DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs
Abstract
1. Introduction
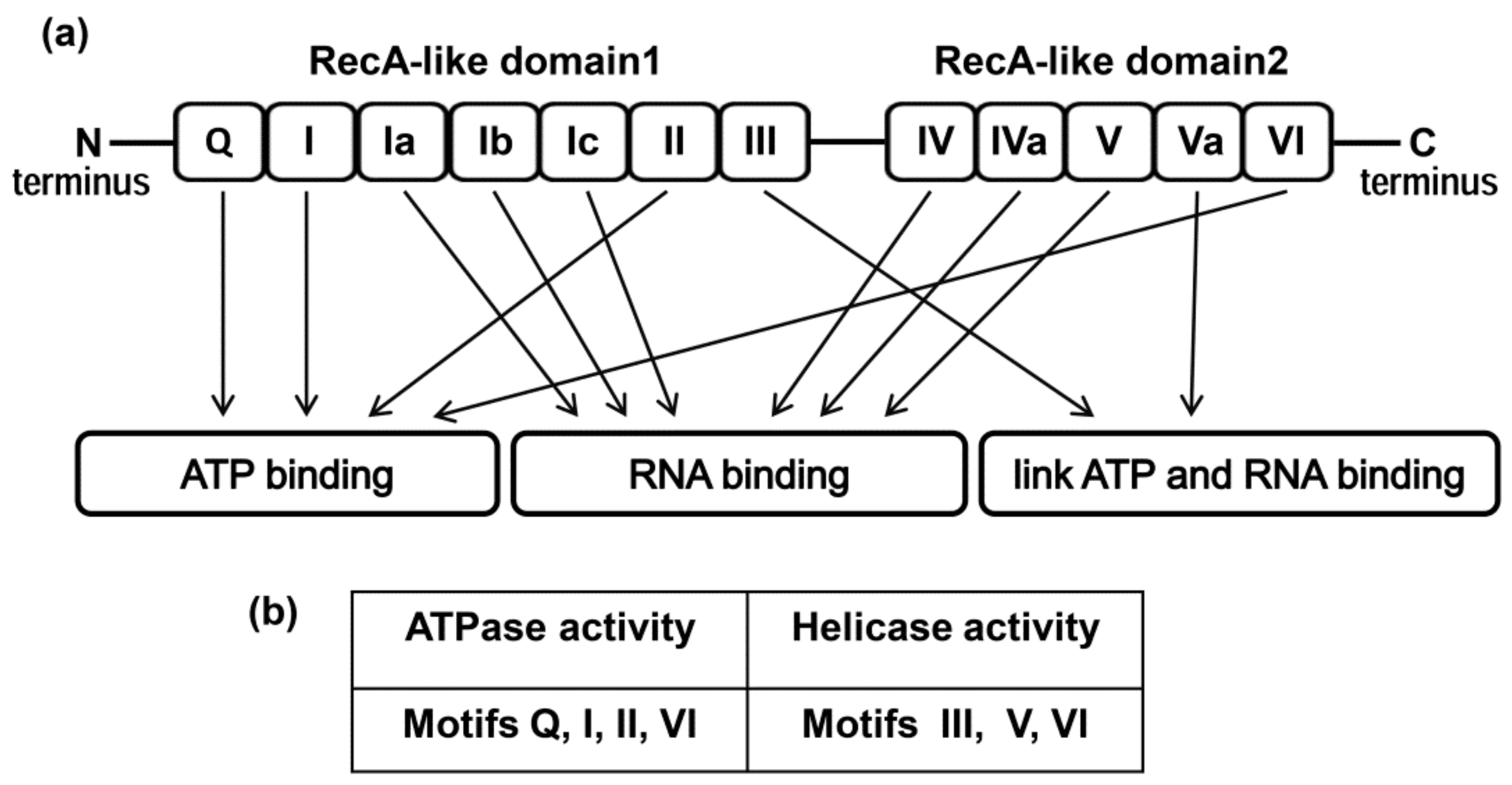
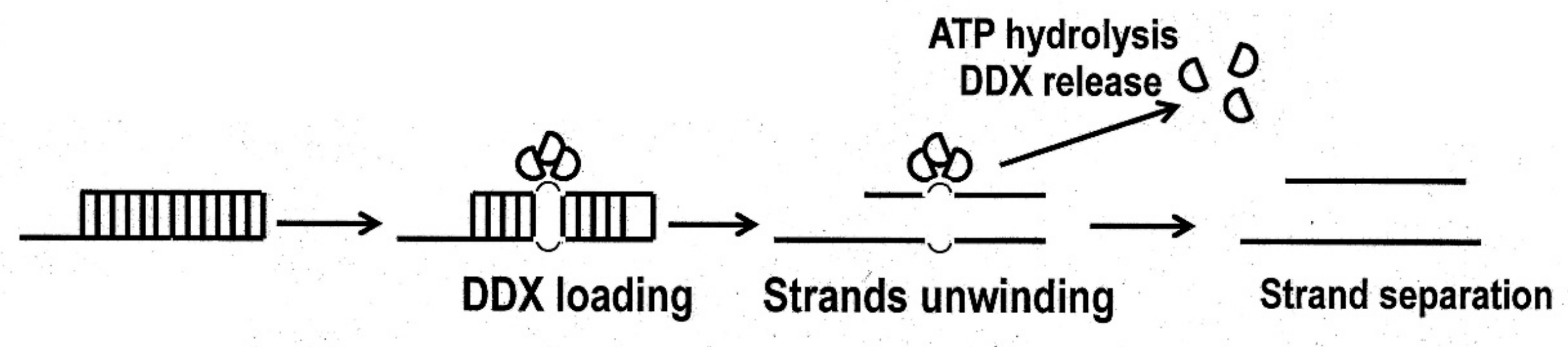
2. Structure of the DDX3 Helicase and Its Enzymatic Properties
3. Hypothetical Mechanisms of the DDX3 Helicase Role in Viral Replication
4. Inhibitors of the DDX3 Helicase
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Xi, X.G. Helicases as antiviral and anticancer drug targets. Curr. Med. Chem. 2007, 14, 883–915. [Google Scholar] [PubMed]
- Maga, G.; Falchi, F.; Radi, M.; Botta, L.; Casaluce, G.; Bernardini, M.; Irannejad, H.; Manetti, F.; Garbelli, A.; Samuele, A.; et al. Toward the discovery of novel anti-HIV drugs. Second-generation inhibitors of the cellular ATPase DDX3 with improved anti-HIV activity: Synthesis, structure-activity relationship analysis, cytotoxicity studies, and target validation. ChemMedChem 2011, 6, 1371–1389. [Google Scholar] [CrossRef] [PubMed]
- Konig, R.; Zhou, Y.; Elleder, D.; Diamond, T.L.; Bonamy, G.M.; Irelan, J.T.; Chiang, C.Y.; Tu, B.P.; De Jesus, P.D.; Lilley, C.E.; et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 2008, 135, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.W.; Holland, J.J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 1999, 96, 13910–13913. [Google Scholar] [CrossRef]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Tintori, C.; Brai, A.; Fallacara, A.L.; Fazi, R.; Schenone, S.; Botta, M. Protein-protein interactions and human cellular cofactors as new targets for HIV therapy. Curr. Opin. Pharmacol. 2014, 18, 1–8. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, S.G.; Kim, Y.; Song, K. Assignment of a human putative RNA helicase gene, DDX3, to human X chromosome bands p11.3-->p11.23. Cytogenet. Cell Genet. 1998, 81, 178–179. [Google Scholar] [CrossRef]
- Brai, A.; Fazi, R.; Tintori, C.; Zamperini, C.; Bugli, F.; Sanguinetti, M.; Stigliano, E.; Este, J.; Badia, R.; Franco, S.; et al. Human DDX3 protein is a valuable target to develop broad spectrum antiviral agents. Proc. Natl. Acad. Sci. USA 2016, 113, 5388–5393. [Google Scholar] [CrossRef]
- Ariumi, Y. Multiple functions of DDX3 RNA helicase in gene regulation, tumorigenesis, and viral infection. Front Genet. 2014, 5, 423. [Google Scholar] [CrossRef]
- Bol, G.M.; Xie, M.; Raman, V. DDX3, a potential target for cancer treatment. Mol. Cancer 2015, 14, 188. [Google Scholar] [CrossRef]
- Schroder, M. Viruses and the human DEAD-box helicase DDX3: Inhibition or exploitation? Biochem. Soc. Trans. 2011, 39, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Upadya, M.H.; Aweya, J.J.; Tan, Y.J. Understanding the interaction of hepatitis C virus with host DEAD-box RNA helicases. World J. Gastroenterol. 2014, 20, 2913–2926. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Singh, N.; Abdin, M.Z.; Patel, A.H.; Medigeshi, G.R. Dengue Virus Capsid Interacts with DDX3X-A Potential Mechanism for Suppression of Antiviral Functions in Dengue Infection. Front Cell Infect. Microbiol. 2017, 7, 542. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Feng, T.; Pan, W.; Shi, X.; Dai, J. DEAD-box RNA helicase DDX3X inhibits DENV replication via regulating type one interferon pathway. Biochem. Biophys. Res. Commun. 2015, 456, 327–332. [Google Scholar] [CrossRef]
- Li, C.; Ge, L.L.; Li, P.P.; Wang, Y.; Dai, J.J.; Sun, M.X.; Huang, L.; Shen, Z.Q.; Hu, X.C.; Ishag, H.; et al. Cellular DDX3 regulates Japanese encephalitis virus replication by interacting with viral un-translated regions. Virology 2014, 449, 70–81. [Google Scholar] [CrossRef]
- Brai, A.; Martelli, F.; Riva, V.; Garbelli, A.; Fazi, R.; Zamperini, C.; Pollutri, A.; Falsitta, L.; Ronzini, S.; Maccari, L.; et al. DDX3X Helicase Inhibitors as a New Strategy to Fight the West Nile Virus Infection. J. Med. Chem. 2019, 62, 2333–2347. [Google Scholar] [CrossRef]
- Yedavalli, V.S.; Neuveut, C.; Chi, Y.H.; Kleiman, L.; Jeang, K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef]
- Lorgeoux, R.P.; Guo, F.; Liang, C. From promoting to inhibiting: Diverse roles of helicases in HIV-1 Replication. Retrovirology 2012, 9, 79. [Google Scholar] [CrossRef]
