Host-Guest Complexation of Oxaliplatin and Para-Sulfonatocalix[n]Arenes for Potential Use in Cancer Therapy
Abstract
1. Introduction
2. Experimental and Computational Studies
2.1. Chemicals and Reagents
2.2. Instrumentation
2.3. Construction of Calibration Graphs (DD1 Method)
2.4. Cell Viability Assay
Sulforhodamine B Colorimetric Assay (SRB)
2.5. Computational Details
3. Results
3.1. H NMR Spectroscopy
3.2. UV-Vis Spectroscopy
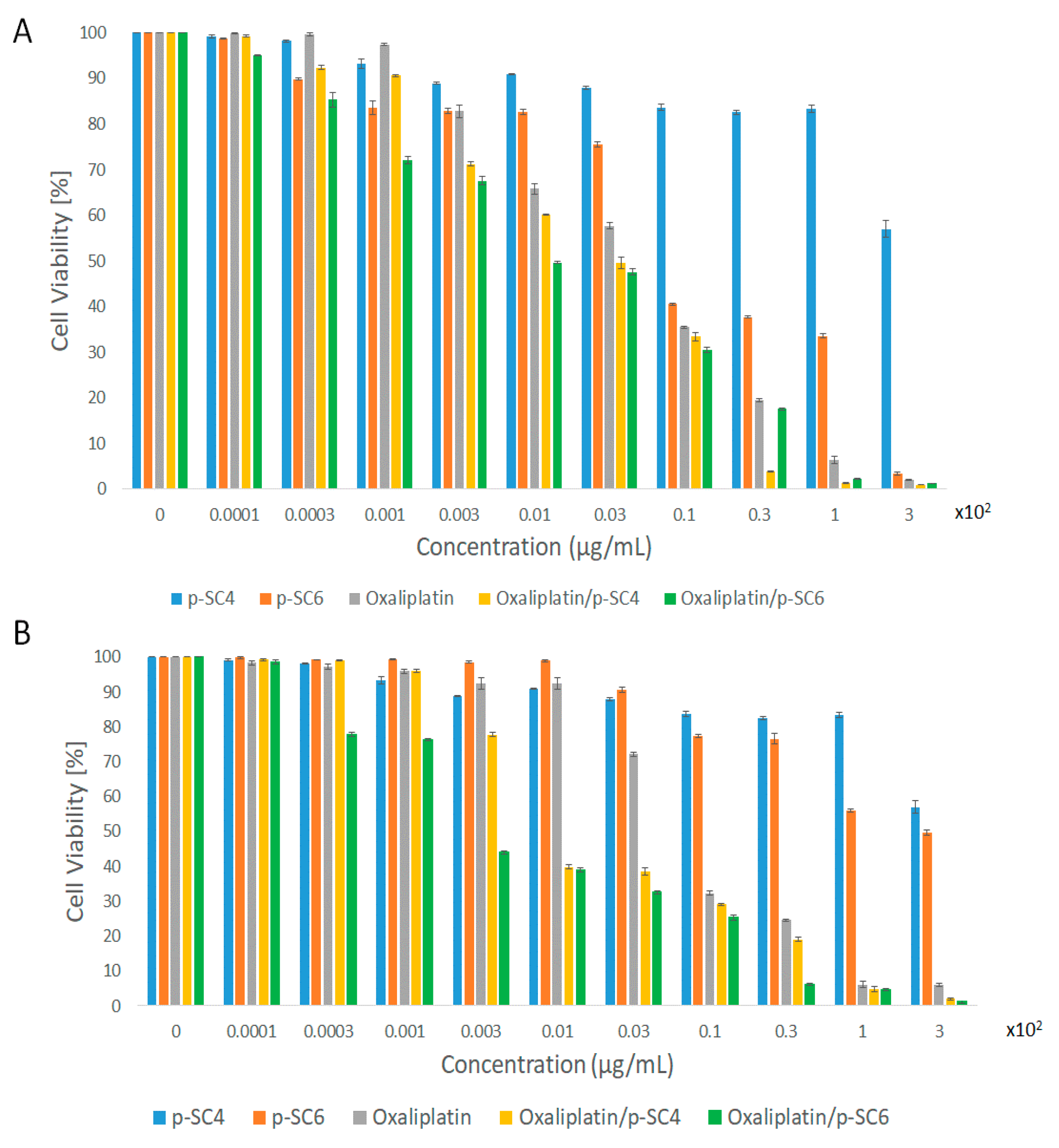
3.3. In Vitro Cell Viability Assay
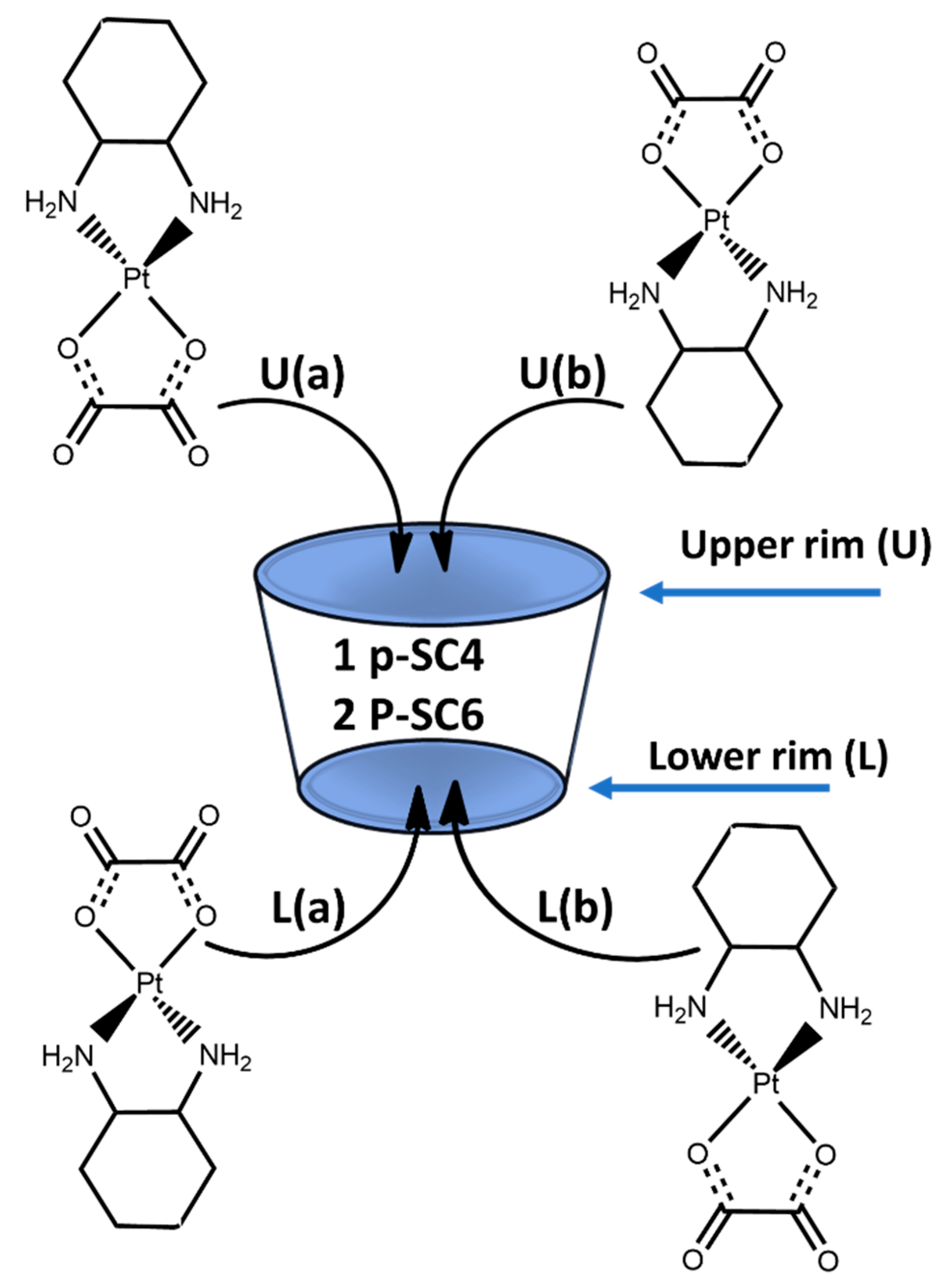
3.4. Computational Results
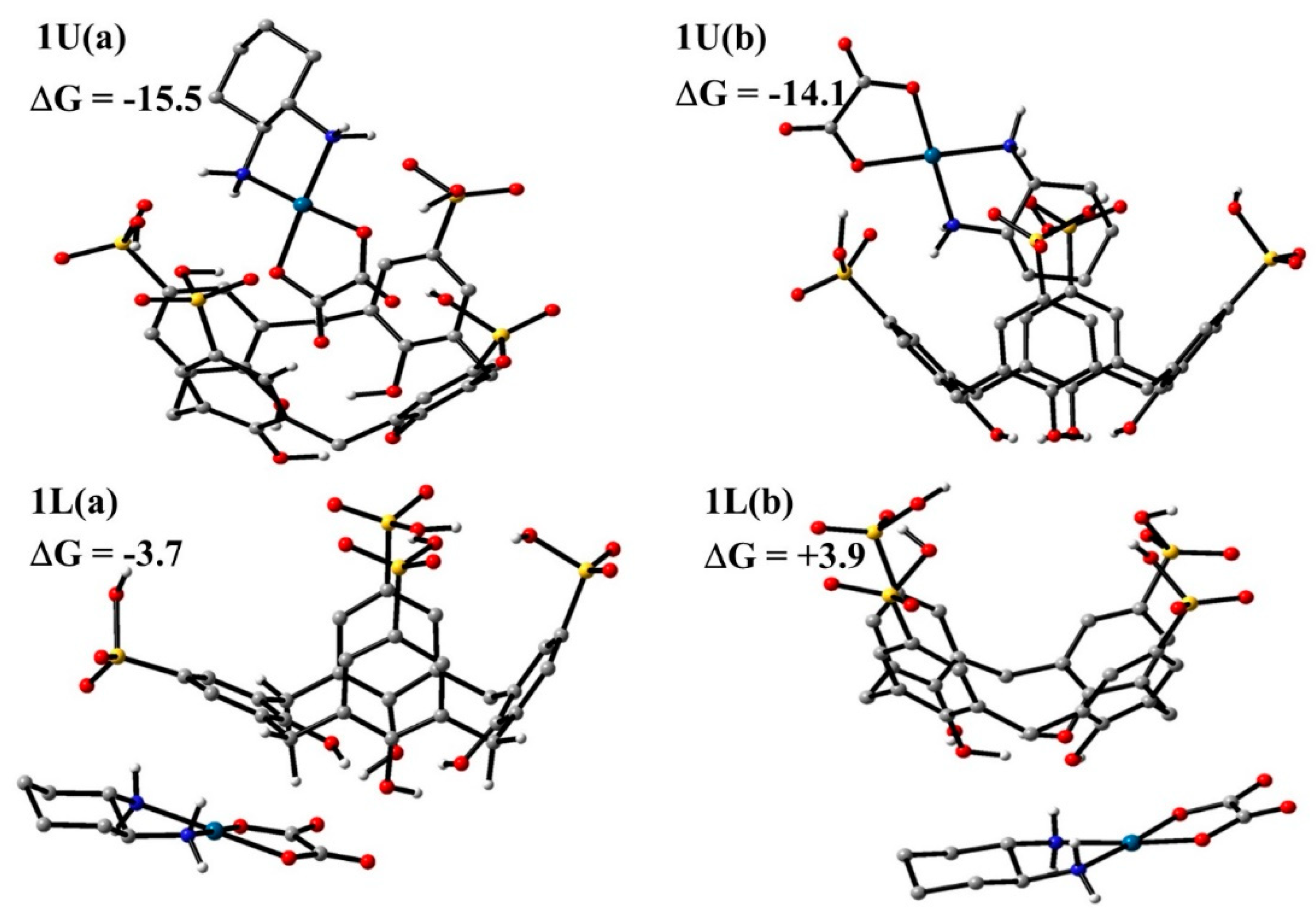
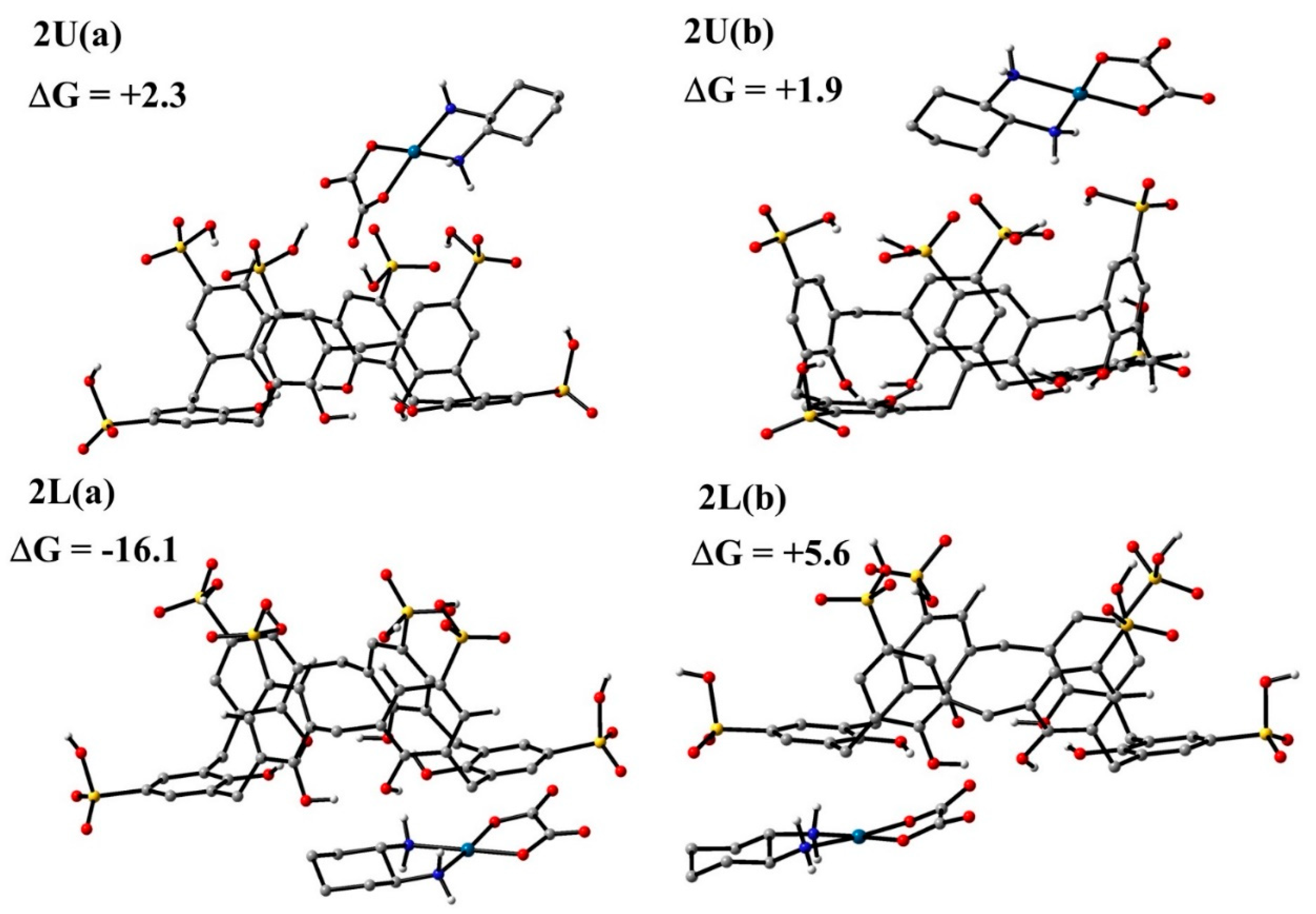
3.4.1. DFT Calculations
3.4.2. ADMET Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ritacco, I.; Al Assy, M.; Abd El-Rahman, M.K.; Fahmy, S.A.; Russo, N.; Shoeib, T.; Sicilia, E. Hydrolysis in Acidic Environment and Degradation of Satraplatin: A Joint Experimental and Theoretical Investigation. Inorg. Chem. 2017, 56, 6013–6026. [Google Scholar] [CrossRef] [PubMed]
- Harper, B.W.; Krause-Heuer, A.M.; Grant, M.P.; Manohar, M.; Garbutcheon-Singh, K.B.; Aldrich-Wright, J.R. Advances in platinum chemotherapeutics. Chem. A Eur. J. 2010, 16, 7064–7077. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.; Giandomenico, C.M. Current Status of Platinum-Based Antitumor Drugs. Chem. Rev. 1999, 99, 2451–2466. [Google Scholar] [CrossRef]
- Jakupec, M.A.; Galanski, M.; Keppler, B.K. Tumour-inhibiting platinum complexes-state of the art and future perspectives. Rev. Physiol. Biochem. Pharmacol. 2003, 146, 1–54. [Google Scholar] [PubMed]
