Quantum Dot Labelling of Tepary Bean (Phaseolus acutifolius) Lectins by Microfluidics
Abstract
1. Introduction
2. Results
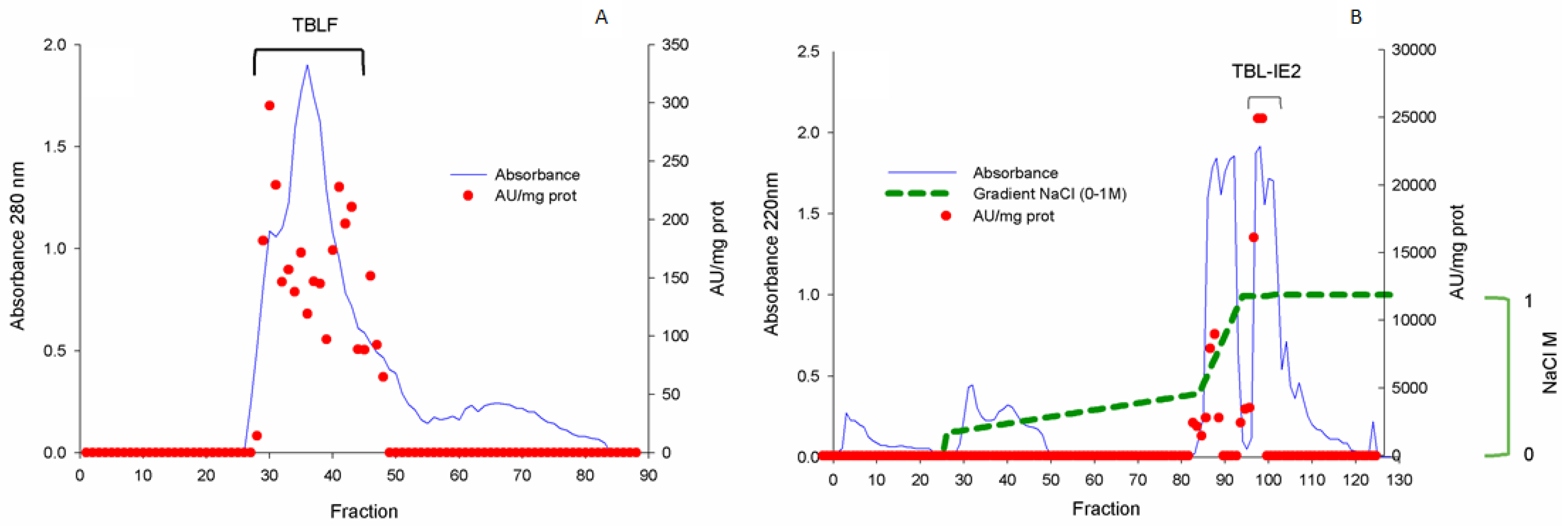
2.1. Lectin Purification
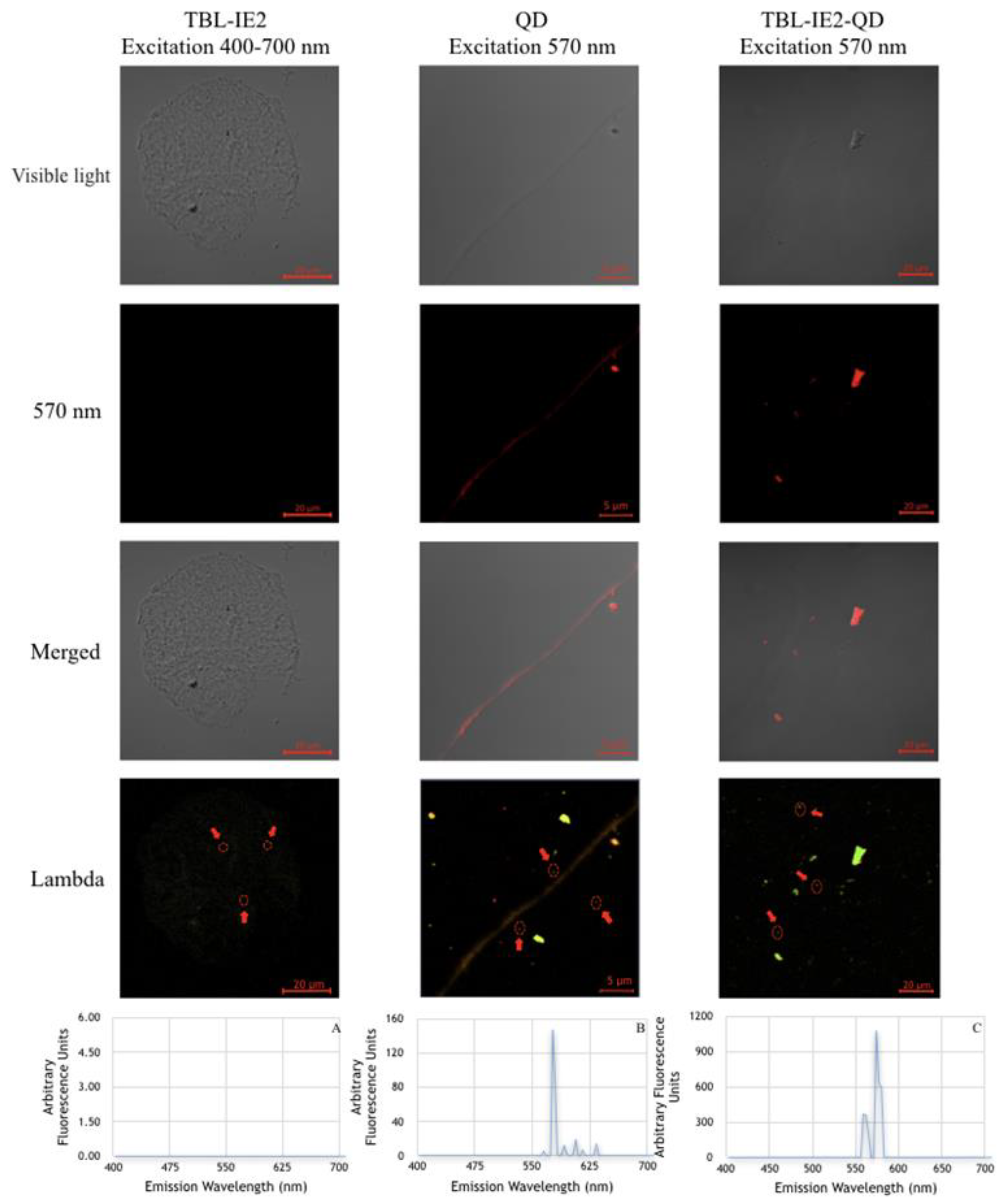
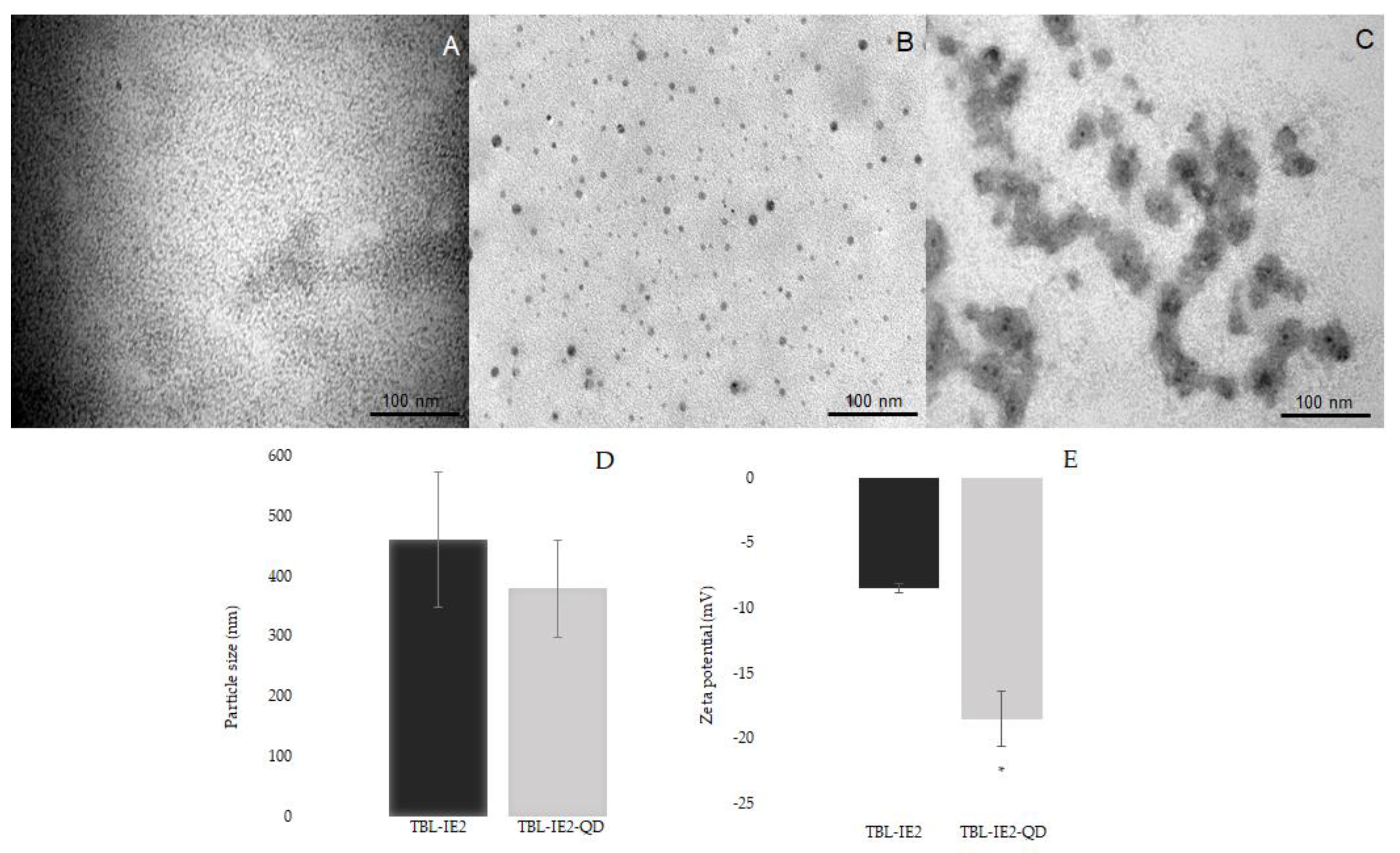
2.2. Lectin Labelling and Analysis
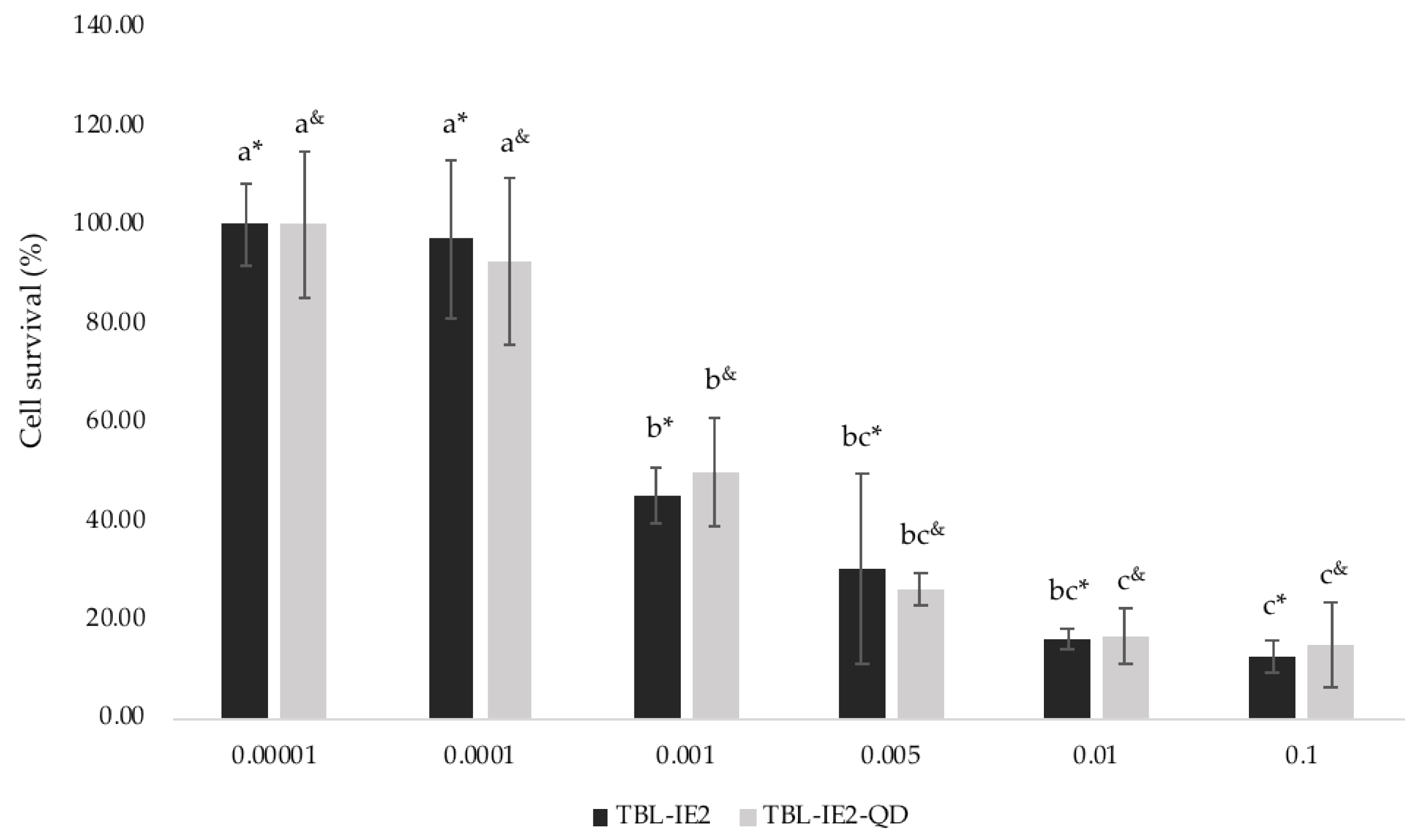
2.3. Cytotoxic Effects of TBL-IE2 and TBL-IE2-QD on HT-29 Cell Line
3. Discussion
4. Materials and Methods
4.1. Biological Materials and Quantum Dots
4.2. Lectin Extraction and Purification
4.3. One- and Two-Dimensional Electrophoretic Separations and Immunoblotting Determination
4.4. TBL-IE2-QD Coupling by Microfluidic Technique
4.5. Determination of the TBL-IE2, QD, and TBL-IE2-QD Fluorescence Emission by Multiphoton Microscopy
4.6. Morphological Analysis by Transmission Electron Microscopy (TEM)
4.7. TBL-IE2-QD Zeta Potential and Hydrodynamic Diameter Determination
4.8. Cytotoxicity Assay
4.9. Statistics
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Damme, E.J.; Lannoo, N.; Fouquaert, E.; Peumans, W.J. The identification of inducible cytoplasmic/nuclear carbohydrate-binding proteins urges to develop novel concepts about the role of plant lectins. Glycoconj. J. 2003, 20, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Luo, Y.; Zhou, T.T.; Zhang, W.Z. Could plant lectins become promising anti-tumour drugs for causing autophagic cell death? Cell Prolif. 2013, 46, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J. History of plant lectin research. In Lectins; Humana Press: New York, NY, USA, 2014; pp. 3–13. [Google Scholar] [CrossRef]
- Jiang, Q.L.; Zhang, S.; Tian, M.; Zhang, S.Y.; Xie, T.; Chen, D.Y.; Chen, Y.J.; He, J.; Liu, J.; Ouyang, L.; et al. Plant lectins, from ancient sugar-binding proteins to emerging anti-cancer drugs in apoptosis and autophagy. Cell Prolif. 2015, 48, 17–28. [Google Scholar] [CrossRef]
- Manning, J.C.; Romero, A.; Habermann, F.A.; Caballero, G.G.; Kaltner, H.; Gabius, H.J. Lectins: A primer for histochemists and cell biologists. Histochem. Cell Biol. 2017, 147, 199–222. [Google Scholar] [CrossRef]
- Dan, X.; Liu, W.; Ng, T.B. Development and applications of lectins as biological tools in biomedical research. Med. Res. Rev. 2016, 36, 221–247. [Google Scholar] [CrossRef]
- Coulibaly, F.S.; Youan, B.B.C. Current status of lectin based cancer diagnosis and therapy. AIMS Mol. Sci. 2017, 4, 1–27. [Google Scholar] [CrossRef]
- Lagarda-Diaz, I.; Guzman-Partida, A.M.; Vazquez-Moreno, L. Legume lectins: Proteins with diverse applications. Int. J. Mol. Sci. 2017, 18, 1242. [Google Scholar] [CrossRef]