- Wang, H.; Kim, S.; Ryu, W.S. DDX3 DEAD-Box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J. Virol. 2009, 83, 5815–5824. [Google Scholar] [CrossRef]
- Kalverda, A.P.; Thompson, G.S.; Vogel, A.; Schroder, M.; Bowie, A.G.; Khan, A.R.; Homans, S.W. Poxvirus K7 protein adopts a Bcl-2 fold: Biochemical mapping of its interactions with human DEAD box RNA helicase DDX3. J. Mol. Biol. 2009, 385, 843–853. [Google Scholar] [CrossRef]
- Vashist, S.; Urena, L.; Chaudhry, Y.; Goodfellow, I. Identification of RNA-protein interaction networks involved in the norovirus life cycle. J. Virol. 2012, 86, 11977–11990. [Google Scholar] [CrossRef] [PubMed]
- Thulasi Raman, S.N.; Liu, G.; Pyo, H.M.; Cui, Y.C.; Xu, F.; Ayalew, L.E.; Tikoo, S.K.; Zhou, Y. DDX3 Interacts with Influenza A Virus NS1 and NP Proteins and Exerts Antiviral Function through Regulation of Stress Granule Formation. J. Virol. 2016, 90, 3661–3675. [Google Scholar] [CrossRef] [PubMed]
- Ishaq, M.; Hu, J.; Wu, X.; Fu, Q.; Yang, Y.; Liu, Q.; Guo, D. Knockdown of cellular RNA helicase DDX3 by short hairpin RNAs suppresses HIV-1 viral replication without inducing apoptosis. Mol. Biotechnol. 2008, 39, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Lee, Y.H.; Tarn, W.Y. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell 2008, 19, 3847–3858. [Google Scholar] [CrossRef]
- Tantravedi, S.; Vesuna, F.; Winnard, P.T., Jr.; Martin, A.; Lim, M.; Eberhart, C.G.; Berlinicke, C.; Raabe, E.; van Diest, P.J.; Raman, V. Targeting DDX3 in Medulloblastoma Using the Small Molecule Inhibitor RK-33. Transl. Oncol. 2019, 12, 96–105. [Google Scholar] [CrossRef]
- Zhao, L.; Mao, Y.; Zhou, J.; Zhao, Y.; Cao, Y.; Chen, X. Multifunctional DDX3: Dual roles in various cancer development and its related signaling pathways. Am. J. Cancer Res. 2016, 6, 387–402. [Google Scholar]
- Botlagunta, M.; Vesuna, F.; Mironchik, Y.; Raman, A.; Lisok, A.; Winnard, P., Jr.; Mukadam, S.; Van Diest, P.; Chen, J.H.; Farabaugh, P.; et al. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene 2008, 27, 3912–3922. [Google Scholar] [CrossRef]
- Andreou, A.Z.; Klostermeier, D. Conformational changes of DEAD-box helicases monitored by single molecule fluorescence resonance energy transfer. Methods Enzymol. 2012, 511, 75–109. [Google Scholar]
- Linder, P.; Jankowsky, E. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011, 12, 505–516. [Google Scholar] [CrossRef]
- Hilbert, M.; Karow, A.R.; Klostermeier, D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol. Chem. 2009, 390, 1237–1250. [Google Scholar] [CrossRef]
- Hogbom, M.; Collins, R.; van den Berg, S.; Jenvert, R.M.; Karlberg, T.; Kotenyova, T.; Flores, A.; Karlsson Hedestam, G.B.; Schiavone, L.H. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J. Mol. Biol. 2007, 372, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Ji, X. The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X. Nat. Commun. 2019, 10, 3085. [Google Scholar] [CrossRef] [PubMed]
- Sengoku, T.; Nureki, O.; Nakamura, A.; Kobayashi, S.; Yokoyama, S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 2006, 125, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Schutz, P.; Karlberg, T.; van den Berg, S.; Collins, R.; Lehtio, L.; Hogbom, M.; Holmberg-Schiavone, L.; Tempel, W.; Park, H.W.; Hammarstrom, M.; et al. Comparative structural analysis of human DEAD-box RNA helicases. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The DEAD-box protein family of RNA helicases. Gene 2006, 367, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Potratz, J.P.; Tijerina, P.; Del Campo, M.; Lambowitz, A.M.; Russell, R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc. Natl. Acad. Sci. USA 2008, 105, 20203–20208. [Google Scholar] [CrossRef] [PubMed]
- Putnam, A.; Jankowsky, E. Analysis of duplex unwinding by RNA helicases using stopped-flow fluorescence spectroscopy. Methods Enzymol. 2012, 511, 1–27. [Google Scholar]
- Pause, A.; Methot, N.; Sonenberg, N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol. 1993, 13, 6789–6798. [Google Scholar] [CrossRef][Green Version]
