Parallel Strategy Increases the Thermostability and Activity of Glutamate Decarboxylase
Abstract
1. Introduction
2. Results
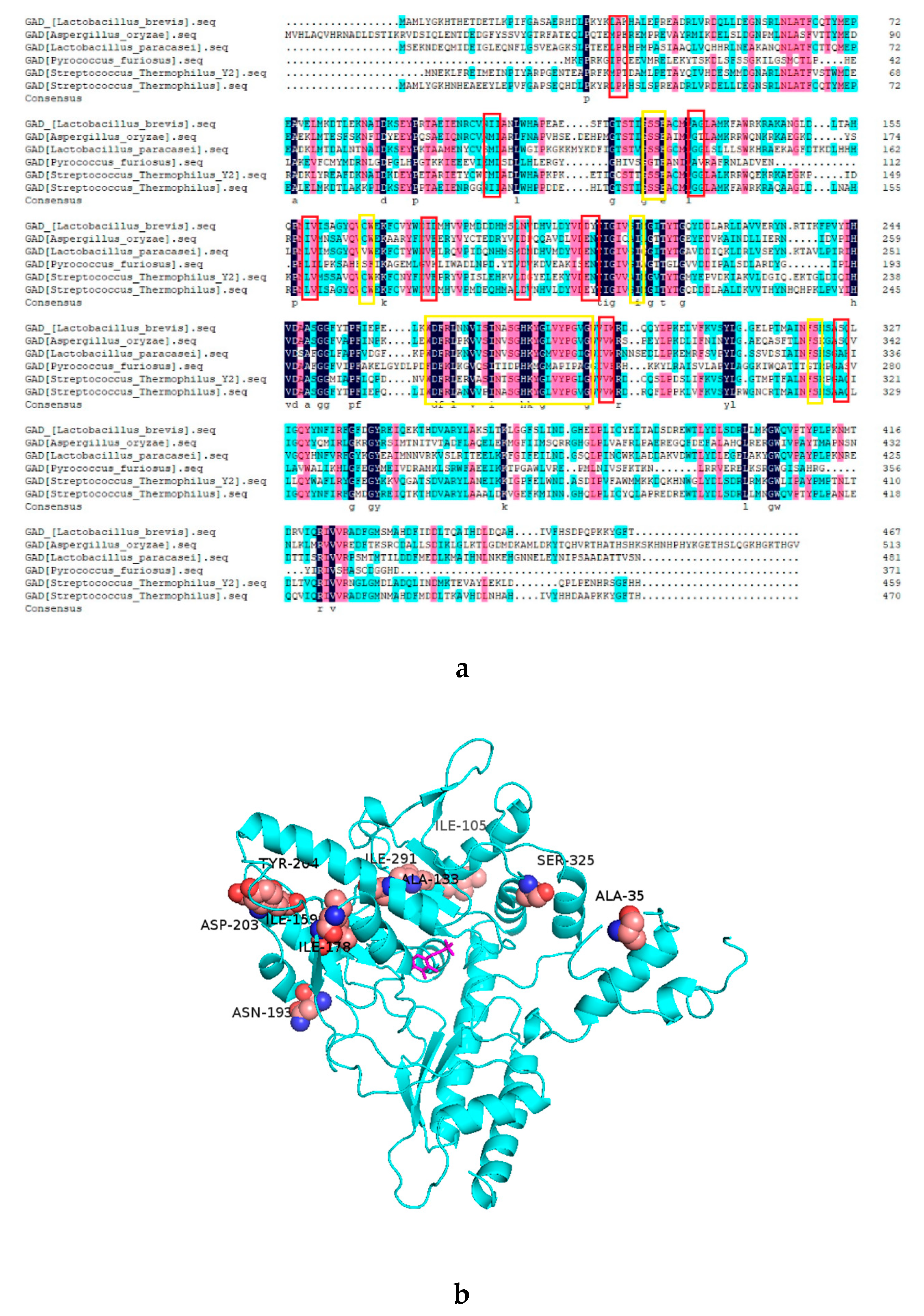
2.1. Analysis of the Mutant GAD by Sequence Alignment
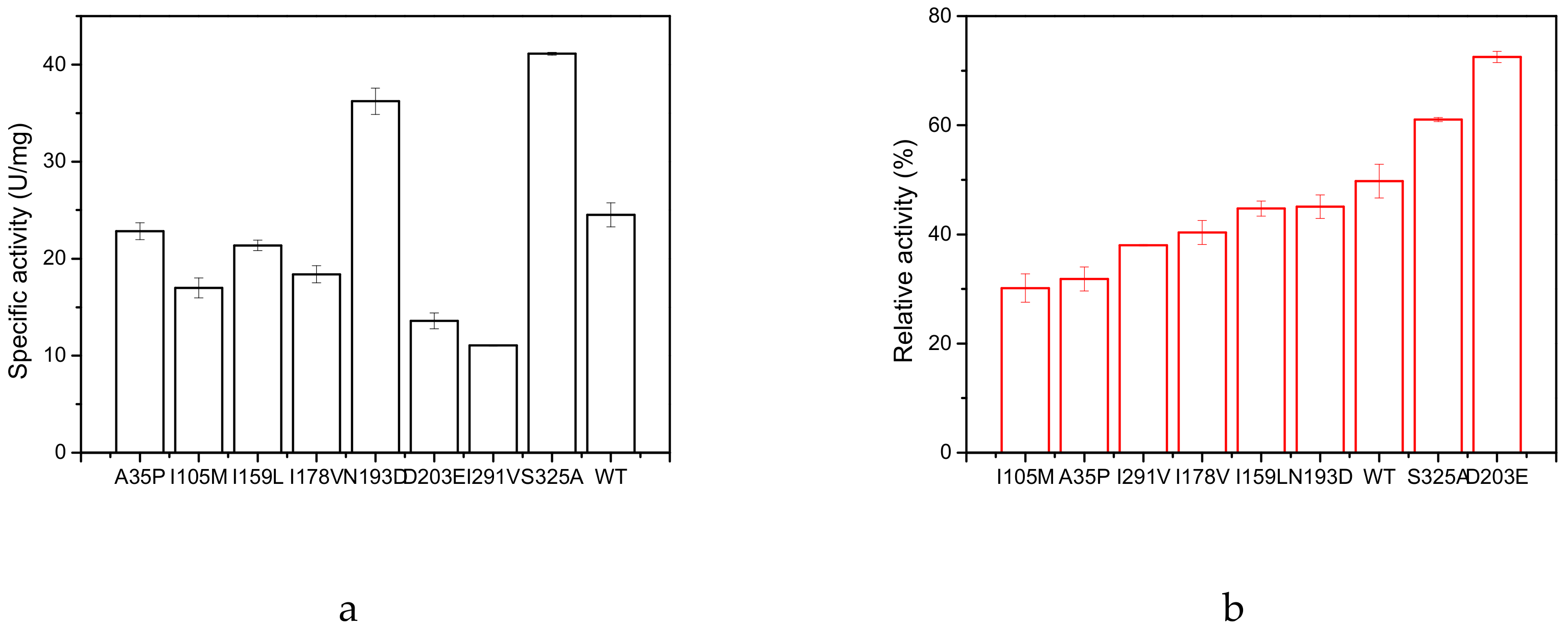
2.2. Specific Activity and Kinetic Constants of GAD and its Mutants
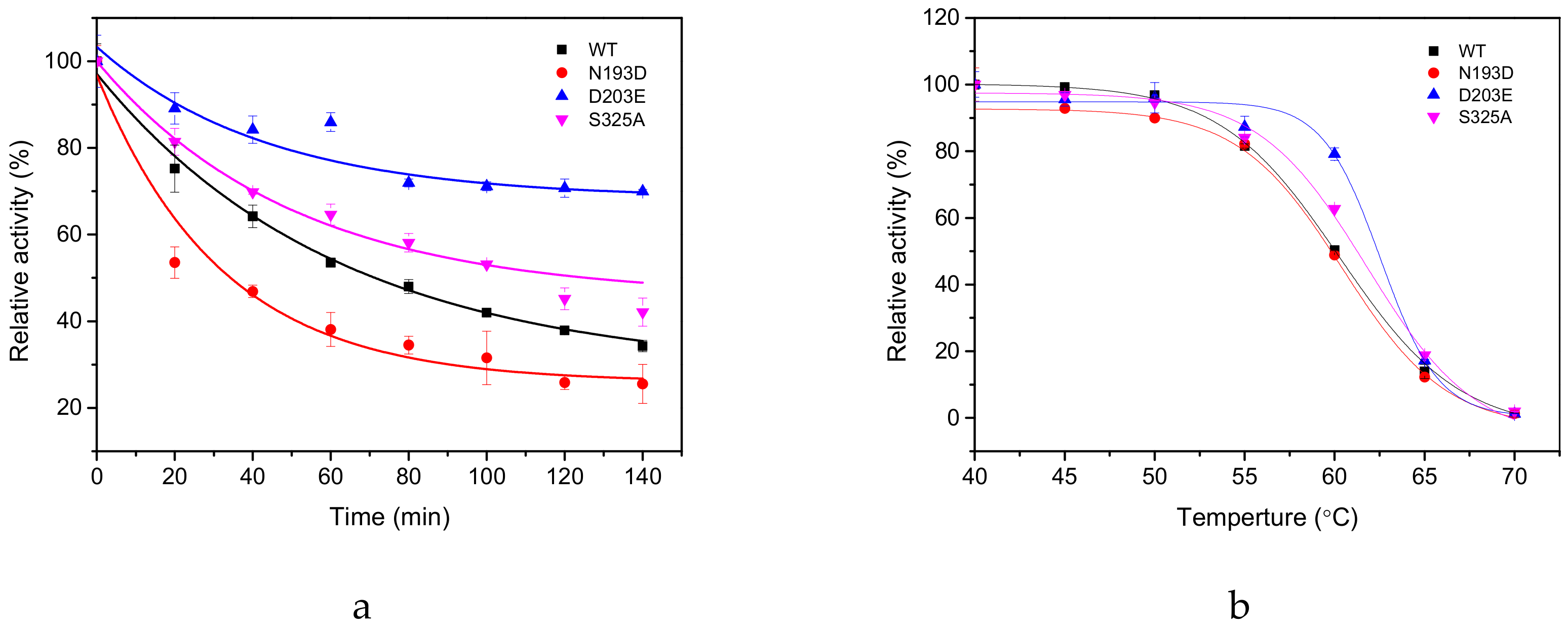
2.3. Thermal Stability of GAD and its Mutants
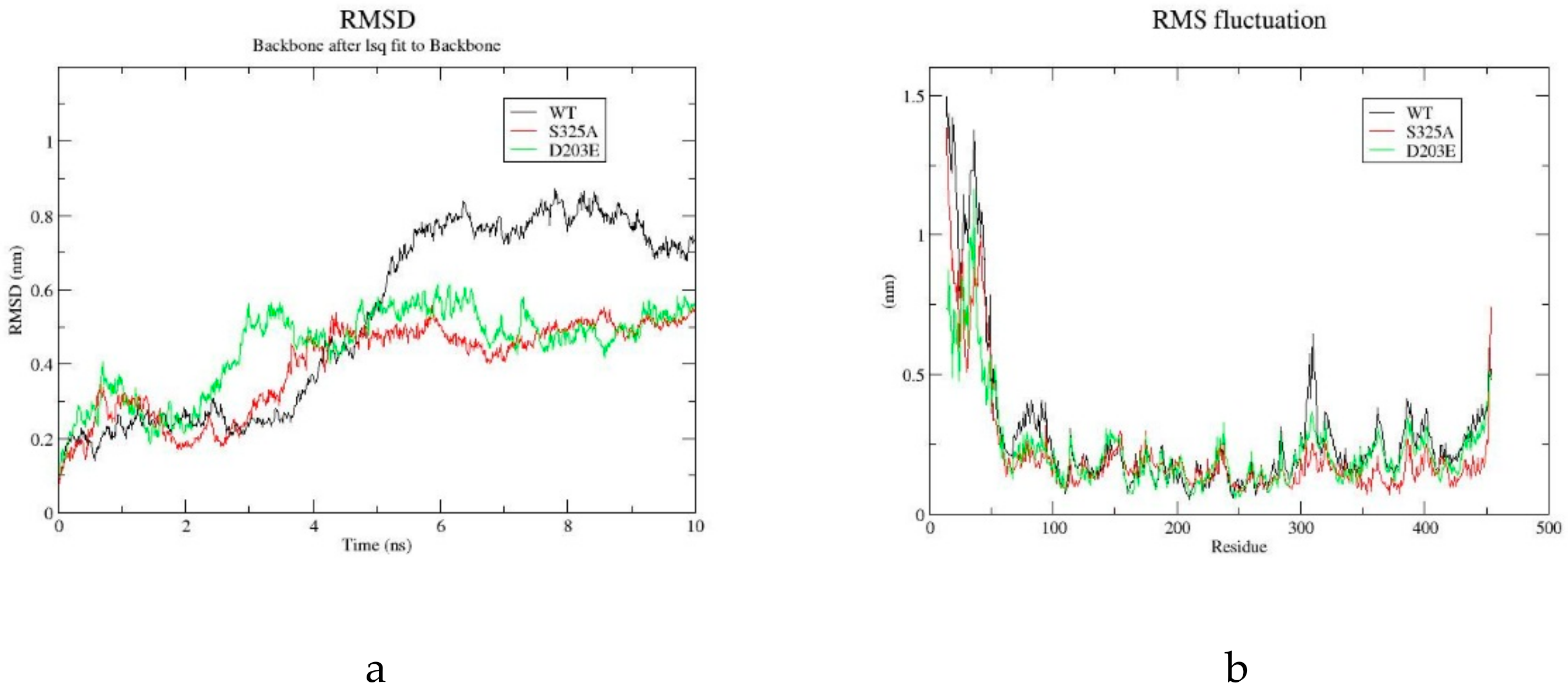
2.4. Molecular Dynamics Simulation of GAD and its Mutants
3. Discussion
4. Materials and Methods
4.1. Strains and Plasmids
4.2. Rational Design
4.3. Construction of Mutants
4.4. Enzyme Expression and Purification
4.5. Enzymatic Parameters of Wild-Type Enzyme and Mutant Enzymes
4.6. Molecular Dynamics Simulation of WT and its Mutants
Author Contributions
Funding
Conflicts of Interest
References
- Roberts, E.; Frankel, S. Glutamic Acid Decarboxylase in Brain. J. Biol. Chem. 1951, 188, 789–795. [Google Scholar]
- Sze, P. L-Glutamate Decarboxylase. Adv. Exp. Med. Biol. 1979, 123, 59–78. [Google Scholar] [PubMed]
- Wong, C.G.; Bottiglieri, T. GABA, Gamma-Hydroxybutyric Acid, and Neurological Disease. Ann. Neurol. 2003, 54, S3–S12. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Shirai, T.; Ochiai, H.; Kasao, M.; Hayakawa, K.; Kimura, M.; Sansawa, H. Blood-Pressure-Lowering Effect of A Novel Fermented Milk Containing Gamma-Aminobutyric Acid (GABA) in Mild Hypertensives. Eur. J. Clin. Nutr. 2003, 57, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Leventhal, A.G.; Yongchang, W.; Mingliang, P.; Yifeng, Z.; Yuanye, M. GABA and its Agonists Improved Visual Cortical Function in Senescent Monkeys. Science 2003, 300, 812–815. [Google Scholar] [CrossRef] [PubMed]