- Munkley, J.; Elliott, D.J. Hallmarks of glycosylation in cancer. Oncotarget 2016, 7, 35478. [Google Scholar] [CrossRef]
- Brooks, S.A. Lectin Histochemistry: Historical Perspectives, State of the Art, and the Future. Methods Mol. Biol. 2017, 1560, 93–107. [Google Scholar] [CrossRef]
- Hashim, O.H.; Jayapalan, J.J.; Lee, C.S. Lectins: An effective tool for screening of potential cancer biomarkers. PeerJ 2017, 5, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Celis, U.; López-Martínez, J.; Blanco-Labra, A.; Cervantes-Jiménez, R.; Estrada-Martínez, L.E.; García-Pascalin, A.E.; García-Gasca, T. Phaseolus acutifolius Lectin Fractions Exhibit Apoptotic Effects on Colon Cancer: Preclinical Studies Using Dimethilhydrazine or Azoxi-Methane as Cancer Induction Agents. Molecules 2017, 22, 1670. [Google Scholar] [CrossRef]
- Liu, B.; Bian, H.J.; Bao, J.K. Plant lectins: Potential antineoplastic drugs from bench to clinic. Cancer Lett. 2010, 287, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pervin, M.; Koyama, Y.; Isemura, M.; Nakamura, Y. Plant lectins in therapeutic and diagnostic cancer research. Int. J. Plant Biol. Res. 2015, 3, 1030–1035. [Google Scholar]
- Fu, L.L.; Zhou, C.C.; Yao, S.; Yu, J.Y.; Liu, B.; Bao, J.K. Plant lectins: Targeting programmed cell death pathways as antitumor agents. Int. J. Biochem. Cell Biol. 2011, 43, 1442–1449. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; An, N.; Zhao, S.; Li, X.; Bao, J.K.; Yue, B.S. In silico analysis of molecular mechanisms of legume lectin-induced apoptosis in cancer cells. Cell Prolif. 2013, 46, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Yau, T.; Dan, X.; Ng, C.C.W.; Ng, T.B. Lectins with potential for anti-cancer therapy. Molecules 2015, 20, 3791–3810. [Google Scholar] [CrossRef] [PubMed]
- De Mejía, E.G.; Prisecaru, V.I. Lectins as bioactive plant proteins: A potential in cancer treatment. Crit. Rev. Food Sci. Nutr. 2005, 45, 425–445. [Google Scholar] [CrossRef] [PubMed]
- Coelho, L.C.B.B.; Silva, P.M.D.S.; Lima, V.L.D.M.; Pontual, E.V.; Paiva, P.M.G.; Napoleão, T.H.; Correia, M.T.D.S. Lectins, interconnecting proteins with biotechnological/pharmacological and therapeutic applications. Evid. Based Complementary Altern. Med. 2017, 2017, 1594074. [Google Scholar] [CrossRef]
- Estrada-Martínez, L.E.; Moreno-Celis, U.; Cervantes-Jiménez, R.; Ferriz-Martínez, R.A.; Blanco-Labra, A.; García-Gasca, T. Plant lectins as medical tools against digestive system cancers. Int. J. Mol. Sci. 2017, 18, 1403. [Google Scholar] [CrossRef]