- Rogers, G.W., Jr.; Komar, A.A.; Merrick, W.C. eIF4A: The godfather of the DEAD box helicases. Prog Nucleic Acid Res. Mol. Biol. 2002, 72, 307–331. [Google Scholar]
- Putnam, A.A.; Jankowsky, E. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim. Biophys. Acta 2013, 1829, 884–893. [Google Scholar] [CrossRef]
- Radi, M.; Falchi, F.; Garbelli, A.; Samuele, A.; Bernardo, V.; Paolucci, S.; Baldanti, F.; Schenone, S.; Manetti, F.; Maga, G.; et al. Discovery of the first small molecule inhibitor of human DDX3 specifically designed to target the RNA binding site: Towards the next generation HIV-1 inhibitors. Bioorg. Med. Chem. Lett. 2012, 22, 2094–2098. [Google Scholar] [CrossRef] [PubMed]
- Shadrick, W.R.; Ndjomou, J.; Kolli, R.; Mukherjee, S.; Hanson, A.M.; Frick, D.N. Discovering new medicines targeting helicases: Challenges and recent progress. J. Biomol. Screen 2013, 18, 761–781. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Vesuna, F.; Botlagunta, M.; Bol, G.M.; Irving, A.; Bergman, Y.; Hosmane, R.S.; Kato, Y.; Winnard, P.T., Jr.; Raman, V. NZ51, a ring-expanded nucleoside analog, inhibits motility and viability of breast cancer cells by targeting the RNA helicase DDX3. Oncotarget 2015, 6, 29901–29913. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, P.; Baier, A.; Cova, L.; Hosmane, R.S. Dual inhibition of HCV and HIV by ring-expanded nucleosides containing the 5:7-fused imidazo [4,5-e][1,3]diazepine ring system. In vitro results and implications. Bioorg. Med. Chem. Lett. 2014, 24, 1154–1157. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sharma, D.; Putnam, A.A.; Jankowsky, E. Biochemical Differences and Similarities between the DEAD-Box Helicase Orthologs DDX3X and Ded1p. J. Mol. Biol. 2017, 429, 3730–3742. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Putnam, A.; Jankowsky, E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. USA 2008, 105, 20209–20214. [Google Scholar] [CrossRef]
- Liu, F.; Putnam, A.A.; Jankowsky, E. DEAD-box helicases form nucleotide-dependent, long-lived complexes with RNA. Biochemistry 2014, 53, 423–433. [Google Scholar] [CrossRef]
- Garbelli, A.; Beermann, S.; Di Cicco, G.; Dietrich, U.; Maga, G. A motif unique to the human DEAD-box protein DDX3 is important for nucleic acid binding, ATP hydrolysis, RNA/DNA unwinding and HIV-1 replication. PLoS ONE 2011, 6, e19810. [Google Scholar] [CrossRef]
- Franca, R.; Belfiore, A.; Spadari, S.; Maga, G. Human DEAD-box ATPase DDX3 shows a relaxed nucleoside substrate specificity. Proteins 2007, 67, 1128–1137. [Google Scholar] [CrossRef]
- Fullam, A.; Schroder, M. DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication. Biochim. Biophys. Acta 2013, 1829, 854–865. [Google Scholar] [CrossRef]
- Ranji, A.; Boris-Lawrie, K. RNA helicases: Emerging roles in viral replication and the host innate response. RNA Biol. 2010, 7, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Kubota, S.; Yang, B.; Zhou, N.; Zhang, H.; Godbout, R.; Pomerantz, R.J. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology 2004, 330, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, A.; Rojas-Araya, B.; Pereira-Montecinos, C.; Dellarossa, A.; Toro-Ascuy, D.; Prades-Perez, Y.; Garcia-de-Gracia, F.; Garces-Alday, A.; Rubilar, P.S.; Valiente-Echeverria, F.; et al. DEAD-box RNA helicase DDX3 connects CRM1-dependent nuclear export and translation of the HIV-1 unspliced mRNA through its N-terminal domain. Biochim. Biophys. Acta 2016, 1859, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Jeang, K.T.; Yedavalli, V. Role of RNA helicases in HIV-1 replication. Nucleic Acids Res. 2006, 34, 4198–4205. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Wang, S.W.; Cheng, L.; Tarn, W.Y.; Tsai, S.J.; Sun, H.S. Human DDX3 interacts with the HIV-1 Tat protein to facilitate viral mRNA translation. PLoS ONE 2013, 8, e68665. [Google Scholar] [CrossRef]
- Naji, S.; Ambrus, G.; Cimermancic, P.; Reyes, J.R.; Johnson, J.R.; Filbrandt, R.; Huber, M.D.; Vesely, P.; Krogan, N.J.; Yates, J.R.; et al. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell Proteomics 2012, 11, M111.015313. [Google Scholar] [CrossRef]