- Kaila, K.; Ruusuvuori, E.; Seja, P.; Voipio, J.; Puskarjov, M. GABA Actions and Ionic Plasticity in Epilepsy. Curr. Opin. Neurobiol. 2014, 26, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cao, Y. Lactic Acid Bacterial Cell Factories for Gamma-Aminobutyric Acid. Amino Acids 2010, 39, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Fan, E.; Huang, J.; Hu, S.; Mei, L. Cloning, Sequencing and Expression of a Glutamate Decarboxylase Gene from the GABA-Producing Strain Lactobacillus brevis CGMCC 1306. Ann. Microbiol. 2012, 62, 689–698. [Google Scholar] [CrossRef]
- Chen, K.Q.; Arnold, F.H. Enzyme Engineering for Nonaqueous Solvents: Random Mutagenesis to Enhance Activity of Subtilisin E in Polar Organic Media. Biotechnology 1991, 9, 1073–1077. [Google Scholar] [CrossRef]
- Arnold, F.H. Engineering Proteins for Nonnatural Environments. The FASEB Journal 1993, 7, 744–749. [Google Scholar] [CrossRef]
- Stemmer, C.W.P. Rapid Evolution of a Protein in Vitro by DNA Shuffling. Nature 1994, 370, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Paredes, D.I.; Watters, K.; Pitman, D.J.; Bystroff, C.; Dordick, J.S. Comparative Void-Volume Analysis of Psychrophilic and Mesophilic Enzymes: Structural Bioinformatics of Psychrophilic Enzymes Reveals Sources of Core Flexibility. BMC Struct. Biol. 2011, 11, 42. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N. Contribution of the Hydrophobic Effect to Globular Protein Stability. J. Mol. Biol. 1992, 226, 29–35. [Google Scholar] [CrossRef]
- Li, W.F.; Zhou, X.X.; Lu, P. Structural Features of Thermozymes. Biotechnol. Adv. 2005, 23, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, S.; Varadarajan, R. Elucidation of Factors Responsible for Enhanced Thermal Stability of Proteins: A Structural Genomics Based Study. Biochemistry 2002, 41, 8152–8161. [Google Scholar] [CrossRef] [PubMed]