- Poiroux, G.; Barre, A.; van Damme, E.J.; Benoist, H.; Rougé, P. Plant Lectins Targeting O-Glycans at the Cell Surface as Tools for Cancer Diagnosis, Prognosis and Therapy. Int. J. Mol. Sci. 2017, 18, 1232. [Google Scholar] [CrossRef]
- García-Gasca, T.; García-Cruz, M.; Hernandez-Rivera, E.; López-Matínez, J.; Castaneda-Cuevas, A.L.; Yllescas-Gasca, L.; Rodríguez-Méndez, A.J.; Mendiola-Olaya, E.; Castro-Guillén, J.L.; Blanco-Labra, A. Effects of Tepary bean (Phaseolus acutifolius) protease inhibitor and semipure lectin fractions on cancer cells. Nutr. Cancer 2012, 64, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Valadez-Vega, C.; Morales-González, J.; Sumaya-Martínez, M.; Delgado-Olivares, L.; Cruz-Castañeda, A.; Bautista, M.; Zuñiga-Pérez, C. Cytotoxic and antiproliferative effect of tepary bean lectins on C33-A, MCF-7, SKNSH, and SW480 cell lines. Molecules 2014, 19, 9610–9627. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Celis, U.; López-Martínez, F.J.; Cervantes-Jiménez, R.; Ferríz-Martínez, R.A.; Blanco-Labra, A.; García-Gasca, T. Tepary Bean (Phaseolus acutifolius) Lectins Induce Apoptosis and Cell Arrest in G0/G1 by P53(Ser46) Phosphorylation in Colon Cancer Cells. Molecules 2020, 25, 1021. [Google Scholar] [CrossRef]
- Ferriz-Martínez, R.; García-García, K.; Torres-Arteaga, I.; Rodriguez-Mendez, A.J.; de Jesús Guerrero-Carrillo, M.; Moreno-Celis, U.; Mendiola-Olaya, E. Tolerability assessment of a lectin fraction from Tepary bean seeds (Phaseolus acutifolius) orally administered to rats. Toxicol. Rep. 2015, 2, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Alatorre-Cruz, J.M.; Pita-López, W.; López-Reyes, R.G.; Ferriz-Martínez, R.A.; Cervantes-Jiménez, R.; Carrillo, M.D.J.G.; Vargas, P.J.A.; López-Herrera, G.; Rodríguez-Méndez, A.J.; Zamora-Arroyo, A.; et al. Effects of intragastrically-administered Tepary bean lectins on digestive and immune organs: Preclinical evaluation. Toxicol. Rep. 2018, 5, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, I.T.; Guillen, J.C.; Olaya, E.M.; Gasca, T.G.; Zaragoza, M.V.Á.; García-Santoyo, V.; Castillo, J.A.T.; Aguirre, C.; Phinney, B.; Blanco-Labra, A. Characterization of two non-fetuin binding lectins from tepary bean (Phaseolus acutifolius) seeds with differential cytotoxicity on colon cancer cells. J. Glycobiol. 2016, 5, 117. [Google Scholar] [CrossRef]
- Cong, Y.; Katipamula, S.; Trader, C.D.; Orton, D.J.; Geng, T.; Baker, E.S.; Kelly, R.T. Mass spectrometry-based monitoring of millisecond protein–ligand binding dynamics using an automated microfluidic platform. Lab Chip 2016, 16, 1544–1548. [Google Scholar] [CrossRef]