- Liu, J.; Henao-Mejia, J.; Liu, H.; Zhao, Y.; He, J.J. Translational regulation of HIV-1 replication by HIV-1 Rev cellular cofactors Sam68, eIF5A, hRIP, and DDX3. J. Neuroimmune Pharmacol. 2011, 6, 308–321. [Google Scholar] [CrossRef]
- Geissler, R.; Golbik, R.P.; Behrens, S.E. The DEAD-box helicase DDX3 supports the assembly of functional 80S ribosomes. Nucleic Acids Res. 2012, 40, 4998–5011. [Google Scholar] [CrossRef]
- Yasuda-Inoue, M.; Kuroki, M.; Ariumi, Y. DDX3 RNA helicase is required for HIV-1 Tat function. Biochem. Biophys. Re.s Commun. 2013, 441, 607–611. [Google Scholar] [CrossRef]
- Schroder, M.; Baran, M.; Bowie, A.G. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008, 27, 2147–2157. [Google Scholar] [CrossRef]
- Soulat, D.; Burckstummer, T.; Westermayer, S.; Goncalves, A.; Bauch, A.; Stefanovic, A.; Hantschel, O.; Bennett, K.L.; Decker, T.; Superti-Furga, G. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 2008, 27, 2135–2146. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Fullam, A.; Brennan, R.; Schroder, M. Human DEAD box helicase 3 couples IkappaB kinase epsilon to interferon regulatory factor 3 activation. Mol. Cell Biol. 2013, 33, 2004–2015. [Google Scholar] [CrossRef] [PubMed]
- Ariumi, Y.; Kuroki, M.; Abe, K.; Dansako, H.; Ikeda, M.; Wakita, T.; Kato, N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 2007, 81, 13922–13926. [Google Scholar] [CrossRef] [PubMed]
- Owsianka, A.M.; Patel, A.H. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 1999, 257, 330–340. [Google Scholar] [CrossRef]
- Mamiya, N.; Worman, H.J. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J. Biol. Chem. 1999, 274, 15751–15756. [Google Scholar] [CrossRef]
- You, L.R.; Chen, C.M.; Yeh, T.S.; Tsai, T.Y.; Mai, R.T.; Lin, C.H.; Lee, Y.H. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J. Virol. 1999, 73, 2841–2853. [Google Scholar] [CrossRef]
- Angus, A.G.; Dalrymple, D.; Boulant, S.; McGivern, D.R.; Clayton, R.F.; Scott, M.J.; Adair, R.; Graham, S.; Owsianka, A.M.; Targett-Adams, P.; et al. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J. Gen. Virol. 2010, 91, 122–132. [Google Scholar] [CrossRef]
- Sun, C.; Pager, C.T.; Luo, G.; Sarnow, P.; Cate, J.H. Hepatitis C virus core-derived peptides inhibit genotype 1b viral genome replication via interaction with DDX3X. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Oshiumi, H.; Ikeda, M.; Matsumoto, M.; Watanabe, A.; Takeuchi, O.; Akira, S.; Kato, N.; Shimotohno, K.; Seya, T. Hepatitis C virus core protein abrogates the DDX3 function that enhances IPS-1-mediated IFN-beta induction. PLoS ONE 2010, 5, e14258. [Google Scholar] [CrossRef]
- Kang, J.I.; Kwon, Y.C.; Ahn, B.Y. Modulation of the type I interferon pathways by culture-adaptive hepatitis C virus core mutants. FEBS Lett. 2012, 586, 1272–1278. [Google Scholar] [CrossRef]
- Sumpter, R., Jr.; Loo, Y.M.; Foy, E.; Li, K.; Yoneyama, M.; Fujita, T.; Lemon, S.M.; Gale, M., Jr. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 2005, 79, 2689–2699. [Google Scholar] [CrossRef] [PubMed]
- Anggakusuma; Frentzen, A.; Gurlevik, E.; Yuan, Q.; Steinmann, E.; Ott, M.; Staeheli, P.; Schmid-Burgk, J.; Schmidt, T.; Hornung, V.; et al. Control of hepatitis C virus replication in mouse liver-derived cells by MAVS-dependent production of type I and type III interferons. J. Virol. 2015, 89, 3833–3845. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, C.; Tan, C.Y.; Horner, S.M. Hepatitis C Virus Infection Is Inhibited by a Noncanonical Antiviral Signaling Pathway Targeted by NS3-NS4A. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Pene, V.; Krishnamurthy, S.; Cha, H.; Liang, T.J. Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly. Nat. Med. 2013, 19, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Vieyres, G.; Pietschmann, T. HCV Pit Stop at the Lipid Droplet: Refuel Lipids and Put on a Lipoprotein Coat before Exit. Cells 2019, 8, 233. [Google Scholar] [CrossRef]