- Pantoliano, M.W.; Whitlow, M.; Wood, J.F.; Dodd, S.W.; Hardman, K.D.; Rollence, M.L.; Bryan, P.N. Large Increases in General Stability for Subtilisin BPN’ through Incremental Changes in the Free Energy of Unfolding. Biochemistry 1989, 28, 7205–7213. [Google Scholar] [CrossRef] [PubMed]
- Zhizhuang, X.; Hélène, B.; Stephan, G.; Manon, B.; Marie-Line, G.; David, S.; Traian, S.; Miroslaw, C.; Lau, P.C.K. Improvement of the Thermostability and Activity of a Pectate Lyase by Single Amino Acid Substitutions, Using a Strategy Based on Melting-Temperature-Guided Sequence Alignment. Appl. Environ. Microbiol. 2008, 74, 1183–1189. [Google Scholar]
- Max, K.E.; Wunderlich, M.; Roske, Y.; Schmid, F.X.; Heinemann, U. Optimized Variants of the Cold Shock Protein from in Vitro Selection: Structural Basis of their High Thermostability. J. Mol. Biol. 2007, 369, 1087–1097. [Google Scholar] [CrossRef]
- Lehmann, M.; Pasamontes, L.; Lassen, S.F.; Wyss, M. The Consensus Concept for Thermostability Engineering of Proteins. Biochim. Biophys. Acta 2000, 1543, 408–415. [Google Scholar] [CrossRef]
- Kazlauskas, R. Engineering More Stable Proteins. Chem. Soc. Rev. 2018, 47, 9026–9045. [Google Scholar] [CrossRef]
- Bommarius, A.S.; Blum, J.K.; Abrahamson, M.J. Status of Protein Engineering for Biocatalysts: How to Design an Industrially Useful Biocatalyst. Curr. Opin. Chem. Biol. 2011, 15, 194–200. [Google Scholar] [CrossRef]
- Pikkemaat, M.G.; Linssen, A.B.M.; Berendsen, H.J.C.; Janssen, D.B. Molecular Dynamics Simulations as A Tool for Improving Protein Stability. Protein Eng. 2002, 15, 185–192. [Google Scholar] [CrossRef]
- Matthews, B.W.; Nicholson, H.; Becktel, W.J. Enhanced Protein Thermostability from Site-Directed Mutations that Decrease the Entropy of Unfolding. Proc. Natl. Acad. Sci. USA 1987, 84, 6663–6667. [Google Scholar] [CrossRef]
