPs were found to be in the range of 35±10 nm as measured by TEM and were larger from its hydrodynamic sizes in water (1004 ± 134 nm), PBS (3373 ± 249 nm), serum free culture media (1735 ± 305 nm) and complete culture media (542 ± 108 nm). Surface reactivity of Y2O3 NPs with bovine serum albumin (BSA) was found significantly higher than for CeO2 and ZnO NPs. The displacement studies clearly suggested that adsorption to either BSA, filtered serum or serum free media was quite stable, and was dependent on whichever component interacted first with the Y2O3 NPs. Enzyme mimetic activity, like that of CeO2 NPs, was not detected for the NPs of Y2O3 or ZnO. Cell viability measured by MTT and neutral red uptake (NRU) assays suggested Y2O3 NPs were not toxic in human breast carcinoma MCF-7 and fibroblast HT-1080 cells up to the concentration of 200 μg/mL for a 24 h treatment period. Oxidative stress markers suggested Y2O3 NPs to be tolerably non-oxidative and biocompatible. Moreover, mitochondrial potential determined by JC-1 as well as lysosomal activity determined by lysotracker (LTR) remained un-affected and intact due to Y2O3 and CeO2 NPs whereas, as expected, were significantly induced by ZnO NPs. Hoechst-PI dual staining clearly suggested apoptotic potential of only ZnO NPs. With high surface reactivity and biocompatibility, NPs of Y2O3 could be a promising agent in the field of nanomedicine.
1. Introduction
Screening of nanoparticles (NPs; defined as particles having, at least, one dimension below 100 nm) to be exploited in nanomedicine has been unprecedented in recent years since they provide multiple avenues in diagnosis and therapy of diseases. NPs could be engineered to target cancerous cells and release anticancer drugs therein, thus, avoiding normal cell toxicity that is a bigger obstacle in conventional chemotherapy [1,2]. NPs originating from lanthanide group exhibit remarkable properties owing to their high optical activity, surface reactivity and superior photo stability [3,4,5]. High refractory properties, good thermal conductivity, superior stability and excellent mechanical property make yttrium oxide (Y2O3) NPs a fascinating material [6]. Engineering NPs for active targeting to cancer cells while sparing normal cells, however, requires NPs with certain inherent properties. Obviously, biocompatibility and surface reactivity are some of the important criteria. Due to their intrinsic physical properties, inorganic NPs, especially of lanthanide group, offer highly tunable surface properties and stability over polymeric NPs [7,8]. NPs of antioxidant CeO2 and quite a different pro-oxidant (toxic) NPs of ZnO, for example, have received much attention for their potential usefulness in medicine though both NPs exhibit different biological effects [9]. Some investigators have reported that chemically related NPs of CeO2 and Y2O3 possess redox potential that enables them to mimic the activity of antioxidant enzymes such as superoxide dismutases (SODs) [10]. Moreover, as a novel treatment strategy, Y2O3 NPs could be used in the therapy of fulminant hepatic failure and oxidative stress-related diseases [11].
The present study was designed seeking a higher surface reactivity of NPs that could be exploited in nanomedicine. Since surface reactivity gives information about the extent to which NPs surface can be tuned, therefore, determining surface reactivity is considered a pre-requisite in the field of nanomedicine. Understanding the surface reactivity of protein molecules such as bovine serum albumin (BSA) is crucial towards predictability of drug interaction with a NP [12,13]. Quantification of BSA reactivity towards NPs have been one of the reliable methods in the evaluation of the degree of NPs activity towards biological models and molecules [14]. In this study, surface reactivity of BSA with Y2O3 NPs was studied and compared with the surface reactivity of CeO2 NPs and ZnO NPs. To the best of our knowledge, we could not find any report on BSA reactivity with that of Y2O3 NPs. There are, however, reports on BSA reactivity with the NPs of europium-doped Y2O3 (Y2O3:Eu3+) and ZnO NPs [14]. As will be discussed later, BSA affinity with Y2O3 NPs was found to be highest among all of the three NPs chosen in this study. Strong BSA reactivity suggested Y2O3 NPs could be a potential candidate in drug loading and delivery. In addition to BSA powder suspended in aqueous solution, surface reactivity of Y2O3 NPs towards culture media ingredient was also compared with that of CeO2 and ZnO NPs since surface modification can significantly reduce the toxicity of NPs [15].
Due to aforementioned properties, NPs of Y2O3, if found to be biocompatible, could serve the purpose of nano-carrier for various drugs. Biocompatibility of Y2O3 NP was examined in the two human cell lines- human breast (MCF-7) cells and fibroblast (HT-1080). These cells are a proven model for bio-medical testing [9,16,17]. This study, therefore, reports a comparative finding in terms of surface reactivity and biocompatibility of Y2O3 NPs with the chemically related NPs of CeO2 and chemically un-related NPs of ZnO. It is notable that NPs of Y2O3, like that of CeO2 NPs, have been reported to be antioxidant and protective in cells of neural [18,19] and retinal [20], whereas some investigators have reported this NP as oxidative and toxic [21,22]. In this study, we have provided a comparative data on surface reactivity with BSA and media components which is not known. Moreover, detailed mechanisms of potential toxicity carried out in the two human cell lines could further shed light on the discrepancy in toxicity level that exists for Y2O3 NP [18,19,20,21,22].
2. Results
2.1. Y2O3 NPs TEM Sizes, Hydrodynamic Sizes and Zeta Potential
NPs size calculated by TEM imaging at different resolution came to be 35 ± 10 nm while shapes appeared mostly cuboidal (Figure 1A,B). The matte texture observed in high-resolution (HR) TEM images (Figure 1C,D) confirms the crystallinity of Y2O3 NPs.
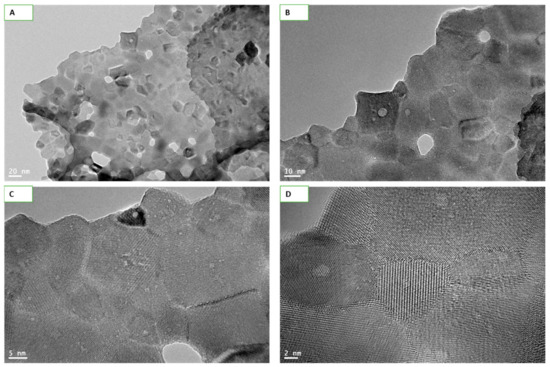
Figure 1.
Transmission electron microscopy (TEM) images of Y2O3 nanoparticles (NPs) captured at different resolutions. Images taken at 20 nm (A) and 10 nm (B) clearly depicts the sizes and shapes of NPs. Size of NPs was calculated to be 35 ± 10 nm while shapes were mostly cuboidal. Matte texture, an evidence of crystallinity of the materials, was observed at the high resolutions of 5 nm (C) and 2 nm (D).
Sizes and zeta potential of NPs in distilled water, phosphate buffer saline, serum free culture medium and complete culture media were measured to see the potential differences caused by serum and media components. Dynamic light scattering (DLS) measurements taken at 200 μg/mL show a differential dispersion of Y2O3 NPs in distilled water, phosphate buffer saline, serum free culture media and complete culture media as given in Table 1.

Table 1.
Y2O3 NP’s properties in powder form and its behavior in relevant aqueous media.