- Lee, J.Y.; Cortese, M.; Haselmann, U.; Tabata, K.; Romero-Brey, I.; Funaya, C.; Schieber, N.L.; Qiang, Y.; Bartenschlager, M.; Kallis, S.; et al. Spatiotemporal Coupling of the Hepatitis C Virus Replication Cycle by Creating a Lipid Droplet- Proximal Membranous Replication Compartment. Cell Rep. 2019, 27, 3602–3617.e3605. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Tunitskaya, V.L.; Ivanova, O.N.; Mitkevich, V.A.; Prassolov, V.S.; Makarov, A.A.; Kukhanova, M.K.; Kochetkov, S.N. Hepatitis C virus NS5A protein modulates template selection by the RNA polymerase in in vitro system. FEBS Lett. 2009, 583, 277–280. [Google Scholar] [CrossRef]
- Masaki, T.; Suzuki, R.; Murakami, K.; Aizaki, H.; Ishii, K.; Murayama, A.; Date, T.; Matsuura, Y.; Miyamura, T.; Wakita, T.; et al. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 2008, 82, 7964–7976. [Google Scholar] [CrossRef]
- Dolde, C.; Bischof, J.; Gruter, S.; Montada, A.; Halekotte, J.; Peifer, C.; Kalbacher, H.; Baumann, U.; Knippschild, U.; Suter, B. A CK1 FRET biosensor reveals that DDX3X is an essential activator of CK1epsilon. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Quintavalle, M.; Sambucini, S.; Di Pietro, C.; De Francesco, R.; Neddermann, P. The alpha isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylation. J. Virol. 2006, 80, 11305–11312. [Google Scholar] [CrossRef]
- Masaki, T.; Matsunaga, S.; Takahashi, H.; Nakashima, K.; Kimura, Y.; Ito, M.; Matsuda, M.; Murayama, A.; Kato, T.; Hirano, H.; et al. Involvement of hepatitis C virus NS5A hyperphosphorylation mediated by casein kinase I-alpha in infectious virus production. J. Virol. 2014, 88, 7541–7555. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.T.; Tsai, T.Y.; Chao, C.H.; Lai, B.Y.; Wu Lee, Y.H. Y-Box Binding Protein 1 Stabilizes Hepatitis C Virus NS5A via Phosphorylation-Mediated Interaction with NS5A To Regulate Viral Propagation. J. Virol. 2015, 89, 11584–11602. [Google Scholar] [CrossRef] [PubMed]
- Stoecklin, G.; Kedersha, N. Relationship of GW/P-bodies with stress granules. Adv. Exp. Med. Biol. 2013, 768, 197–211. [Google Scholar] [PubMed]
- Shih, J.W.; Wang, W.T.; Tsai, T.Y.; Kuo, C.Y.; Li, H.K.; Wu Lee, Y.H. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 2012, 441, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Hess, D.; Eglinger, J.; Fritsch, A.W.; Kreysing, M.; Weinert, B.T.; Choudhary, C.; Matthias, P. Acetylation of intrinsically disordered regions regulates phase separation. Nat. Chem. Biol. 2019, 15, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Garaigorta, U.; Heim, M.H.; Boyd, B.; Wieland, S.; Chisari, F.V. Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress. J. Virol. 2012, 86, 11043–11056. [Google Scholar] [CrossRef] [PubMed]
- Pene, V.; Li, Q.; Sodroski, C.; Hsu, C.S.; Liang, T.J. Dynamic Interaction of Stress Granules, DDX3X, and IKK-alpha Mediates Multiple Functions in Hepatitis C Virus Infection. J. Virol. 2015, 89, 5462–5477. [Google Scholar] [CrossRef]
- Valentin-Vega, Y.A.; Wang, Y.D.; Parker, M.; Patmore, D.M.; Kanagaraj, A.; Moore, J.; Rusch, M.; Finkelstein, D.; Ellison, D.W.; Gilbertson, R.J.; et al. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci. Rep. 2016, 6, 25996. [Google Scholar] [CrossRef]
- Epling, L.B.; Grace, C.R.; Lowe, B.R.; Partridge, J.F.; Enemark, E.J. Cancer-associated mutants of RNA helicase DDX3X are defective in RNA-stimulated ATP hydrolysis. J. Mol. Biol. 2015, 427, 1779–1796. [Google Scholar] [CrossRef]
- Oh, S.; Flynn, R.A.; Floor, S.N.; Purzner, J.; Martin, L.; Do, B.T.; Schubert, S.; Vaka, D.; Morrissy, S.; Li, Y.; et al. Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress. Oncotarget 2016, 7, 28169–28182. [Google Scholar] [CrossRef]
- Hilliker, A.; Gao, Z.; Jankowsky, E.; Parker, R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell 2011, 43, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Yu, H.I.; Yang, M.H.; Tarn, W.Y. DDX3 Activates CBC-eIF3-Mediated Translation of uORF-Containing Oncogenic mRNAs to Promote Metastasis in HNSCC. Cancer Res. 2018, 78, 4512–4523. [Google Scholar] [CrossRef] [PubMed]