- Wheate, N.J.; Walker, S.; Craig, G.E.; Craig, G.E.; Oun, R. The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 2010, 39, 8113–8127. [Google Scholar] [CrossRef]
- Arango, D.; Wilson, A.J.; Shi, Q.; Corner, G.A.; Arañes, M.J.; Nicholas, C.; Lesser, M.; Mariadason, J.M.; Augenlicht, L.H. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br. J. Cancer 2004, 91, 1931–1946. [Google Scholar] [CrossRef]
- Boulikas, T.; Pantos, A.; Bellis, E.; Christofis, P. Designing platinum compounds in cancer: Structures and mechanisms (review). Cancer Therapy 2007, 5, 537–583. [Google Scholar]
- Alcindor, T.; Beauger, N. Oxaliplatin: A review in the era of molecularly targeted therapy. Curr. Oncol. 2011, 18, 18–25. [Google Scholar] [CrossRef]
- Scheeff, E.D.; Briggs, J.M.; Howell, S.B. Molecular modeling of the intrastrand guanine-guanine DNA adducts produced by cisplatin and oxaliplatin. Mol. Pharmacol. 1999, 56, 633–643. [Google Scholar] [CrossRef]
- Messori, L.; Marzo, T.; Merlino, A. Protein metalation by metal-based drugs: X-ray crystallography and mass spectrometry studies. J. Inorg. Biochem. 2015, 153, 136–142. [Google Scholar] [CrossRef]
- Sciortino, G.; Sánchez-Aparicio, J.E.; Rodríguez-Guerra Pedregal, J.; Garribba, E.; Maréchal, J.D. Computational insight into the interaction of oxaliplatin with insulin. Metallomics 2019, 11, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Kweekel, D.; Gelderblom, H.; Guchelaar, H. Pharmacology of oxaliplatin and the use of pharmacogenomics to individualize therapy. Cancer Treat. Rev. 2005, 31, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Katherine, S.; Lovejoy, J.E.; Shima, L.L.; Lagpacan, Y.S.; Lapuk, A.; Chen, Y.; Komori, T.; Gray, J.W.; Chen, X.; et al. Organic Cation Transporters Are Determinants of Oxaliplatin Cytotoxicity. Cancer Res. 2006, 66, 8847–8857. [Google Scholar] [CrossRef] [PubMed]