2.2. Y2O3 NPs Strongly React with BSA, Media and Non-Protein Serum Components
Surface reactivity of Y2O3 NPs with BSA was examined and compared with surface reactivity of two other NPs—CeO2 and ZnO—known to exert opposite responses in a wide variety of biological models. Among these NPs, Y2O3 NPs showed significantly higher BSA adsorption (Figure 2A). Control optical density of BSA (500 μg/mL in water) was taken as a reference point. BSA adsorption study with all three NPs was performed in water. BSA adsorbed, at 6 h of incubation, by NPs of Y2O3, CeO2, ZnO was found to be 330 ± 8, 145 ± 6 and 17 ± 4 μg, respectively. Similar is the trend at 24 h of incubation, though, with a slight decrease in BSA adsorption. To see the alteration in protein adsorption, if any, caused by proteins from other source (fetal bovine serum for example), or components of culture media, the Y2O3 NPs were suspended first in the liquid containing the respective adsorbent and incubated for 6 h or 24 h. After the respective period of incubation, NPs were centrifuged and in BSA containing water. Results clearly indicate that serum or media component caused significant shielding effect to BSA adsorption onto NPs surface (Figure 2B). Time dependent study shows that shielding effect due to serum or media component is quite stable. Data shows that Y2O3 NPs bind with serum proteins at rate of 30 ± 4 μg/mg NP when incubated for 6 h. BSA pre-treated NPs, however, did not show any noticeable amount of serum adsorption. Moreover, reactivity towards either BSA or media component is stable; neither BSA adsorption occurred to media treated Y2O3 NPs nor serum free media caused BSA displacement from Y2O3 NPs pre-treated with BSA (see summary in Table 2).
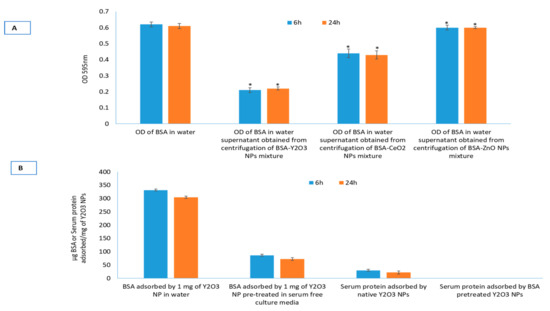
Figure 2.
Surface reactivity of the three NPs with relevant biomolecules. Adsorption of bovine serum albumin (BSA) on to Y2O3, CeO2 and ZnO NPs (A). Adsorption to BSA was highest for Y2O3 NPs while least for ZnO NPs. Y2O3 NPs resulted in highest surface reactivity, therefore, displacement experiment was further extended to Y2O3 NPs to understand the relative stability of adsorption of BSA, serum albumin and media ingredients with respect to each other (B). Triplicates of each treatment group were used in each independent experiment. * Denotes a significant difference from the control (p < 0.05).

Table 2.
Comparison of serum adsorption to NPs in media and serum with that of BSA adsorption to pristine and media treated NPs.
2.3. Y2O3 NPs Do Not Possess Inherent SOD-Like or CAT-Like Activity
Redox active NPs such as CeO2 are known to possess reactive oxygen species (ROS) scavenging capability. This ROS scavenging capacity plays critical role in the antioxidant activity reported of these NPs, though other factors might also exist [23,24]. In this study, SOD-like and CAT-like activity of Y2O3 NPs was measured along with NPs of CeO2 and ZnO. Table 3 shows that no significant enzyme-like activity of Y2O3 NPs was detected except for the NPs of CeO2.

Table 3.
Reactive oxygen species (ROS) scavenging potential of NPs in respective buffer.
2.4. Y2O3 Nps Did Not Cause Significant Decrease in Cell Viability
There was no difference found in the cell viability data due to BSA treated or non-treated Y2O3 NPs in MCF-7 and HT-1080 cells (see Supplementary Figure S1). The rest of the biological parameters were, therefore, conducted using naïve NPs of Y2O3, CeO2 and ZnO. As could be observed in Figure 3A–C, NPs of Y2O3 (and chemically similar CeO2 NPs at 100 μg/mL) did not cause inhibition to cell viability whereas ZnO caused 50% inhibition to cell viability (defined as IC50) at 41 ± 4 μg/mL to MCF-7 cells and 33 ± 4 μg/mL to HT-1080 cells.
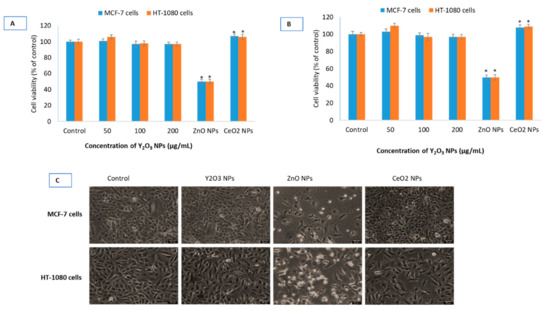
Figure 3.
Cell viability in human breast (MCF-7) cells and human fibroblast (HT-1080) cells treated with the three NPs of Y2O3, CeO2 and ZnO determined by MTT (A) and neutral red uptake (NRU) (B) assay at indicated concentrations. It should be noted that a single concentration of CeO2 NPs (100 μg/mL) and ZnO NPs (IC50) have been used in all of the biological experiments since these two NPs have been taken as primarily for comparison purposes with that of Y2O3 NPs. Phase-contrast images (20×, Leica DMi8, Germany) of MCF-7 cells and HT-1080 cells treated with highest concentration (i.e., 200 μg/mL) of Y2O3 NPs, IC50 of ZnO and CeO2 (100 μg/mL) have been given (C). For IC50 calculation, a scatter plot in Microsoft Excel was inserted followed by setting the Y-axis to logarithmic. Then a trend line was selected and ‘exponential’ picked. Then ‘display equation’ was used in calculating ICs. IC50s calculations were further verified and confirmed from the online IC50 calculator (https://www.aatbio.com/tools/ic50-calculator) provided by AAT Bioquest, Inc. (CA 94085, USA). Scale-bar represents 40 μm (micron) in each image. Triplicates (n = 3) of each treatment group were used in each independent experiment. * Denotes a significant difference from the control (p < 0.05).
2.5. Y2O3 NPs Did Not Induce Oxidative Stress
Potential oxidative damage induced by NPs was assessed by cellular integrity directly affected by ROS. Lactate dehydrogenase (LDH) release, a marker of cell membrane damage, was measured in cells at concentrations of 50, 100 and 200 μg/mL exposed for 24 h (Figure 4A). Y2O3 NPs did not cause any LDH release in MCF-7 and HT-1080 cells at concentrations of 50, 100 and 200 μg/mL. Another marker of membrane damage, lipid peroxidation (LPO), followed similar trend in both cells (Figure 4B). It should be noted that statistically significance membrane damage caused by NPs of ZnO at IC50. As expected, antioxidant CeO2 NPs did not elevate markers of membrane damage. Similarly, Y2O3 NPs did not induce ROS that was measured by DCF probe and H2O2 specific sensor (Figure 4C,D). A standard oxidant, H2O2, was always included as a positive control of ROS during 2′, 7′-dichlorofluorescin diacetate (DCFH-DA) measurement (see Figure S2).
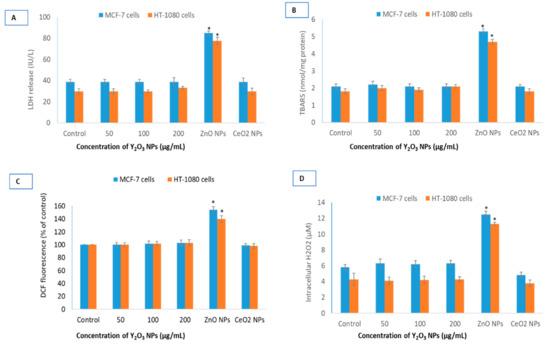
Figure 4.