- Takano, K.; Higashi, R.; Okada, J.; Mukaiyama, A.; Tadokoro, T.; Koga, Y.; Kanaya, S. Proline Effect on the Thermostability and Slow Unfolding of a Hyperthermophilic Protein. J. Biochem. 2009, 145, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Borjesson, U.; Engkvist, O.; Kogej, T.; Svensson, M.A.; Blomberg, N.; Weigelt, D.; Burrows, J.N.; Lange, T. ProSAR: A New Methodology for Combinatorial Library Design. J. Chem. Inf. Model. 2009, 49, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Cadet, F.; Fontaine, N.; Li, G.; Sanchis, J.; Ng Fuk Chong, M.; Pandjaitan, R.; Vetrivel, I.; Offmann, B.; Reetz, M.T. A Machine Learning Approach for Reliable Prediction of Amino Acid Interactions and its Application in the Directed Evolution of Enantioselective Enzymes. Sci. Rep. 2018, 8, 16757. [Google Scholar] [CrossRef] [PubMed]
- Jun, C.; Joo, J.C.; Lee, J.H.; Kim, Y.H. Thermostabilization of Glutamate Decarboxylase B from Escherichia Coli by Structure-Guided Design of its pH-Responsive N-Terminal Interdomain. J. Biotechnol. 2014, 174, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Ke, P.; Huang, J.; Hu, S.; Zhao, W.; Changjiang, L.; Yu, K.; Lei, Y.; Wang, J.; Mei, L. Enhancing Glutamate Decarboxylase Activity by Site-Directed Mutagenesis: An Insight from Ramachandran Plot. Chin. J. Biotechnol. 2016, 32, 31. [Google Scholar]
- Fang, H.; Lu, C.; Hua, Y.; Hu, S.; Zhao, W.; Fang, W.; Song, K.; Huang, J.; Mei, L. Increasing the Thermostability of Glutamate Decarboxylase from Lactobacillus Brevis by Introducing Proline. Sheng Wu Chin. J. Biotechnol. 2019, 35, 636–646. [Google Scholar]
- Pete, H.; Snow, C.D.; Smith, M.A.; Xinlin, Y.; Arvind, K.; Kevin, B.; Alan, V.; Sridhar, G.; Jeremy, M.; Arnold, F.H. SCHEMA Recombination of a Fungal Cellulase Uncovers a Single Mutation that Contributes Markedly to Stability. J. Biol. Chem. 2009, 284, 26229. [Google Scholar]