- Bokemeyer, C.; Kollmannsberger, C.; Harstrick, A.; Beyer, J.; Gerl, A.; Casper, J.; Metzner, B.; Hartmann, J.T.; Clemm, C.; Schmoll, H.-J.; et al. Treatment of patients with cisplatin-refractory testicular germ-cell cancer. Int. J. Cancer 1999, 83, 848–851. [Google Scholar] [CrossRef]
- Stordal, B.; Pavlakis, N.; Davey, R. Oxaliplatin for the treatment of cisplatin-resistant cancer: A systematic review. Cancer Treat. Rev. 2007, 33, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Lévi, F.; Metzger, G.; Massari, C.; Milano, G. Oxaliplatin: Pharmacokinetics and Chronopharmacological Aspects. Clin. Pharmacokinet. 2000, 38, 1–21. [Google Scholar] [PubMed]
- Montagnani, F.; Turrisi, G.; Marinozzi, C.; Aliberti, C.; Fiorentini, G. Effectiveness and safety of oxaliplatin compared to cisplatin for advanced, unresectable gastric cancer: A systematic review and meta-analysis. Gastric Cancer 2011, 14, 50–55. [Google Scholar] [CrossRef]
- Dragovich, T.; Mendelson, D.; Kurtin, S.; Richardson, K.; Von Hoff, D.; Hoos, A. A Phase 2 trial of the liposomal DACH platinum L-NDDP in patients with therapy-refractory advanced colorectal cancer. Cancer Chemother. Pharmacol. 2006, 58, 759–764. [Google Scholar] [CrossRef]
- Lu, C.; Perez-Soler, R.; Piperdi, B.; Walsh, G.L.; Swisher, S.G.; Smythe, W.R.; Shin, H.J.; Ro, J.; Feng, L.; Truong, M.; et al. Phase II study of a liposome-entrapped cisplatin analog (L-NDDP) administered intrapleurally and pathologic response rates in patients with malignant pleural mesothelioma. J. Clin. Oncol. 2005, 23, 3495–3501. [Google Scholar] [CrossRef]