Comparative oxidative stress parameters for the three NPs. Potential lactate dehydrogenase (LDH) release (A), lipid peroxidation (LPO) induction (B), DCF (fluorescent part of the non-fluorescent DCFH-DA molecule that is formed inside cells after ROS induced cleavage) fluorescence (C) and intracellular H2O2 induction (D) in MCF-7 and HT-1080 cells by NPs of Y2O3, ZnO and CeO2. Triplicates (n = 3) of each treatment group were used in each independent experiment. * Denotes a significant difference from the control (p < 0.05).
2.6. Y2O3 NPs Did Not Cause Mitochondrial Membrane Potential Induction
To test further biocompatibility of Y2O3 NPs, mitochondrial membrane potential (MMP) was determined qualitatively and quantitatively using combined advantage of a fluorescence microscope (Leica DMi8, Wetzlar, Germany) and ImageJ software. Qualitative and quantitative results clearly suggest that Y2O3 NPs exposure at 200 μg/mL, highest concentration of NPs taken in this study, to both cells did not exert negative impact (Figure 5A,B). Punctate red fluorescence of JC-1 aggregate coming from intact mitochondria is a qualitative evidence of non-toxicity of Y2O3 NPs. In essence, there was no appreciable change in MMP after 24 h of incubation with any concentration of Y2O3 NPs relative to control cells.
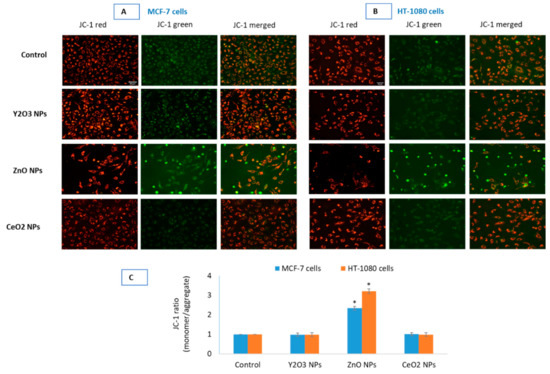
Figure 5.
Comparative mitochondrial outer membrane potential (MOMP or simply MMP) was detected by JC-1 in control and in the cells treated with the NPs of Y2O3 (200 μg/mL), ZnO (IC50) and CeO2 (100 μg/mL) in MCF-7 (A) and HT-1080 (B). Images were captured in tandem for JC-1 aggregate (red) and JC-1 monomer (green). Quantification of MOMP (C) in the two cells is given as JC-1 ratio (monomer/aggregate) of corrected total cell fluorescence (CTCF) analyzed in open source ImageJ software from NIH. Fluorescence intensities and cell morphology suggest no significant induction of MOMP in cells treated with 200 μg/mL of Y2O3 NPs and CeO2 NPs with that of control cells. NPs of ZnO, as expected, induced significant MOMP in the two cells. Images (captured by Leica DFC450C camera fitted in Leica DMi8 microscope, Germany, using 20× objectives) are representative of the three independent experiments with similar results. Scale-bar represents 40 μm (micron) and given in the first image of each parameter. Data represented are mean ± SD of three identical experiments (n = 3). * statistically significant difference as compared to the controls (p < 0.05).
2.7. Y2O3 NPs Did Not Cause Appreciable Change in Autophagy Activity
Finally, a comparative potential of autophagy modulation due to the three NPs was evaluated to understand the mechanism of biocompatibility in greater depth because many biocompatible NPs established by cytotoxicity and oxidative stress later turned to be inducer of autophagy. Above mitochondrial data provide sufficient set of evidence that suggests Y2O3 NPs are biocompatible in MCF-7 and HT-1080 cells. Similar to the role of mitochondria in cell survival and death, lysosomes are thought to play critical part in determining the fate of the cells. The two related NPs-Y2O3 and CeO2, did not induce changes in lysosomal activity in MCF-7 (Figure 6A) and HT-1080 (Figure 6B) cells. LTR fluorescence, however, suggested a definite increase in lysosomal or acidic autolysosomes that occurred in ZnO NPs treated cells. A higher LTR fluorescence, quantified for both cells in Figure 6C, is equivalent to extensive acidic vesicle formation within cells.
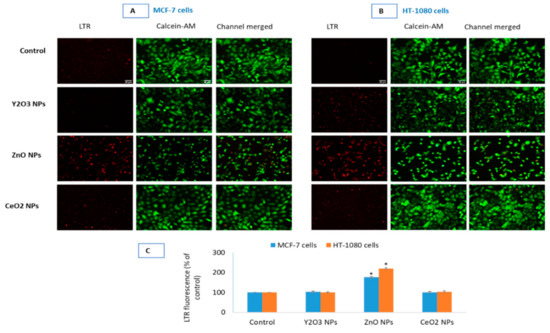
Figure 6.
Lysosomal activity due to the NPs of Y2O3, ZnO and CeO2 was evaluated in MCF-7 cells (A) and HT-1080 cells (B). CalceinAM was used as control of live cell fluorescence. Quantification of LTR fluorescence (C) in the two cells is given as corrected total cell fluorescence (CTCF) of control cells vs. treated. CTCF was analyzed in open source ImageJ software from NIH. Triplicates (n = 3) of each treatment group were used in each independent experiment. * Denotes a significant difference from the control (p < 0.05).
2.8. Y2O3 NPs Did Not Exhibit Apoptotic-Necrotic Potential
Hoechst and PI dual staining clearly shows NPs of Y2O3 and CeO2 to be non-cytotoxic (Figure 7). However, ZnO NP treated cell depicts IC50 toxicity. Moreover, an apoptotic mode of cells death in the case of ZnO NPs is evident by the fragmented and clumped pattern of chromatin condensation (see Figure S3).
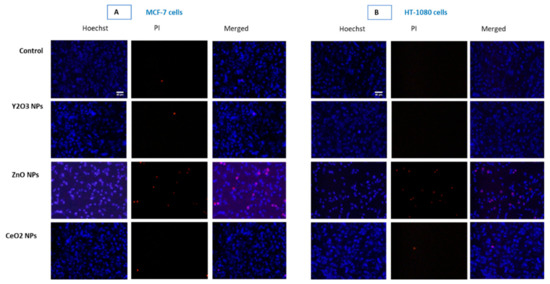
Figure 7.
Propidium iodide (PI) staining clearly shows NPs of Y2O3 and CeO2 to be non-cytotoxic in MCF-7 cells (A) and HT-1080 cells (B). However, ZnO NP treated cell depicts IC50 toxicity. Moreover, an apoptotic mode of cells death in the case of ZnO NPs is evident by the fragmented and clumped pattern of chromatin condensation (see Figure S3). Triplicates (n = 3) of each treatment group were used in each independent experiment.