- Huang, J.; Fang, H.; Gai, Z.C.; Mei, J.Q.; Li, J.N.; Hu, S.; Lv, C.J.; Zhao, W.R.; Mei, L.H. Lactobacillus Brevis CGMCC 1306 Glutamate Decarboxylase: Crystal Structure and Functional Analysis. Biochem. Biophys. Res. Commun. 2018, 503, 1703–1709. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX Web Server: An Online Force Field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Kellis, J.T.; Nyberg, K.; S˘ail, D.a.; Fersht, A.R. Contribution of Hydrophobic Interactions to Protein Stability. Nature 1988, 333, 784–786. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Fu, H.; Fryar, K.L.; Landua, J.; Trevino, S.R.; Shirley, B.A.; Hendricks, M.M.; Iimura, S.; Gajiwala, K.; Scholtz, J.M.; et al. Contribution of Hydrophobic Interactions to Protein Stability. J. Mol. Biol. 2011, 408, 514–528. [Google Scholar] [CrossRef] [PubMed]
- Gumulya, Y.; Baek, J.-M.; Wun, S.-J.; Thomson, R.E.S.; Harris, K.L.; Hunter, D.J.B.; Behrendorff, J.B.Y.H.; Kulig, J.; Zheng, S.; Wu, X.; et al. Engineering Highly Functional Thermostable Proteins Using Ancestral Sequence Reconstruction. Nat. Catal. 2018, 1, 878–888. [Google Scholar] [CrossRef]
- Warden, A.C.; Williams, M.; Peat, T.S.; Seabrook, S.A.; Newman, J.; Dojchinov, G.; Haritos, V.S. Rational Engineering of A Mesohalophilic Carbonic Anhydrase to An Extreme Halotolerant Biocatalyst. Nat. Commun. 2015, 6, 10278. [Google Scholar] [CrossRef]
- Lin, L.; Hu, S.; Yu, K.; Huang, J.; Yao, S.; Lei, Y.; Hu, G.; Mei, L. Enhancing the Activity of Glutamate Decarboxylase from Lactobacillus brevis by Directed Evolution. Chin. J. Chem. Eng. 2014, 22, 1322–1327. [Google Scholar] [CrossRef]
- Komatsuzaki, N.; Nakamura, T.; Kimura, T.; Shima, J. Characterization of Glutamate Decarboxylase from a High Gamma-Aminobutyric Acid (GABA)-Producer, Lactobacillus Paracasei. Biosci. Biotechnol. Biochem. 2008, 72, 278–285. [Google Scholar] [CrossRef]