- El-Shafie, S.; Fahmy, S.A.; Ziko, L.; Elzahed, N.; Shoeib, T.; Kakarougkas, A. Encapsulation of Nedaplatin in Novel PEGylated Liposomes Increases Its Cytotoxicity and Genotoxicity against A549 and U2OS Human Cancer Cells. Pharmaceutics 2020, 12, 863. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Mamdouh, W. Garlic oil–loaded PLGA nanoparticles with controllable size and shape and enhanced antibacterial activities. J. Appl. Polym. Sci. 2018, 135. [Google Scholar] [CrossRef]
- Koshkaryev, A.; Sawant, R.; Deshpande, M.; Torchilin, V. Immunoconjugates and long circulating systems: Origins, current state of the art and future directions. Adv. Drug Deliv. Rev. 2013, 65, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Alexis, F.; Pridgen, E.M.; Langer, R.; Farokhzad, O.C. Nanoparticle Technologies for Cancer Therapy. In Drug Delivery. Handbook of Experimental Pharmacology; Schäfer-Korting, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 197. [Google Scholar]
- Haag, R. Supramolecular Drug-Delivery Systems Based on Polymeric Core–Shell. Architectures. Angew. Chem. Int. Ed. 2004, 43, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Loh, X.J. Supramolecular host–guest polymeric materials for biomedical applications. Mater. Horiz. 2014, 1, 185–195. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Brüßler, J.; Alawak, M.; El-Sayed, M.M.H.; Bakowsky, U.; Shoeib, T. Chemotherapy Based on Supramolecular Chemistry: A Promising Strategy in Cancer Therapy. Pharmaceutics 2019, 11, 292. [Google Scholar] [CrossRef]