- Bilan, R.; Nabiev, I.; Sukhanova, A. Quantum Dot-Based Nanotools for Bioimaging, Diagnostics, and Drug Delivery. ChemBioChem 2016, 17, 2103–2114. [Google Scholar] [CrossRef]
- Chan, W.C.; Maxwell, D.J.; Gao, X.; Bailey, R.E.; Han, M.; Nie, S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr. Opin. Biotechnol. 2002, 13, 40–46. [Google Scholar] [CrossRef]
- Mo, D.; Hu, L.; Zeng, G.; Chen, G.; Wan, J.; Yu, Z.; Cheng, M. Cadmium-containing quantum dots: Properties, applications, and toxicity. Appl. Microbiol. Biotechnol. 2017, 101, 2713–2733. [Google Scholar] [CrossRef]
- Cunha, C.R.A.; Oliveira, A.D.P.R.; Firmino, T.V.C.; Tenório, D.P.L.A.; Pereira, G.; Carvalho, L.B., Jr.; Santos, B.S.; Correia, M.T.S.; Fontes, A. Biomedical applications of glyconanoparticles based on quantum dots. BBA Gen. Subj. 2018, 1862, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Moura, I.M.; Cabral Filho, P.E.; Seabra, M.A.; Pereira, G.; Pereira, G.A.; Fontes, A.; Santos, B.S. Highly fluorescent positively charged ZnSe quantum dots for bioimaging. J. Lumin. 2018, 201, 284–289. [Google Scholar] [CrossRef]
- Palomo, V.; Cistrone, P.A.; Zhan, N.; Palui, G.; Mattoussi, H.; Dawson, P.E. Efficient Assembly of Quantum Dots with Homogenous Glycans Derived from Natural N-Linked Glycoproteins. Bioconjug. Chem. 2018, 29, 3144–3153. [Google Scholar] [CrossRef] [PubMed]
- Pinaud, F.; Clarke, S.; Sittner, A.; Dahan, M. Probing cellular events, one quantum dot at a time. Nat. Methods 2010, 7, 275. [Google Scholar] [CrossRef] [PubMed]
- Fontes, A.; de Lira, R.B.; Seabra, M.A.B.L.; da Silva, T.G.; de Castro Neto, A.G.; Santos, B.S. Quantum dots in biomedical research. In Biomedical Engineering-Technical Applications in Medicine, 1st ed.; InTec: Pernambuco, Brazil, 2012; pp. 269–290. [Google Scholar] [CrossRef]
- Pierobon, P.; Cappello, G. Quantum dots to tail single bio-molecules inside living cells. Adv. Drug Deliv. Rev. 2012, 64, 167–178. [Google Scholar] [CrossRef]
- Yao, J.; Li, P.; Li, L.; Yang, M. Biochemistry and biomedicine of quantum dots: From biodetection to bioimaging, drug discovery, diagnosis, and therapy. Acta Biomater. 2018, 74, 36–55. [Google Scholar] [CrossRef]
- Higuchi, Y.; Oka, M.; Kawakami, S.; Hashida, M. Mannosylated semiconductor quantum dots for the labeling of macrophages. J. Controlled Release 2008, 125, 131–136. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, S.; Yuan, D.; Alici, G.; Nguyen, N.T.; Warkiani, M.E.; Li, W. Fundamentals and applications of inertial microfluidics: A review. Lab Chip 2016, 16, 10–34. [Google Scholar] [CrossRef]