- Adjibade, P.; Grenier St-Sauveur, V.; Bergeman, J.; Huot, M.E.; Khandjian, E.W.; Mazroui, R. DDX3 regulates endoplasmic reticulum stress-induced ATF4 expression. Sci. Rep. 2017, 7, 13832. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.S.; Tsai, A.H.; Ho, Y.F.; Huang, S.Y.; Liu, Y.C.; Hwang, L.H. Stimulation of the Internal Ribosome Entry Site (IRES)-Dependent Translation of Enterovirus 71 by DDX3X RNA Helicase and Viral 2A and 3C Proteases. Front Microbiol. 2018, 9, 1324. [Google Scholar] [CrossRef] [PubMed]
- Linsalata, A.E.; He, F.; Malik, A.M.; Glineburg, M.R.; Green, K.M.; Natla, S.; Flores, B.N.; Krans, A.; Archbold, H.C.; Fedak, S.J.; et al. DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation. EMBO Rep. 2019, 20, e47498. [Google Scholar] [CrossRef] [PubMed]
- Chahar, H.S.; Chen, S.; Manjunath, N. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology 2013, 436, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.; Lee, S.; Windisch, M.P.; Ryu, W.S. DDX3 DEAD-box RNA helicase is a host factor that restricts hepatitis B virus replication at the transcriptional level. J. Virol. 2014, 88, 13689–13698. [Google Scholar] [CrossRef]
- Wang, H.; Ryu, W.S. Hepatitis B virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3: Implications for immune evasion. PLoS Pathog. 2010, 6, e1000986. [Google Scholar] [CrossRef]
- Yu, S.; Chen, J.; Wu, M.; Chen, H.; Kato, N.; Yuan, Z. Hepatitis B virus polymerase inhibits RIG-I- and Toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKepsilon and DDX3. J. Gen. Virol. 2010, 91, 2080–2090. [Google Scholar]
- Loret, S.; Guay, G.; Lippe, R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 2008, 82, 8605–8618. [Google Scholar] [CrossRef]
- Stegen, C.; Yakova, Y.; Henaff, D.; Nadjar, J.; Duron, J.; Lippe, R. Analysis of virion-incorporated host proteins required for herpes simplex virus type 1 infection through a RNA interference screen. PLoS ONE 2013, 8, e53276. [Google Scholar] [CrossRef] [PubMed]
- Khadivjam, B.; Stegen, C.; Hogue-Racine, M.A.; El Bilali, N.; Dohner, K.; Sodeik, B.; Lippe, R. The ATP-Dependent RNA Helicase DDX3X Modulates Herpes Simplex Virus 1 Gene Expression. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Cavignac, Y.; Lieber, D.; Laib Sampaio, K.; Madlung, J.; Lamkemeyer, T.; Jahn, G.; Nordheim, A.; Sinzger, C. The Cellular Proteins Grb2 and DDX3 Are Increased upon Human Cytomegalovirus Infection and Act in a Proviral Fashion. PLoS ONE 2015, 10, e0131614. [Google Scholar] [CrossRef] [PubMed]
- DeFilippis, V.R.; Alvarado, D.; Sali, T.; Rothenburg, S.; Fruh, K. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J. Virol. 2010, 84, 585–598. [Google Scholar] [CrossRef]
- Jorba, N.; Juarez, S.; Torreira, E.; Gastaminza, P.; Zamarreno, N.; Albar, J.P.; Ortin, J. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 2008, 8, 2077–2088. [Google Scholar] [CrossRef]
- Park, E.S.; Byun, Y.H.; Park, S.; Jang, Y.H.; Han, W.R.; Won, J.; Cho, K.C.; Kim, D.H.; Lee, A.R.; Shin, G.C.; et al. Co-degradation of interferon signaling factor DDX3 by PB1-F2 as a basis for high virulence of 1918 pandemic influenza. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Niu, Q.; Cheng, Y.; Wang, H.; Yan, Y.; Sun, J. Chicken DDX3X Activates IFN-beta via the chSTING-chIRF7-IFN-beta Signaling Axis. Front Immunol. 2019, 10, 822. [Google Scholar] [CrossRef]
- Diot, C.; Fournier, G.; Dos Santos, M.; Magnus, J.; Komarova, A.; van der Werf, S.; Munier, S.; Naffakh, N. Influenza A Virus Polymerase Recruits the RNA Helicase DDX19 to Promote the Nuclear Export of Viral mRNAs. Sci. Rep. 2016, 6, 33763. [Google Scholar] [CrossRef]
- Tsai, T.Y.; Wang, W.T.; Li, H.K.; Chen, W.J.; Tsai, Y.H.; Chao, C.H.; Wu Lee, Y.H. RNA helicase DDX3 maintains lipid homeostasis through upregulation of the microsomal triglyceride transfer protein by interacting with HNF4 and SHP. Sci. Rep. 2017, 7, 41452. [Google Scholar] [CrossRef]
- Xiang, N.; He, M.; Ishaq, M.; Gao, Y.; Song, F.; Guo, L.; Ma, L.; Sun, G.; Liu, D.; Guo, D.; et al. The DEAD-Box RNA Helicase DDX3 Interacts with NF-kappaB Subunit p65 and Suppresses p65-Mediated Transcription. PLoS ONE 2016, 11, e0164471. [Google Scholar] [CrossRef]