- Cao, L.; Hettiarachchi, G.; Briken, V.; Isaacs, L. Cucurbit[7]uril containers for targeted delivery of oxaliplatin to cancer cells. Angew. Chem. 2013, 52, 12033–12037. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, Z.; Zhao, H.; Xu, J.-F.; Sun, Z.; Zhang, X. Supramolecular Chemotherapy: Cooperative Enhancement of Antitumor Activity by Combining Controlled Release of Oxaliplatin and Consuming of Spermine by Cucurbit[7]uril. ACS Appl. Mater. Interfaces 2017, 9, 8602–8608. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Kim, S.Y.; Ko, Y.H.; Sakamoto, S.; Yamaguchi, K.; Kim, K. Novel molecular drug carrier: Encapsulation of oxaliplatin in cucurbit[7]uril and its effects on stability and reactivity of the drug. Org. Biomol. Chem. 2005, 3, 2122–2125. [Google Scholar] [CrossRef]
- Plumb, J.A.; Venugopal, B.; Oun, R.; Gomez-Roman, N.; Kawazoe, Y.; Venkataramanan, N.S.; Wheate, N.J. Cucurbit[7]uril encapsulated cisplatin overcomes cisplatin resistance via a pharmacokinetic effect. Met. Integr. Biometal Sci. 2012, 4, 561–567. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Ponte, F.; Abd El-Rahman, M.K.; Russo, N.; Sicilia, E.; Shoeib, T. Investigation of the host-guest complexation between 4-sulfocalix[4]arene and nedaplatin for potential use in drug delivery. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 193, 528–536. [Google Scholar] [CrossRef]
- El-Rahman, M.K.A.; Mahmoud, A.M. A novel approach for spectrophotometric determination of succinylcholine in pharmaceutical formulation via host–guest complexation with water-soluble p-sulfonatocalixarene. RSC Adv. 2015, 5, 62469. [Google Scholar] [CrossRef]