- Chen, K.; Fan, Z.H. Introduction to Microfluidics. In Circulating Tumor Cells: Isolation and Analysis; Fan, Z.H., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 33–50. [Google Scholar] [CrossRef]
- Liao, Z.; Zhang, Y.; Li, Y.; Miao, Y.; Gao, S.; Lin, F.; Deng, Y.; Geng, L. Microfluidic chip coupled with optical biosensors for simultaneous detection of multiple analytes: A review. Biosens. Bioelectron. 2019, 126, 697–706. [Google Scholar] [CrossRef]
- Zhelev, Z.; Ohba, H.; Bakalova, R.; Jose, R.; Fukuoka, S.; Nagase, T.; Ishikawa, M.; Baba, Y. Fabrication of quantum dot–lectin conjugates as novel fluorescent probes for microscopic and flow cytometric identification of leukemia cells from normal lymphocytes. Chem. Commun. 2005, 15, 1980–1982. [Google Scholar] [CrossRef]
- Liu, S.L.; Zhang, Z.L.; Sun, E.Z.; Peng, J.; Xie, M.; Tian, Z.Q.; Pang, D.W. Visualizing the endocytic and exocytic processes of wheat germ agglutinin by quantum dot-based single-particle tracking. Biomaterials 2011, 32, 7616–7624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, L.; Liang, R.P.; Huang, J.; Qiu, J.D. Simultaneous determination of Concanavalin A and Peanut agglutinin by dual-color quantum dots. Anal. Chem. 2013, 85, 10969–10976. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Li, Y.Q.; Zhang, H.L.; Wang, H.Q.; Lin, S.; Chen, J.; Zhao, Y.D.; Luo, Q.M. Bioconjugation of concanavalin and CdTe quantum dots and the detection of glucose. Colloid. Surf. A Physicochem. Eng. Asp. 2010, 364, 82–86. [Google Scholar] [CrossRef]
- Hu, B.; Zhang, L.P.; Chen, M.L.; Chen, M.L.; Wang, J.H. The inhibition of fluorescence resonance energy transfer between quantum dots for glucose assay. Biosens. Bioelectron. 2012, 32, 82–88. [Google Scholar] [CrossRef]
- Xie, M.; Hu, J.; Long, Y.-M.; Zhang, Z.L.; Xie, H.-Y.; Pang, D.W. Lectin-modified trifunctional nanobiosensors for mapping cell surface glycoconjugates. Biosens. Bioelectron. 2009, 24, 1311–1317. [Google Scholar] [CrossRef]
- Akca, O.; Unak, P.; Medine, E.I.; Sakarya, S.; Kilcar, A.Y.; Ichedef, C.; Bekis, R.; Timur, S. Radioiodine labeled CdSe/CdS quantum dots: Lectin targeted dual probes. Radiochim. Acta 2014, 102, 849–859. [Google Scholar] [CrossRef]
- Andrade, C.G.; Cabral Filho, P.E.; Tenório, D.P.; Santos, B.S.; Beltrão, E.I.; Fontes, A.; Carvalho, L.B., Jr. Evaluation of glycophenotype in breast cancer by quantum dot-lectin histochemistry. Int. J. Nanomed. 2013, 8, 4623. [Google Scholar] [CrossRef]