- Yang, S.-Y.; Lin, Q.; Lu, Z.-X.; Lü, F.-X.; Bie, X.-M.; Zou, X.-K.; Sun, L.-J. Characterization of A Novel Glutamate Decarboxylase from Streptococcus Salivarius ssp. Thermophilus Y2. J. Chem. Technol. Biotechnol. 2008, 83, 855–861. [Google Scholar] [CrossRef]
- Lee, E.-S.; Kim, H.-W.; Kim, D.-E.; Kim, Y.-H.; Nam, S.-W.; Kim, B.-W.; Jeon, S.-J. Gene Expression and Characterization of Thermostable Glutamate Decarboxylase from Pyrococcus Furiosus. Biotechnol. Bioprocess Eng. 2013, 18, 375–381. [Google Scholar] [CrossRef]
- Tsuchiya, K.; Nishimura, K.; Iwahara, M. Purification and Characterization of Glutamate Decarboxylase from Aspergillus Oryzae. Food Sci. Technol. Res. 2003, 9, 283–287. [Google Scholar] [CrossRef]
- Qian, L.; Shengyuan, Y.; Fengxia, L.; Zhaoxin, L.; Xiaomei, B.; Yang, J.; Xiaokui, Z. Cloning and Expression of Glutamate Decarboxylase Gene from Streptococcus Thermophilus Y2. J. Gen. Appl. Microbiol. 2009, 55, 305. [Google Scholar]
- Chen, X.; Li, D.; Lü, J.; Fang, F. Determination of Gamma-Aminobutyric Acid and Glutamic Acid in Human Cerebrospinal Fluid by high Performance Liquid Chromatography. Chin. J. Chromatogr. 1997, 15, 237–239. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |

| Mutants | ΔΔG unfoldT = 323 k | ΔΔG unfoldT = 333 k | ΔΔG unfoldT = 343 k | Mutants | ΔΔG unfoldT = 323 k | ΔΔG unfoldT = 333 k | ΔΔG unfoldT = 343 k |
|---|---|---|---|---|---|---|---|
| D203E | −1.33 | −1.34 | −1.35 | I178V | 0.24 | 0.22 | 0.20 |
| N193D | −0.86 | −0.97 | −1.13 | I291V | 0.84 | 0.81 | 0.79 |
| S325A | −0.88 | −0.88 | −0.87 | I159L | 0.89 | 0.84 | 0.81 |
| A35P | −0.76 | −0.81 | −0.82 | A133G | 1.59 | 1.61 | 1.63 |
| I105M | −0.53 | −0.53 | −0.52 | Y204N | 2.17 | 2.22 | 2.19 |