- Samir, P.; Kesavardhana, S.; Patmore, D.M.; Gingras, S.; Malireddi, R.K.S.; Karki, R.; Guy, C.S.; Briard, B.; Place, D.E.; Bhattacharya, A.; et al. DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 2019, 573, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Ivanova, O.N.; Kochetkov, S.N.; Starodubova, E.S.; Bartosch, B.; Isaguliants, M.G. Oxidative Stress during HIV Infection: Mechanisms and Consequences. Oxid. Med. Cell Longev. 2016, 2016, 8910396. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Tyurina, D.A.; Ivanova, O.N.; Kochetkov, S.N.; Bartosch, B.; Isaguliants, M.G. Oxidative stress, a trigger of hepatitis C and B virus-induced liver carcinogenesis. Oncotarget 2017, 8, 3895–3932. [Google Scholar] [CrossRef] [PubMed]
- Khomich, O.A.; Kochetkov, S.N.; Bartosch, B.; Ivanov, A.V. Redox Biology of Respiratory Viral Infections. Viruses 2018, 10, 392. [Google Scholar] [CrossRef] [PubMed]
- Aksenova, M.; Sybrandt, J.; Cui, B.; Sikirzhytski, V.; Ji, H.; Odhiambo, D.; Lucius, M.D.; Turner, J.R.; Broude, E.; Pena, E.; et al. Inhibition of the Dead Box RNA Helicase 3 Prevents HIV-1 Tat and Cocaine-Induced Neurotoxicity by Targeting Microglia Activation. J. Neuroimmune Pharmacol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Bol, G.M.; Vesuna, F.; Xie, M.; Zeng, J.; Aziz, K.; Gandhi, N.; Levine, A.; Irving, A.; Korz, D.; Tantravedi, S.; et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 2015, 7, 648–669. [Google Scholar] [CrossRef] [PubMed]
- Maga, G.; Falchi, F.; Garbelli, A.; Belfiore, A.; Witvrouw, M.; Manetti, F.; Botta, M. Pharmacophore modeling and molecular docking led to the discovery of inhibitors of human immunodeficiency virus-1 replication targeting the human cellular aspartic acid-glutamic acid-alanine-aspartic acid box polypeptide 3. J. Med. Chem. 2008, 51, 6635–6638. [Google Scholar] [CrossRef]
- Fazi, R.; Tintori, C.; Brai, A.; Botta, L.; Selvaraj, M.; Garbelli, A.; Maga, G.; Botta, M. Homology Model-Based Virtual Screening for the Identification of Human Helicase DDX3 Inhibitors. J. Chem. Inf. Model 2015, 55, 2443–2454. [Google Scholar] [CrossRef]
- Yedavalli, V.S.; Zhang, N.; Cai, H.; Zhang, P.; Starost, M.F.; Hosmane, R.S.; Jeang, K.T. Ring expanded nucleoside analogues inhibit RNA helicase and intracellular human immunodeficiency virus type 1 replication. J. Med. Chem. 2008, 51, 5043–5051. [Google Scholar] [CrossRef]
- Heerma van Voss, M.R.; van Diest, P.J.; Raman, V. Targeting RNA helicases in cancer: The translation trap. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 510–520. [Google Scholar] [CrossRef]
- Zhang, N.; Chen, H.M.; Koch, V.; Schmitz, H.; Liao, C.L.; Bretner, M.; Bhadti, V.S.; Fattom, A.I.; Naso, R.B.; Hosmane, R.S.; et al. Ring-expanded (“fat”) nucleoside and nucleotide analogues exhibit potent in vitro activity against flaviviridae NTPases/helicases, including those of the West Nile virus, hepatitis C virus, and Japanese encephalitis virus. J. Med. Chem. 2003, 46, 4149–4164. [Google Scholar] [CrossRef] [PubMed]
- Hosmane, R.S. Ring-expanded (“Fat”) nucleosides as broad-spectrum anticancer and antiviral agents. Curr. Top Med. Chem. 2002, 2, 1093–1109. [Google Scholar] [CrossRef] [PubMed]
- Riva, V.; Maga, G. From the magic bullet to the magic target: Exploiting the diverse roles of DDX3X in viral infections and tumorigenesis. Future Med. Chem. 2019, 11, 1357–1381. [Google Scholar] [CrossRef] [PubMed]
- Heerma van Voss, M.R.; Schrijver, W.A.; Ter Hoeve, N.D.; Hoefnagel, L.D.; Manson, Q.F.; van der Wall, E.; Raman, V.; van Diest, P.J. The prognostic effect of DDX3 upregulation in distant breast cancer metastases. Clin. Exp. Metastasis 2017, 34, 85–92. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, D.; Yang, Y.; Wang, X.; Zhao, X.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. A double-edged function of DDX3, as an oncogene or tumor suppressor, in cancer progression (Review). Oncology Rep. 2018, 39, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.C.; Chi, C.W.; Chau, G.Y.; Li, F.Y.; Tsai, Y.H.; Wu, J.C.; Wu Lee, Y.H. DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control. Oncogene 2006, 25, 1991–2003. [Google Scholar] [CrossRef]