- Oliveira, W.F.; Cabrera, M.P.; Santos, N.R.; Napoleão, T.H.; Paiva, P.M.; Neves, R.P.; Silva, M.V.; Santos, B.S.; Coelho, L.C.; Cabral Filho, P.E.; et al. Evaluating Glucose and Mannose pro Fi Les in Candida Species Using Quantum Dots Conjugated with Cramoll Lectin as Fluorescent Nanoprobes. Microbiol. Res. 2020, 230, 126330. [Google Scholar] [CrossRef]
- Zhang, J.T.; Cai, Z.; Kwak, D.H.; Liu, X.; Asher, S.A. Two-dimensional photonic crystal sensors for visual detection of lectin concanavalin A. Anal. Chem. 2014, 86, 9036–9041. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; Ganganboina, A.B.; Tsai, Y.C.; Chiu, H.C.; Doong, R.A. Multifunctional GQDs-Concanavalin A Fe3O4 nanocomposites for cancer cells detection and targeted drug delivery. Anal. Chim. Acta. 2018, 1027, 109–120. [Google Scholar] [CrossRef]
- Nascimento, K.S.; Cunha, A.I.; Nascimento, K.S.; Cavada, B.S.; Azevedo, A.M.; Aires-Barros, M.R. An overview of lectins purification strategies. J. Mol. Recognit. 2012, 25, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Alarcón, D.; Mora-Avilés, A.; Espinoza-Núñez, A.; Cruz-Hernández, A.; Rodríguez-Torres, A.; Castro-Guillen, J.L.; Blanco-Labra, A.; García-Gasca, T. Rhizosecretion of a cisgenic lectin by genetic manipulation of Tepary bean plants (Phaseolus acutifolius). J. Biotechno. X 2019, 3, 100013. [Google Scholar] [CrossRef]
- Vetri, V.; Librizzi, F.; Militello, V.; Leone, M. Effects of succinylation on thermal induced amyloid formation in Concanavalin A. Eur. Biophys. J. 2007, 36, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Sperling, R.A.; Parak, W.J. Surface modification, functionalization and bioconjugation of colloidal inorganic nanoparticles. Philos. Trans. R. Soc. A 2010, 368, 1333–1383. [Google Scholar] [CrossRef]
- Carvalho, M.E.T.; Oliveira, W.F.; Cunha, C.R.; Coelho, L.C.; Silva, M.V.; Junior, L.B.C.; Santos, B.S.; Filho, P.E.C.; Fontes, A.; Correia, M.T.S. Evaluating the glycophenotype on breast cancer tissues with quantum dots-Cramoll lectin conjugates. Int. J. Biol. Macromol. 2019, 138, 302–308. [Google Scholar] [CrossRef]
- Albani, J.R. Fluorophores: Descriptions and Properties. In Structure and Dynamics of Macromolecules: Absorption and Fluorescence Studies, 1st ed.; Elsevier: Amsterdam, The Netherland, 2004; pp. 99–140. [Google Scholar] [CrossRef]
- Joglekar, S.S.; Gholap, H.M.; Alegaonkar, P.S.; Kale, A.A. The interactions between CdTe quantum dots and proteins: Understanding nano-bio interface. J. Mater. Sci. 2017, 4, 209–222. [Google Scholar] [CrossRef]