3. Discussion
A variety of potential interacting component in relevant aqueous fluids may affect NPs native property prior to biological interaction [25,26]. Zeta potential and hydrodynamic sizes of NPs play crucial role in their secondary surface formation or coronization [27,28]. In this study, as determined by DLS, zeta potential of Y2O3 NPs suspension in complete culture media (−27.0 ± 1.2 mV) was much better than that in water (−16.0 ± 4.2 mV), PBS (−6.0 ± 2.4mV) and serum free culture media (−10.0 ± 4.0 mV). Interestingly, in a previous study, zeta potential of all aqueous (in deionized water) Y2O3 NPs preparations were highly negative with spherical Y2O3 NPs having an average zeta potential of −13 mV, platelets shaped NPs having an average zeta potential of −27 mV and the rod shaped NPs having an average zeta potential of −28 mV [29]. The respective mean hydrodynamic sizes for the spherical NPs (TEM size; 3 nm) was 700 nm, the platelet-shaped NPs (TEM size; 4 nm) was 700 nm and the rod-shaped NPs (TEM size; 11 nm) was 800 nm [29]. It should be recalled that in the present study the shape of the Y2O3 NP (TEM size; 35 ± 10 nm) was cubic in shape. In the present study, hydrodynamic size of Y2O3 NPs in water came out to be 1004 ± 134 nm while in complete culture media it was 542 ± 108 nm. The more negative zeta potential and lesser agglomeration observed in complete culture media for Y2O3 NPs is partially attributable to high surface reactivity of Y2O3 NPs with protein molecules as is evident by next experiment on Y2O3 NPs interaction with BSA proteins. In addition, BSA-treated Y2O3 NPs exhibited better dispersion in all of the tested aqueous media than native NPs (see Table 1).
Protein adsorption to NPs is a dynamic process in fluids be it culture media, blood or other bio fluids independent of nature of the NPs either polymeric [28] or metallic [30]. One of the most notable alterations when NP is under biological fluids is the formation of NP-protein corona [28]. In addition to protein coating, there are reports about non-protein coating on Y2O3 NPs to achieve desirable outcome. Y2O3 NPs coating with glycerol citrate polymer, for example, caused increased antibacterial activity [31]. Realizing the importance of surface reactivity, in this study protein adsorption on to NPs in its native form was quantified using bovine serum albumin (BSA) in a time-dependent manner (see Figure 2). From the results, it can be safely concluded that the cellular response of Y2O3 NPs is actually the response of complete-media-component (that include serum proteins as well media ingredients) coated-Y2O3 NPs rather than only serum protein coated NPs. Thus, living systems mostly interact with the surface modified NPs rather than with bare NPs only when the NPs are surface reactive [32,33]. Bio-molecular corona formed on surface reactive NPs can provide stealth effect during NP uptake by immune cells [34]. Corona formation can also aid in biocompatibility [25]. Of the three NPs tested for surface reactivity, NPs of Y2O3 emerge with highest surface reactivity. The ROS scavenging capacity plays critical role in the antioxidant activity reported for CeO2 NPs [23,24]. Several reports have shown NPs of Y2O3 and CeO2 with similar characteristics to offer protection against induced toxicity by pro-oxidants [9,22]. Antioxidant enzyme-like activity exhibited by certain NPs such as that of CeO2 has been implicated in its potential protective effect against induced toxicity. Only CeO2 NPs, however, were found to exhibit SOD-like and catalase-like activity in this report (Table 3).
Cell viability in cells treated with the NPs of Y2O3 (35 ± 10 nm) and CeO2 up to the concentration of 200 μg/mL was find equal to that of control in the MCF-7 and HT-1080 cells. Some investigators, however, have found NPs of Y2O3 (41 ± 5 nm) to inhibit 50% cell growth (i.e., IC50) at the concentration of 108 μg/mL in human embryonic kidney (HEK293) cells [35]. Other researchers have examined NPs of Y2O3 and yttrium ions simultaneously in yeast and have concluded the yttrium ions derived from NPs responsible for the toxicity observed in yeast cells [36]. In mouse normal hepatocyte cell line, Y2O3 NPs (7–8 nm) coated with polymer of acrylic acid have been reported to cause toxicity at the concentration of 10 μg/mL [21]. On the other hand, NPs of Y2O3 have been reported to be antioxidant and protective in cells of neural [18,19] and retinal [20] origin. In this study, no significant difference in cell viability was observed for BSA-coated and native NPs of Y2O3 (See Figure S1). ROS such as H2O2 and O2•−, can damage cellular components when produced in large amounts but can initiate a diverse array of signaling pathways leading to control cell division, differentiation and migration [37]. Depending on its intracellular concentration and localization, H2O2 initiates either pro-apoptotic or anti-apoptotic response [38]. Levels of H2O2 in the range of 20–50 μM seem to have limited cytotoxicity in many cell types [39]. NPs of Y2O3 and CeO2 did not cause significant ROS induction in the two cell types. NPs of ZnO, however, induced H2O2 levels over 2-folds in both cells. MMP is the result of ROS that may contribute in inducing death activating pathways [38]. Therefore, NPs are reported to be strong modulator of MMP that can bring the onset of apoptosis, necrosis and autophagy [40,41]. Data suggested that there was no appreciable change in MMP after 24 h of incubation with any concentration of Y2O3 NPs relative to control cells.
The tendency of autophagy modulation is considered as a survival strategy under short-term nutrient stress. However, if the depletion of antioxidants and nutrients is prolonged that might happen under toxicant exposure; the same autophagy may lead to course of cell death. Interestingly, NPs composed of rare earth oxide have been reported as a new class of autophagy inducers [42,43]. Certain NPs, as those of silica, have been widely used in nanomedicine and other applications due to their biocompatibility [44,45]. However, when examined on the parameter of autophagy, these NPs came to cause impairment in lysosomal activity and cellular protein clearance [46,47]. Given the importance of autophagy and lysosomal protein degradation in maintaining cellular homeostasis, therefore, in this report biocompatibility test was extended to analyzing the potential impact Y2O3 NPs might have on the activity of lysosomes. Recall that lysosomes are the major cellular organelle involved in the regulation of autophagy. In our study, the two related NPs-Y2O3 and CeO2, did not induce changes in lysosomal activity in both MCF-7 and HT-1080 cells. NPs of ZnO, used as toxic form of NPs, however, increased the number of cellular acidic vesicles and apoptotic mode of cell death in both cell types (see Figure S3 for the apoptotic mode of cell death induced by ZnO NPs).
4. Materials and Methods
4.1. Chemicals and Reagents
Fetal bovine serum, penicillin–streptomycin, Lysotracker Red-DND, CalceinAM were purchased from Thermo Fisher Scientific (Eugene, OR, USA). DMEM F-12K, Neutral red dye, MTT [3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide], Cacodylic acid, Pyrogallol, Bradford reagent, DCFH-DA, JC-1, H2O2, TBA, NADPH, pyruvic acid, H2O2 measuring kit, Hoechst, PI, all NPs were obtained from Sigma–Aldrich, USA. Ultrapure deionized-water was prepared using a Milli-Q system (Millipore, Bedford, MA, USA). All other chemicals used were of reagent grade.
4.2. Yttrium Oxide NPs
NPs of Y2O3 were commercially obtained from Sigma-Aldrich, USA. As per the information provided by supplier, TEM size of Y2O3 was below 50 nm. Color of nanopowder was white.
4.3. Transmission Electron Microscopy of Y2O3 NPs
Size of Y2O3 NPs was determined by field emission transmission electron microscopy (FETEM) (JEM-2100F, JEOL, Inc., Tokyo, Japan) using an accelerated voltage of 200 kV [48]. Normal and high resolution (HR) TEM images was also taken to observe the crystallinity of NPs as it happens in HR-TEM texture of typical crystals. Suspension of ultra-sonicated Y2O3 NPs was placed onto a carbon-coated copper grid, air dried and observed with FETEM. Purity of Y2O3 NPs was determined by energy dispersive spectrum (EDS) analysis.