- Bol, G.M.; Raman, V.; van der Groep, P.; Vermeulen, J.F.; Patel, A.H.; van der Wall, E.; van Diest, P.J. Expression of the RNA helicase DDX3 and the hypoxia response in breast cancer. PLoS ONE 2013, 8, e63548. [Google Scholar] [CrossRef]
- He, T.Y.; Wu, D.W.; Lin, P.L.; Wang, L.; Huang, C.C.; Chou, M.C.; Lee, H. DDX3 promotes tumor invasion in colorectal cancer via the CK1epsilon/Dvl2 axis. Sci. Rep. 2016, 6, 21483. [Google Scholar] [CrossRef]
- Heerma van Voss, M.R.; Vesuna, F.; Trumpi, K.; Brilliant, J.; Berlinicke, C.; de Leng, W.; Kranenburg, O.; Offerhaus, G.J.; Burger, H.; van der Wall, E.; et al. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget 2015, 6, 28312–28326. [Google Scholar]
- Cruciat, C.M.; Dolde, C.; de Groot, R.E.; Ohkawara, B.; Reinhard, C.; Korswagen, H.C.; Niehrs, C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science 2013, 339, 1436–1441. [Google Scholar] [CrossRef]
- Chen, H.H.; Yu, H.I.; Cho, W.C.; Tarn, W.Y. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene 2015, 34, 2790–2800. [Google Scholar] [CrossRef] [PubMed]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.H.; Chen, C.M.; Cheng, P.L.; Shih, J.W.; Tsou, A.P.; Lee, Y.H. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006, 66, 6579–6588. [Google Scholar] [CrossRef] [PubMed]
- Heerma van Voss, M.R.; Vesuna, F.; Bol, G.M.; Meeldijk, J.; Raman, A.; Offerhaus, G.J.; Buerger, H.; Patel, A.H.; van der Wall, E.; van Diest, P.J.; et al. Nuclear DDX3 expression predicts poor outcome in colorectal and breast cancer. Onco. Targets Ther. 2017, 10, 3501–3513. [Google Scholar] [CrossRef] [PubMed]
- Heerma van Voss, M.R.; Kammers, K.; Vesuna, F.; Brilliant, J.; Bergman, Y.; Tantravedi, S.; Wu, X.; Cole, R.N.; Holland, A.; van Diest, P.J.; et al. Global Effects of DDX3 Inhibition on Cell Cycle Regulation Identified by a Combined Phosphoproteomics and Single Cell Tracking Approach. Transl. Oncol. 2018, 11, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Wilky, B.A.; Kim, C.; McCarty, G.; Montgomery, E.A.; Kammers, K.; DeVine, L.R.; Cole, R.N.; Raman, V.; Loeb, D.M. RNA helicase DDX3: A novel therapeutic target in Ewing sarcoma. Oncogene 2016, 35, 2574–2583. [Google Scholar] [CrossRef] [PubMed]
- Heerma van Voss, M.R.; Vesuna, F.; Bol, G.M.; Afzal, J.; Tantravedi, S.; Bergman, Y.; Kammers, K.; Lehar, M.; Malek, R.; Ballew, M.; et al. Targeting mitochondrial translation by inhibiting DDX3: A novel radiosensitization strategy for cancer treatment. Oncogene 2018, 37, 63–74. [Google Scholar] [CrossRef]
- Xie, M.; Vesuna, F.; Tantravedi, S.; Bol, G.M.; Heerma van Voss, M.R.; Nugent, K.; Malek, R.; Gabrielson, K.; van Diest, P.J.; Tran, P.T.; et al. RK-33 Radiosensitizes Prostate Cancer Cells by Blocking the RNA Helicase DDX3. Cancer Res. 2016, 76, 6340–6350. [Google Scholar] [CrossRef]

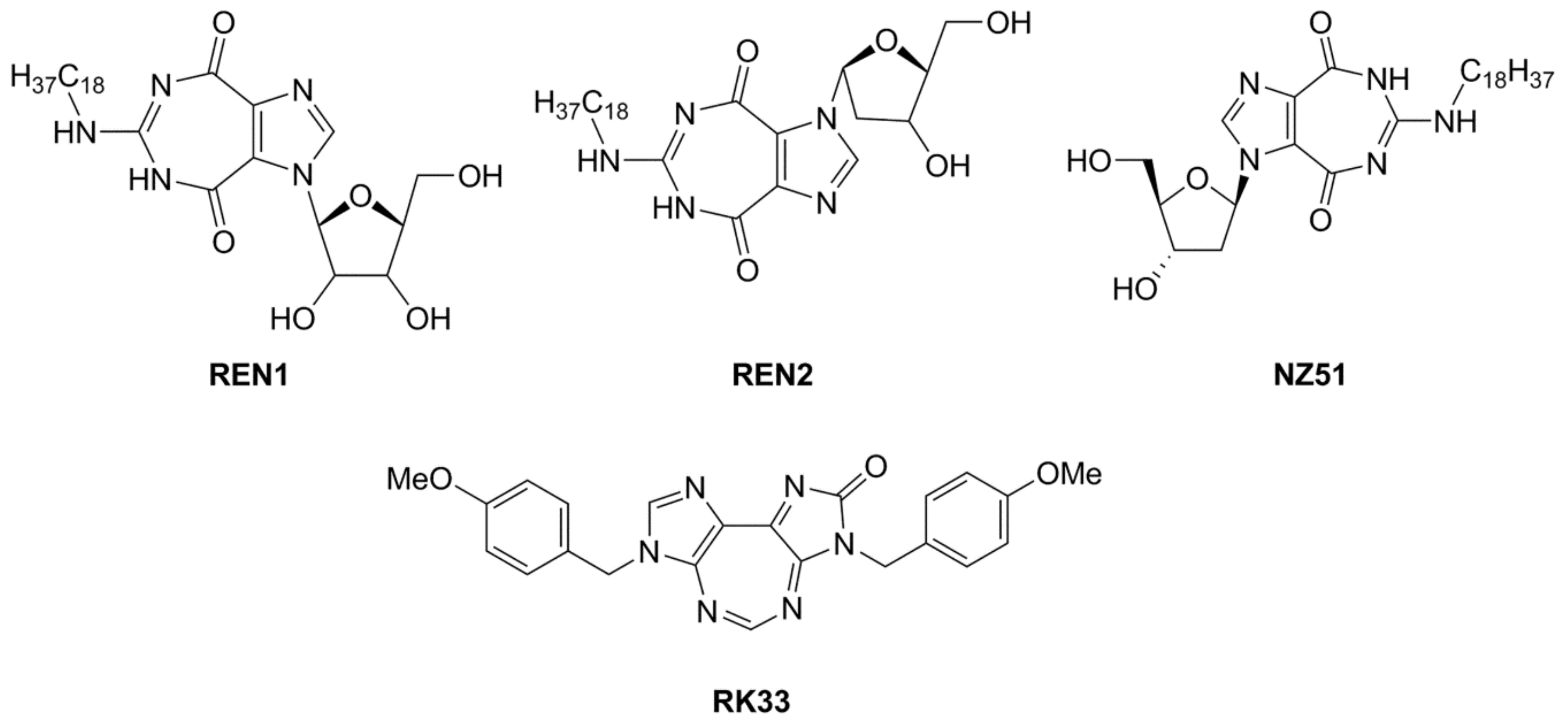
| General Structure | |
|---|---|
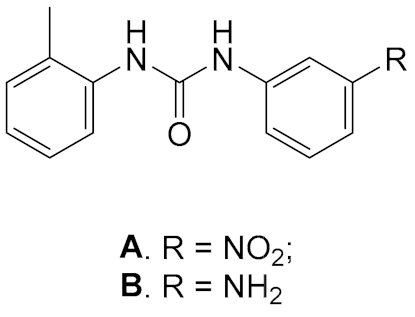 |  |
| A. IC50 1 μM; B. IC50 6 μM | 1. IC50 0.3 μM; 2. IC50 0.98 μM; 3. IC50 3.36 μM |
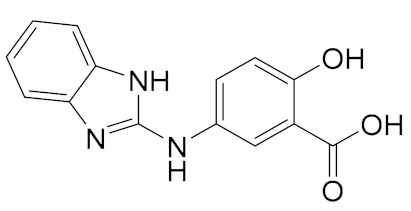 | 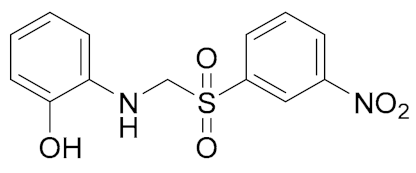 | 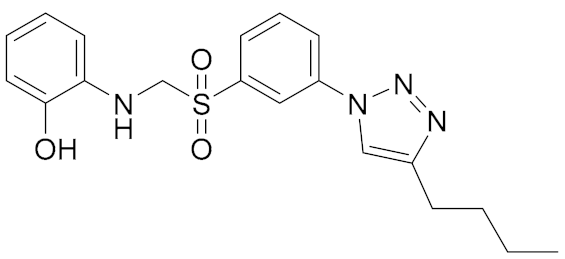 |
| 4. IC50 0.2 μM | 5. IC50 0.3 μM | 6. IC50 0.3 μM |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kukhanova, M.K.; Karpenko, I.L.; Ivanov, A.V. DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs. Molecules 2020, 25, 1015. https://doi.org/10.3390/molecules25041015
Kukhanova MK, Karpenko IL, Ivanov AV. DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs. Molecules. 2020; 25(4):1015. https://doi.org/10.3390/molecules25041015
Chicago/Turabian StyleKukhanova, Marina K., Inna L. Karpenko, and Alexander V. Ivanov. 2020. "DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs" Molecules 25, no. 4: 1015. https://doi.org/10.3390/molecules25041015
APA StyleKukhanova, M. K., Karpenko, I. L., & Ivanov, A. V. (2020). DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs. Molecules, 25(4), 1015. https://doi.org/10.3390/molecules25041015