- Valencia, P.M.; Basto, P.A.; Zhang, L.; Rhee, M.; Langer, R.; Farokhzad, O.C.; Karnik, R. Single-step assembly of homogenous lipid− polymeric and lipid− quantum dot nanoparticles enabled by microfluidic rapid mixing. ACS Nano 2010, 4, 1671–1679. [Google Scholar] [CrossRef]
- Gao, X.; Wang, T.; Wu, B.; Chen, J.; Chen, J.; Yue, Y.; Dai, N.; Chen, H.; Jiang, X. Quantum dots for tracking cellular transport of lectin-functionalized nanoparticles. Biochem. Biophys. Res. Commun. 2008, 377, 35–40. [Google Scholar] [CrossRef]
- Cunha, C.R.; Andrade, C.G.; Pereira, M.I.; Cabral Filho, P.E.; Carvalho, L.B., Jr.; Coelho, L.C.; Santos, B.S.; Fontes, A.; Correia, M.T. Quantum Dot–Cramoll Lectin as Novel Conjugates to Glycobiology. J. Photochem. Photobiol. B 2018, 178, 85–91. [Google Scholar] [CrossRef]
- Tenório, D.P.; Andrade, C.G.; Cabral Filho, P.E.; Sabino, C.P.; Kato, I.T.; Carvalho, L.B., Jr.; Alves, S., Jr.; Ribeiro, M.S.; Fontes, A.; Santos, B.S. CdTe quantum dots conjugated to concanavalin A as potential fluorescent molecular probes for saccharides detection in Candida albicans. J. Photochem. Photobiol. B 2015, 142, 237–243. [Google Scholar] [CrossRef]
- Chen, N.; He, Y.; Su, Y.; Li, X.; Huang, Q.; Wang, H.; Zhang, X.; Tai, R.; Fan, C. The cytotoxicity of cadmium-based quantum dots. Biomaterials 2012, 33, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, R.; Liu, J.; Zhang, B.; Wang, Y.; Liu, X.; Law, W.C.; Liu, L.; Ye, L.; Yong, K.T. Cytotoxicity assessment of functionalized CdSe, CdTe and InP quantum dots in two human cancer cell models. Mater. Sci. Eng. 2015, 57, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Yan, J.; He, Y.; Lu, H.; Weng, L.; Song, S.; Fan, C.; Wang, L. Ultrasensitive, Multiplexed Detection of Cancer Biomarkers Directly in Serum by Using a Quantum Dot-Based Microfluidic Protein Chip. ACS Nano 2010, 4, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.H.; Kim, Y.G.; Jang, S.C.; Yi, H.; Lee, C.S. Profiling surface glycans on live cells and tissues using quantum dot-lectin nanoconjugates. Lab Chip 2012, 12, 3290–3295. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.H.; Liener, I.E. The use of glutaraldehyde-treated erythrocytes for assaying the agglutinating activity of lectins. Anal. Biochem. 1975, 68, 651–653. [Google Scholar] [CrossRef]
- Jaffé, W. Hemagglutinins (lectins). In Toxic Constituents of Plant Foodstuffs; Academic Press: New York, NY, USA, 1980; pp. 73–102. [Google Scholar]