4.4. Agglomeration and Zeta-Potential of Y2O3 NPs
Agglomeration behavior and zeta potential of Y2O3 NPs in water, phosphate buffer saline (PBS) and complete cell culture medium was determined by dynamic light scattering (DLS) system (Nano-ZetaSizer-HT, Malvern Instruments, Malvern, UK) as described by Murdock et al. [49]. NPs were freshly suspended in respective aqueous medium (200 µg/mL) and ultrasonicated for 10 min (Ultrasonic Cleaner-8891, Cole-Parmer, 625 Bunker Court Vernon Hills, IL USA). DLS measurement was carried out in cuvettes supplied with DLS system. For DLS measurement, a phenol red-free medium was used.
4.5. Determination of Surface Reactivity of NPs
To evaluate surface reactivity, NPs were treated with relevant chemicals such as bovine serum albumin (BSA), serum and media components. To determine the degree of protein adsorption, NPs and BSA were mixed in a 15 mL sterile tube. Final concentrations of BSA and NP were kept at 0.5 mg/mL and 1 mg/mL, respectively in a total volume of 10 mL. Then, a mixture of BSA and NP was put on a gentle shaker at room temperature after ultra-sonication for 20 min. From this NP-BSA mixture, 2 mL was taken in tubes and centrifuged for 10 min at 12,000× g at every 6 h and 24 h to measure protein adsorption. From the supernatant obtained, 100 µL each was taken from either NP-BSA mixture or BSA only solution, and added in cuvette containing 2.0 mL of Bradford reagent. A decrease in absorbance at 595 nm in NP-BSA mixture from BSA only solution was indicative of protein adsorption on NPs. Amount of BSA adsorbed on to NP surface was calculated using BSA standards at 595 nm and expressed as μg BSA adsorbed/mg of NP. Calculation method is similar to that of Patil et al. [50]. Protein adsorption to NPs is a dynamic process in liquids be it culture media, blood or other bio fluids. To see the alteration in protein adsorption, if any, caused by components of culture media, mixture of Y2O3 NPs was suspended in serum free (i.e., protein free) culture medium and left for 24 h. Then, BSA was added as above and any shielding effect caused by media ingredients in protein adsorption was measured at 6 h and 24 h as described above. Surface reactivity of Y2O3 NPs was also determined towards serum, a fluid containing diverse molecules such as albumin and other proteins, hormones, lipids, carbohydrates, etc.
4.6. Enzyme-Like Activity of NPs
Enzyme like activity of NPs was determined by the protocol of Marklund and Marklund [51] using autoxidation of pyrogallol. The principle of this method is based on the competition between the pyrogallol autoxidation by O2•− and the dismutation of this radical by enzyme SOD in a 75 mM Tris/cacodylic acid buffer, pH 8.2. For uninhibited reaction, a 0.02 OD/min for 3 min at 420 nm was utilized. Amount of enzyme required to inhibit this reaction by 50% was defined as one-unit of SOD enzyme. Catalase was determined by recording the rate of H2O2 disappearance at 240 nm for 3 min as described by Aebi [52]. For this, a 100 mL phosphate buffer, pH 7.0 was mixed with H2O2 to give an absorbance of 0.54 at 240 nm. For calculation purposes, 0.436 mM extinction co-efficient for H2O2 at 240 nm was taken. Reaction was initiated by 50 μL of 1 mg/mL NPs suspension in one mL of phosphate buffer with H2O2 absorbance of 0.54 at 240 nm. A suitable NP control was used for subtracting absorbance decrease due to NPs settling in reaction cuvette from the absorbance caused by potential enzyme-like activity of NPs.
4.7. Cell Culture
Human breast cancer (MCF-7) cells and fibrosarcoma (HT-1080) cells (ATCC, Manassas, VA, USA) were maintained in DMEM-F12K supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 µg/mL streptomycin, at 37 °C in a humidified 5% CO2 incubator (HERACell 150i, Thermo Fisher Scientific, Waltham, MA, USA). The cells were passaged for every 3–4 days before reaching confluence level.
4.8. Viability Studies
Cell viability was carried out by MTT, NRU and LDH assay as briefly described below. MTT assay was carried out according to the protocol described by Mosmann [53] with minor modifications. Briefly, around 20,000 MCF-7 cells per well were seeded in 96-well plates in a 100 µL of culture medium. Blue formazon formed in viable cells were solubilized and absorbance taken at 570 nm using a plate reader (Synergy HT, Bio-Tek, Winooski, VT, USA). Cell viability of treated and control cells is given as cell viability in percentage of control. To minimize the interference in absorption that may be potentially caused by the NPs, plates were centrifuged to suspend the NPs, and 100 µL supernatant was carefully transferred (without disturbing NPs at the plate bottom in the original experimental plate) to another fresh well in a 96-well plate as previously reported [54]. The neutral red uptake (NRU) assay is based on the method described by Repetto et al. [55]. Absorbance was taken at 540 nm using a plate reader (Synergy HT, Bio-Tek, Winooski, Vermont, US ) and data is given as cell viability in percentage of control. The activity of cytoplasmic LDH released into the culture media was determined with the method described [56]. A 100 µL sample from the centrifuged culture media was collected after the cells were treated for 24 h. The rate of NADH oxidation was determined by following the decrease in absorbance at 340 nm for 3 min at 30-s interval at 25 °C using a spectrophotometer (Genesys 10 Bio, Thermo Fisher Scientific, Madison, WI, USA). The amount of LDH released is represented as LDH activity (IU/L) in culture media.
4.9. Determination of Lipid Peroxidation
LPO was assessed by the thiobarbituric acid reactive substances (TBARS) assay, which detects mainly malondialdehyde (MDA), an end product of the peroxidation of polyunsaturated fatty acids and related esters. MDA was measured by slight modification of the method of Ohkawa et al. [57]. The absorbance of the supernatants was read at 532 nm. Results were calculated as nmol TBARS/mg of cellular protein using 1.56 × 105 M−1cm−1 as molar extinction of MDA-TBA.
4.10. Measurement of Total Reactive Oxygen Species by DCFH-DA and H2O2 by Specific Sensor
The generation of intracellular ROS was measured using 2′, 7′-dichlorofluorescin diacetate (DCFH-DA) probe [58]. DCF fluorescent intensity was measured at emission from 528 nm band pass of plate reader (Synergy HT, Bio-Tek, Winooski, Vermont, USA). A specific H2O2 sensor was employed for the detection of intracellular H2O2 according manufacturer’s protocol (MAK164, Sigma-Aldrich, St. Louis, MO, USA). A standard of H2O2 was run and measured similarly using H2O2 sensor.
4.11. Ratio-Metric Measurement of Mitochondrial Membrane Potential by JC-1
Mitochondrial outer membrane potential (MOMP) in control and treated cells were determined by JC-1. In healthy cells with high mitochondrial membrane potential, JC-1 spontaneously forms complexes known as J-aggregates with intense red fluorescence. On the other hand, in apoptotic or unhealthy cells with low membrane potential, JC-1 remains in the monomeric form showing green fluorescence [59]. Cells were seeded at appropriate densities in 12-well plate. When treatment period was over, media was aspirated off from each well and labeled with 5 µM JC-1 in Hepes buffered HBSS for 20 min. Fluorescence pictures of JC-1 monomer and aggregate in cells were captured by a fluorescence microscope (DMi8, Leica Microsystems, Wetzlar, Germany, using objective 20×, Leica DFC450 C camera and its LAS software version 4.13).