- Joyce, L.A.; Shabbir, S.H.; Anslyn, E.V. The uses of supramolecular chemistry in synthetic methodology development: Examples of anion and neutral molecular recognition. Chem. Soc. Rev. 2010, 39, 3621–3632. [Google Scholar] [CrossRef] [PubMed]
- Strobel, M.; Kita-Tokarczyk, K.; Taubert, A. Self-Assembly of Amphiphilic Calix[4]arenes in Aqueous Solution. Adv. Funct. Mater. 2006, 16, 252–259. [Google Scholar] [CrossRef]
- Trush, V.V.; Cherenok, S.O.; Tanchuk, V.Y.V.; Viacheslav, V.; Trush, S.O.; Cherenok, V.Y.; Tanchuk, V.P.K.; Kalchenko, V.I.; Vovk, A.I. Calix[4]arene methylenebisphosphonic acids as inhibitors of protein tyrosine phosphatase 1B. Bioorg. Med. Chem. Lett. 2013, 23, 5619–5623. [Google Scholar] [CrossRef]
- NasuhiPur, F.; Dilmaghani, K.A. Calixplatin: Novel potential anticancer agent based on the platinum complex with functionalized calixarene. J. Coord. Chem. 2014, 67, 440–448. [Google Scholar] [CrossRef]
- Hulíková, K.; Grobárová, V.; Křivohlavá, R.; Fišerová, A. Antitumor activity of N-acetyl-d-glucosamine-substituted glycoconjugates and combined therapy with keyhole limpet hemocyanin in B16F10 mouse melanoma model. Folia Microbiol. 2010, 55, 528–532. [Google Scholar] [CrossRef]
- Cherenok, S.O.; Yushchenko, O.A.; Tanchuk, V.Y.; Mischenko, I.M.; Samus, N.V.; Ruban, O.V.; Matvieiev, Y.I.; Karpenko, J.A.; Kukhar, V.P.; Vovk, A.I.; et al. Calix[4]arene-α-hydroxyphosphonic acids. Synthesis, stereochemistry, and inhibition of glutathione S-transferase. ARKIVOC 2012, 4, 278–298. [Google Scholar] [CrossRef]
- Cherenok, S.; Vovk, A.; Muravyova, I.; Shivanyuk, A.; Kukhar, V.; Lipkowski, J.; Kalchenko, V. Calix[4]arene α-Aminophosphonic Acids: Asymmetric Synthesis and Enantioselective Inhibition of an Alkaline Phosphatase. Org. Lett. 2006, 8, 549–552. [Google Scholar] [CrossRef]