| Name | Specific Activity (U/mg) | KM (mM) | kcat (s-1) | kcat/KM (s−1·mM−1) |
|---|---|---|---|---|
| N193D | 36.28 | 34.35 | 137.56 | 4.00 |
| D203E | 13.64 | 26.60 | 40.21 | 1.51 |
| S325A | 41.12 | 42.39 | 176.19 | 4.16 |
| WT | 24.57 | 39.72 | 102.12 | 2.57 |
| Source | Optimum Temperature | Optimum pH | KM (mM) | Molecular Mass | Sequence Similarity |
|---|---|---|---|---|---|
| GAD(B1B389) [38] Lactobacillus paracasei | 50 °C | 5.0 | 5.0 | 57 kDa | 48.2% |
| GAD(Q0GE18) [39] Streptococcus salivarius ssp. Thermophilus Y2 | 55 °C | 4.0 | 2.3 | 52.6 kDa | 46.5% |
| GAD(Q8U1P6) [40] Pyrococcus furiosus | 75 °C | 6.0 | 2.22 | 41 kDa | 23.8% |
| GAD [41] Aspergillus oryzae | 60 °C | 5.5 | 13.0 | 48 KDa | 43.0% |
| GAD [42] Streptococcus thermophilus | 52 °C | 4.2 | 5.0 | 53 KDa | 72.1% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.-F.; Hu, S.; Zhao, W.-R.; Huang, J.; Mei, J.-Q.; Mei, L.-H. Parallel Strategy Increases the Thermostability and Activity of Glutamate Decarboxylase. Molecules 2020, 25, 690. https://doi.org/10.3390/molecules25030690
Zhang Q-F, Hu S, Zhao W-R, Huang J, Mei J-Q, Mei L-H. Parallel Strategy Increases the Thermostability and Activity of Glutamate Decarboxylase. Molecules. 2020; 25(3):690. https://doi.org/10.3390/molecules25030690
Chicago/Turabian StyleZhang, Qing-Fei, Sheng Hu, Wei-Rui Zhao, Jun Huang, Jia-Qi Mei, and Le-He Mei. 2020. "Parallel Strategy Increases the Thermostability and Activity of Glutamate Decarboxylase" Molecules 25, no. 3: 690. https://doi.org/10.3390/molecules25030690
APA StyleZhang, Q.-F., Hu, S., Zhao, W.-R., Huang, J., Mei, J.-Q., & Mei, L.-H. (2020). Parallel Strategy Increases the Thermostability and Activity of Glutamate Decarboxylase. Molecules, 25(3), 690. https://doi.org/10.3390/molecules25030690