4.12. Measurement of Lysosomal Activity by LTR
Lysosomal activity predominantly refers to intracellular acidic vesicles (lysosomes or fusion product of autophagosomes with lysosomes) during autophagy [60]. Lysosomal activity in cells can be tracked by LysoTracker (LTR) dyes. LTR probes freely permeate cell membranes and label live cells. One such LTR dyes is red fluorescent LysoTracker™ Red DND-99 (Thermo Fisher Scientific, Eugene, Oregon, US) as was used in our previous report [61]. After the specific treatment period, cells were labeled at the final concentration of 1 µM LTR within ongoing culture media. After 30 min of labeling in 12-well culture plates, cells were carefully washed with cold HBSS with three times to remove excess LTR. CalceinAM, a green fluorescence emitting live cell dye was labeled at 0.5 µM simultaneously in LTR imaging to corroborate live cells from dead ones. An increase in punctate red fluorescence of LTR in treated cells than that in control cells indicates an increase in lysosomes or intact acidic vesicles (autolysosomes) [62].
4.13. Apoptotic-Necrotic Potential Detection by Hoechst/PI Staining
A dual staining based on concurrent use of Hoechst and propidium iodide (PI) was carried out again to confirm the mechanism of toxicity if occurred by the NPs of Y2O3. This approach was based on the ability of Hoechst to label every cell including live as well as dead and ability of PI to penetrate only dead cells. Further, if there is toxicity, a fair idea about potential mechanism of cell death induced by either apoptosis or necrosis can be deduced by applying logical conventions set by many cytologists [63]. In apoptotic cells, most nuclei show clumped, compact and discrete fluorescence due the pattern of nuclear DNA breakdown whereas in necrotic cell, most nuclei show increased in perimeter, diffused and peripheral fluorescence. In control dead cells, nucleus is neither increased in perimeter (as in necrosis), nor show compactness and discrete fluorescence (as in apoptotic cells) but rather a smooth and centrally intense fluorescence [64]. This is an inexpensive and more dependable method which was successfully applied in our previous publication deciphering mode of cell death caused by apoptosis to that from apoptosis-independent [61].
4.14. Protein Estimation
The total protein concentration was measured by using a ready-to-use Bradford reagent with bovine serum albumin as protein standard [65].
4.15. Statistics
One way ANOVA (analysis of variance) followed by Dunnett’s multiple comparison tests were employed for statistical analysis of data. Statistical significance was attributed at p < 0.05. The experiment was repeated three times (n = 3) carried in triplicates. For a particular set of the experiment, a burst of images was captured at the constant exposure of time, gain, saturation and gamma. Images representing three independent experiments were used in this report. Calculation of corrected total cellular fluorescence (CTCF) was conducted in ImageJ software (NIH, Bethesda, MD, USA). Using the region of interest (ROI) manager command, the constant area selection was obtained to each image opened in ImageJ. CTCF was calculated by subtracting the mean of background (without cell) fluorescence from the mean of cellular fluorescence (i.e., mean integrated density). Scale bar in images was set using ImageJ after calibrating the scale in terms of pixels/micron according to the respective objectives used during imaging.
5. Conclusions
NPs of Y2O3 could be desired agent in drug loading and targeted delivery as shown by their high surface reactivity, biocompatibility and their inability to affect cellular autophagy functioning. Higher surface reactivity also provides opportunity to test drugs loading, and in the further fine tuning of the NPs surfaces with diverse array of available biocompatible surfactants [27].
Supplementary Materials
Figure S1: Native (or pristine) Y2O3 NPs were first treated with different relevant adsorbents such as BSA, filtered serum or media ingredients without added serum (i.e., incomplete culture media) to confirm their surface activity. Now differently surface modified NPs were adjusted at the concentration of 200 μg/mL in complete culture media and were exposed to MCF-7 cells and HT-1080 cells for 24 h. Cell viability was not significantly different with that of control or native (unmodified) NPs. Triplicates (n = 3) of each treatment group were used in each independent experiment. Figure S2: ROS was measured using DCFH-DA along with H2O2 that was used as positive control. H2O2 (0.65 mM) was exposed to MCF-7 cells and HT-1080 cells that was sufficient to induce cytotoxicity almost 50% in cell population. Figure S3: An apoptotic mode of cells death in the case of ZnO NPs is evident by the fragmented and clumped pattern of chromatin condensation. All yellow circles meant for zooming the part of images are equal in area.
Author Contributions
Conceptualization, M.J.A.; investigation and methodology, M.J.A., Z.M.A.; writing—original draft preparation, M.J.A., M.A., M.M.R., S.A.A.; writing—review and editing, M.A.; S.A.A. and M.M.R.; funding acquisition, M.J.A. All authors have read and agreed to the published version of the manuscript.
Funding
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group no. RG-1440-097.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sutradhar, K.B.; Amin, M.L. Nanotechnology in Cancer Drug Delivery and Selective Targeting. ISRN Nanotechnol. 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Ahamed, M.; Alhadlaq, H.A.; Alrokayan, S.A.; Kumar, S. Targeted anticancer therapy: Overexpressed receptors and nanotechnology. Clin. Chim. Acta 2014, 436, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Sotiriou, G.A.; Franco, D.; Poulikakos, D.; Ferrari, A. Optically stable biocompatible flame-made SiO2-coated Y2O3: Tb3+ nanophosphors for cell imaging. ACS Nano 2012, 6, 3888–3897. [Google Scholar] [CrossRef] [PubMed]
- Ansari, A.A.; Khan, A.; Labis, J.P.; Alam, M.; Aslam Manthrammel, M.; Ahamed, M.; Akhtar, M.J.; Aldalbahi, A.; Ghaithan, H. Mesoporous multi-silica layer-coated Y2O3:Eu core-shell nanoparticles: Synthesis, luminescent properties and cytotoxicity evaluation. Mater. Sci. Eng. C 2019, 96, 365–373. [Google Scholar] [CrossRef]
- Park, Y.I.; Kim, H.M.; Kim, J.H.; Moon, K.C.; Yoo, B.; Lee, K.T.; Lee, N.; Choi, Y.; Park, W.; Ling, D.; et al. Theranostic Probe Based on Lanthanide-Doped Nanoparticles for Simultaneous In Vivo Dual-Modal Imaging and Photodynamic Therapy. Adv. Mater. 2012, 24, 5755–5761. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Starostin, S.A.; Li, S.; Khan, S.A.; Hessel, V. Synthesis of yttrium oxide nanoparticles via a facile microplasma-assisted process. Chem. Eng. Sci. 2018, 178, 157–166. [Google Scholar] [CrossRef]