- Adamová, L.; Malinovská, L.; Wimmerová, M. New sensitive detection method for lectin hemagglutination using microscopy. Microsc. Res. Tech. 2014, 77, 841–849. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Walker, M.J. Methods in Molecular Biology. In Basic Protein and Peptide Protocols; Humana: Totowa, NJ, USA, 1994; Volume 32, pp. 17–22. [Google Scholar] [CrossRef]
- Packer, N.H.; Ball, M.S.; Devine, P.L. Glycoprotein detection of 2-D separated proteins. In 2-D Proteome Analysis Protocols Methods in Molecular Biology; Link, A.J., Ed.; Humana Press: Totowa, NJ, USA, 1999; Volume 112, pp. 341–352. [Google Scholar] [CrossRef]
- Görg, A.; Boguth, G.; Obermaier, C.; Weiss, W. Two-dimensional electrophoresis of proteins in an immobilized pH 4–12 gradient. Electrophoresis 1998, 19, 1516–1519. [Google Scholar] [CrossRef]
- Blum, H.; Beier, H.; Gross, H.J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 1987, 8, 93–99. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 10 February 2019).
Sample Availability: Samples of the compounds are not available from the authors. |


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cervantes-Jiménez, R.; Sánchez-Segura, L.; Estrada-Martínez, L.E.; Topete-Camacho, A.; Mendiola-Olaya, E.; Rosas-Escareño, A.N.; Saldaña-Gutiérrez, C.; Figueroa-Cabañas, M.E.; Dena-Beltrán, J.L.; Kuri-García, A.; et al. Quantum Dot Labelling of Tepary Bean (Phaseolus acutifolius) Lectins by Microfluidics. Molecules 2020, 25, 1041. https://doi.org/10.3390/molecules25051041
Cervantes-Jiménez R, Sánchez-Segura L, Estrada-Martínez LE, Topete-Camacho A, Mendiola-Olaya E, Rosas-Escareño AN, Saldaña-Gutiérrez C, Figueroa-Cabañas ME, Dena-Beltrán JL, Kuri-García A, et al. Quantum Dot Labelling of Tepary Bean (Phaseolus acutifolius) Lectins by Microfluidics. Molecules. 2020; 25(5):1041. https://doi.org/10.3390/molecules25051041
Chicago/Turabian StyleCervantes-Jiménez, Ricardo, Lino Sánchez-Segura, Laura Elena Estrada-Martínez, Antonio Topete-Camacho, Elizabeth Mendiola-Olaya, Abraham Noé Rosas-Escareño, Carlos Saldaña-Gutiérrez, Mónica Eugenia Figueroa-Cabañas, José Luis Dena-Beltrán, Aarón Kuri-García, and et al. 2020. "Quantum Dot Labelling of Tepary Bean (Phaseolus acutifolius) Lectins by Microfluidics" Molecules 25, no. 5: 1041. https://doi.org/10.3390/molecules25051041
APA StyleCervantes-Jiménez, R., Sánchez-Segura, L., Estrada-Martínez, L. E., Topete-Camacho, A., Mendiola-Olaya, E., Rosas-Escareño, A. N., Saldaña-Gutiérrez, C., Figueroa-Cabañas, M. E., Dena-Beltrán, J. L., Kuri-García, A., Blanco-Labra, A., & García-Gasca, T. (2020). Quantum Dot Labelling of Tepary Bean (Phaseolus acutifolius) Lectins by Microfluidics. Molecules, 25(5), 1041. https://doi.org/10.3390/molecules25051041