- Jiao, M.; Zhang, P.; Meng, J.; Li, Y.; Liu, C.; Luo, X.; Gao, M. Recent advancements in biocompatible inorganic nanoparticles towards biomedical applications. Biomater. Sci. 2018, 6, 726–745. [Google Scholar] [CrossRef]
- Giner-Casares, J.J.; Henriksen-Lacey, M.; Coronado-Puchau, M.; Liz-Marzán, L.M. Inorganic nanoparticles for biomedicine: Where materials scientists meet medical research. Mater. Today 2016, 19, 19–28. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Ahamed, M.; Alhadlaq, H.A.; Khan, M.A.M.; Alrokayan, S.A. Glutathione replenishing potential of CeO2 nanoparticles in human breast and fibrosarcoma cells. J. Colloid Interface Sci. 2015, 453, 21–27. [Google Scholar] [CrossRef]
- Pirmohamed, T.; Dowding, J.M.; Singh, S.; Wasserman, B.; Heckert, E.; Karakoti, A.S.; King, J.E.S.; Seal, S.; Self, W.T. Nanoceria exhibit redox state-dependent catalase mimetic activity. Chem. Commun. 2010, 46, 2736–2738. [Google Scholar] [CrossRef]
- Song, X.; Shang, P.; Sun, Z.; Lu, M.; You, G.; Yan, S.; Chen, G.; Zhou, H. Therapeutic effect of yttrium oxide nanoparticles for the treatment of fulminant hepatic failure. Nanomedicine 2019, 14, 2519–2533. [Google Scholar] [CrossRef] [PubMed]
- Kopp, M.; Kollenda, S.; Epple, M. Nanoparticle-Protein Interactions: Therapeutic Approaches and Supramolecular Chemistry. Acc. Chem. Res. 2017, 50, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Darabi Sahneh, F.; Scoglio, C.; Riviere, J. Dynamics of Nanoparticle-Protein Corona Complex Formation: Analytical Results from Population Balance Equations. PLoS ONE 2013, 8, e64690. [Google Scholar] [CrossRef]
- Oviedo, M.J.; Quester, K.; Hirata, G.A.; Vazquez-Duhalt, R. Determination of conjugated protein on nanoparticles by an adaptation of the Coomassie blue dye method. MethodsX 2019, 6, 2134–2140. [Google Scholar] [CrossRef]
- Ahamed, M.; Akhtar, M.J.; Khan, M.A.M.; Alaizeri, Z.M.; Alhadlaq, H.A. Evaluation of the Cytotoxicity and Oxidative Stress Response of CeO2-RGO Nanocomposites in Human Lung Epithelial A549 Cells. Nanomaterials 2019, 9, 1709. [Google Scholar] [CrossRef]
- Zhang, X.F.; Shen, W.; Gurunathan, S. Silver nanoparticle-mediated cellular responses in various cell lines: An in vitro model. Int. J. Mol. Sci. 2016, 17, 1603. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Alhadlaq, H.A.; Alshamsan, A.; Majeed Khan, M.A.; Ahamed, M. Aluminum doping tunes band gap energy level as well as oxidative stress-mediated cytotoxicity of ZnO nanoparticles in MCF-7 cells. Sci. Rep. 2015, 5, 1–16. [Google Scholar] [CrossRef]
- Ghaznavi, H.; Najafi, R.; Mehrzadi, S.; Hosseini, A.; Tekyemaroof, N.; Shakeri-Zadeh, A.; Rezayat, M.; Sharifi, A.M. Neuro-protective effects of cerium and yttrium oxide nanoparticles on high glucose-induced oxidative stress and apoptosis in undifferentiated PC12 cells. Neurol. Res. 2015, 37, 624–632. [Google Scholar] [CrossRef]
- Khaksar, M.R.; Rahimifard, M.; Baeeri, M.; Maqbool, F.; Navaei-Nigjeh, M.; Hassani, S.; Moeini-Nodeh, S.; Kebriaeezadeh, A.; Abdollahi, M. Protective effects of cerium oxide and yttrium oxide nanoparticles on reduction of oxidative stress induced by sub-acute exposure to diazinon in the rat pancreas. J. Trace Elem. Med. Biol. 2017, 41, 79–90. [Google Scholar] [CrossRef]
- Mitra, R.N.; Merwin, M.J.; Han, Z.; Conley, S.M.; Al-Ubaidi, M.R.; Naash, M.I. Yttrium oxide nanoparticles prevent photoreceptor death in a light-damage model of retinal degeneration. Free Radic. Biol. Med. 2014, 75, 140–148. [Google Scholar] [CrossRef]
- Sayour, H.; Kassem, S.; Canfarotta, F.; Czulak, J.; Mohamed, M.; Piletsky, S. Biocompatibility and biodistribution of surface-modified yttrium oxide nanoparticles for potential theranostic applications. Environ. Sci. Pollut. Res. 2019, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.J.; Ahamed, M.; Alhadlaq, H.A.; Alshamsan, A. Mechanism of ROS scavenging and antioxidant signalling by redox metallic and fullerene nanomaterials: Potential implications in ROS associated degenerative disorders. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Heckert, E.G.; Karakoti, A.S.; Seal, S.; Self, W.T. The role of cerium redox state in the SOD mimetic activity of nanoceria. Biomaterials 2008, 29, 2705–2709. [Google Scholar] [CrossRef] [PubMed]
- Kamada, K.; Soh, N. Enzyme-mimetic activity of ce-intercalated titanate nanosheets. J. Phys. Chem. B 2015, 119, 5309–5314. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zhang, X.; Qian, L.; Yao, N.; Pan, Y.; Zhang, L. Doxorubicin-Loaded Unimolecular Micelle-Stabilized Gold Nanoparticles as a Theranostic Nanoplatform for Tumor-Targeted Chemotherapy and Computed Tomography Imaging. Biomacromolecules 2017, 18, 3869–3880. [Google Scholar] [CrossRef]
- Sabuncu, A.C.; Grubbs, J.; Qian, S.; Abdel-Fattah, T.M.; Stacey, M.W.; Beskok, A. Probing nanoparticle interactions in cell culture media. Colloids Surf. B Biointerfaces 2012, 95, 96–102. [Google Scholar] [CrossRef]
- Bewersdorff, T.; Gruber, A.; Eravci, M.; Dumbani, M.; Klinger, D.; Haase, A. Amphiphilic nanogels: Influence of surface hydrophobicity on protein corona, biocompatibility and cellular uptake. Int. J. Nanomed. 2019, Volume 14, 7861–7878. [Google Scholar] [CrossRef]
- Obst, K.; Yealland, G.; Balzus, B.; Miceli, E.; Dimde, M.; Weise, C.; Eravci, M.; Bodmeier, R.; Haag, R.; Calderón, M.; et al. Protein Corona Formation on Colloidal Polymeric Nanoparticles and Polymeric Nanogels: Impact on Cellular Uptake, Toxicity, Immunogenicity, and Drug Release Properties. Biomacromolecules 2017, 18, 1762–1771. [Google Scholar] [CrossRef]
- Andelman, T.; Gordonov, S.; Busto, G.; Moghe, P.V.; Riman, R.E. Synthesis and cytotoxicity of Y2O3 Nanoparticles of various morphologies. Nanoscale Res. Lett. 2010, 5, 263–273. [Google Scholar] [CrossRef]
- Lira, A.L.; Ferreira, R.S.; Torquato, R.J.S.; Zhao, H.; Oliva, M.L.V.; Hassan, S.A.; Schuck, P.; Sousa, A.A. Binding kinetics of ultrasmall gold nanoparticles with proteins. Nanoscale 2018, 10, 3235–3244. [Google Scholar] [CrossRef]
- Mariano-Torres, J.A.; López-Marure, A.; García-Hernández, M.; Basurto-Islas, G.; Domínguez-Sánchez, M.Á. Synthesis and characterization of glycerol citrate polymer and yttrium oxide nanoparticles as a potential antibacterial material. Mater. Trans. 2018, 59, 1915–1919. [Google Scholar] [CrossRef]
- Zhang, T.; Tang, M.; Yao, Y.; Ma, Y.; Pu, Y. MWCNT interactions with protein: Surface-induced changes in protein adsorption and the impact of protein corona on cellular uptake and cytotoxicity. Int. J. Nanomed. 2019, 14, 993–1009. [Google Scholar] [CrossRef] [PubMed]
- Nierenberg, D.; Khaled, A.R.; Flores, O. Formation of a protein corona influences the biological identity of nanomaterials. Reports Pract. Oncol. Radiother. 2018, 23, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Roach, K.A.; Stefaniak, A.B.; Roberts, J.R. Metal nanomaterials: Immune effects and implications of physicochemical properties on sensitization, elicitation, and exacerbation of allergic disease. J. Immunotoxicol. 2019, 16, 87–124. [Google Scholar] [CrossRef]
- Selvaraj, V.; Bodapati, S.; Murray, E.; Rice, K.M.; Winston, N.; Shokuhfar, T.; Zhao, Y.; Blough, E. Cytotoxicity and genotoxicity caused by yttrium oxide nanoparticles in HEK293 cells. Int. J. Nanomed. 2014, 9, 1379–1391. [Google Scholar] [CrossRef]
- Moriyama, A.; Takahashi, U.; Mizuno, Y.; Takahashi, J.; Horie, M.; Iwahashi, H. The Truth of Toxicity Caused by Yttrium Oxide Nanoparticles to Yeast Cells. J. Nanosci. Nanotechnol. 2019, 19, 5418–5425. [Google Scholar] [CrossRef]
- Holmström, K.M.; Finkel, T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 411–421. [Google Scholar] [CrossRef]
- Veal, E.; Day, A. Hydrogen peroxide as a signaling molecule. Antioxid. Redox Signal. 2011, 15, 147–151. [Google Scholar] [CrossRef]
- Halliwell, B.; Clement, M.V.; Long, L.H. Hydrogen peroxide in the human body. FEBS Lett. 2000, 486, 10–13. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Ahamed, M.; Alhadlaq, H.; Alrokayan, S. Toxicity Mechanism of Gadolinium Oxide Nanoparticles and Gadolinium Ions in Human Breast Cancer Cells. Curr. Drug Metab. 2019, 20, 907–917. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Ahamed, M.; Alhadlaq, H.A. Challenges facing nanotoxicology and nanomedicine due to cellular diversity. Clin. Chim. Acta 2018, 487, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Gong, N.; Zhong, L.; Sun, J.; Liang, X.J. Future of nanotherapeutics: Targeting the cellular sub-organelles. Biomaterials 2016, 97, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Shi, S.S.; Zhang, J.Q.; Zhang, Y.J.; Zhang, L.; Liu, Y.; Jin, P.P.; Wei, P.F.; Shi, R.H.; Zhou, W.; et al. Giant Cellular Vacuoles Induced by Rare Earth Oxide Nanoparticles are Abnormally Enlarged Endo/Lysosomes and Promote mTOR-Dependent TFEB Nucleus Translocation. Small 2016, 12, 5759–5768. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Hableel, G.; Zhao, E.R.; Jokerst, J.V. Multifunctional nanomedicine with silica: Role of silica in nanoparticles for theranostic, imaging, and drug monitoring. J. Colloid Interface Sci. 2018, 521, 261–279. [Google Scholar] [CrossRef]
- Xu, Z.; Ma, X.; Gao, Y.E.; Hou, M.; Xue, P.; Li, C.M.; Kang, Y. Multifunctional silica nanoparticles as a promising theranostic platform for biomedical applications. Mater. Chem. Front. 2017, 1, 1257–1272. [Google Scholar] [CrossRef]
- Schütz, I.; Lopez-Hernandez, T.; Gao, Q.; Puchkov, D.; JaBerlinbs, S.; Nordmeyer, D.; Schmudde, M.; Rühl, E.; Graf, C.M.; Haucke, V. Lysosomal dysfunction caused by cellular accumulation of silica nanoparticles. J. Biol. Chem. 2016, 291, 14170–14184. [Google Scholar] [CrossRef]
- Wang, J.; Yu, Y.; Lu, K.; Yang, M.; Li, Y.; Zhou, X.; Sun, Z. Silica nanoparticles induce autophagy dysfunction via lysosomal impairment and inhibition of autophagosome degradation in hepatocytes. Int. J. Nanomed. 2017, 12, 809–825. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Ahamed, M.; Kumar, S.; Majeed Khan, M.A.; Ahmad, J.; Alrokayan, S.A. Zinc oxide nanoparticles selectively induce apoptosis in human cancer cells through reactive oxygen species. Int. J. Nanomed. 2012, 7, 845–857. [Google Scholar]
- Murdock, R.C.; Braydich-Stolle, L.; Schrand, A.M.; Schlager, J.J.; Hussain, S.M. Characterization of nanomaterial dispersion in solution prior to in vitro exposure using dynamic light scattering technique. Toxicol. Sci. 2008, 101, 239–253. [Google Scholar] [CrossRef]
- Patil, S.; Sandberg, A.; Heckert, E.; Self, W.; Seal, S. Protein adsorption and cellular uptake of cerium oxide nanoparticles as a function of zeta potential. Biomaterials 2007, 28, 4600–4607. [Google Scholar] [CrossRef]
- Marklund, S.; Marklund, G. Involvement of the Superoxide Anion Radical in the Autoxidation of Pyrogallol and a Convenient Assay for Superoxide Dismutase. Eur. J. Biochem. 1974, 47, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Aebi, H. Catalase in vitro. Methods Enzymol. 1984, 105, 121–126. [Google Scholar] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Ahamed, M.; Kumar, S.; Siddiqui, H.; Patil, G.; Ashquin, M.; Ahmad, I. Nanotoxicity of pure silica mediated through oxidant generation rather than glutathione depletion in human lung epithelial cells. Toxicology 2010, 276, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Repetto, G.; del Peso, A.; Zurita, J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Welder, A.A. A primary culture system of adult rat heart cells for the evaluation of cocaine toxicity. Toxicology 1992, 72, 175–187. [Google Scholar] [CrossRef]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef]
- Wang, H.; Joseph, J.A. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic. Biol. Med. 1999, 27, 612–616. [Google Scholar] [CrossRef]
- Smiley, S.T.; Reers, M.; Mottola-Hartshorn, C.; Lin, M.; Chen, A.; Smith, T.W.; Steele, G.D.; Chen, L.B. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc. Natl. Acad. Sci. 1991, 88, 3671–3675. [Google Scholar] [CrossRef]
- Thomé, M.P.; Filippi-Chiela, E.C.; Villodre, E.S.; Migliavaca, C.B.; Onzi, G.R.; Felipe, K.B.; Lenz, G. Ratiometric analysis of Acridine Orange staining in the study of acidic organelles and autophagy. J. Cell Sci. 2016, 129, 4622–4632. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Ahamed, M.; Alhadlaq, H.A.; Kumar, S.; Alrokayan, S.A. Mitochondrial dysfunction, autophagy stimulation and non-apoptotic cell death caused by nitric oxide-inducing Pt-coated Au nanoparticle in human lung carcinoma cells. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129452. [Google Scholar] [CrossRef] [PubMed]
- Bampton, E.T.W.; Goemans, C.G.; Niranjan, D.; Mizushima, N.; Tolkovsky, A.M. The dynamics of autophagy visualized in live cells: From autophagosome formation to fusion with endo/lysosomes. Autophagy 2005, 1, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Atale, N.; Gupta, S.; Yadav, U.C.S.; Rani, V. Cell-death assessment by fluorescent and nonfluorescent cytosolic and nuclear staining techniques. J. Microsc. 2014, 255, 7–19. [Google Scholar] [CrossRef]
- Azizi, M.; Ghourchian, H.; Yazdian, F.; Bagherifam, S.; Bekhradnia, S.; Nyström, B. Anti-cancerous effect of albumin coated silver nanoparticles on MDA-MB 231 human breast cancer cell line. Sci. Rep. 2017, 7, 5178. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).