- Grazia, M.L.; Consoli, G.G.; Galante, E.; Di Silvestro, I.; Salafia, L.; Geraci, C. Synthesis of water-soluble nucleotide-calixarene conjugates and preliminary investigation of their in vitro DNA replication inhibitory activity. Tetrahedron 2007, 63, 10758–10763. [Google Scholar]
- Zhou, H.; Wang, D.A.; Baldini, L.; Ennis, E.; Jain, R.; Carie, A.; Sebti, S.M.; Hamilton, A.M. Structure–activity studies on a library of potent calix[4]arene-based PDGF antagonists that inhibit PDGF-stimulated PDGFR tyrosine phosphorylation. Org. Biomol. Chem. 2006, 4, 2376–2386. [Google Scholar] [CrossRef]
- Dings, R.P.M.; Chen, X.; Hellebrekers, D.M.E.I.; van Eijk, L.I.; Zhang, Y.; Hoye, T.R.; Griffioen, A.W.; Mayo, K.H. Design of Nonpeptidic Topomimetics of Antiangiogenic Proteins With Antitumor Activities. J. Natl. Cancer Inst. 2006, 98, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Dings, R.P.M.; Levine, J.I.; Brown, S.G.; Astorgues-Xerri, L.; MacDonald, J.R.; Hoye, T.R.; Raymond, E.; Mayo, K.H. Polycationic calixarene PTX013, a potent cytotoxic agent against tumors and drug resistant cancer. Investig. New Drugs. 2013, 31, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Kamada, R.; Yoshino, W.; Nomura, T.; Chuman, Y.; Imagawa, T.; Suzuki, T.; Sakaguchi, K. Enhancement of transcriptional activity of mutant p53 tumor suppressor protein through stabilization of tetramer formation by calix[6]arene derivatives. Bioorg. Med. Chem. Lett. 2010, 20, 4412–4415. [Google Scholar] [CrossRef] [PubMed]
- Pelizzaro-Rocha, K.J.; de Jesus, M.B.; Ruela-de-Sousa, R.R.; Nakamura, C.V.; Reis, F.S.; de Fátima, A.; Ferreira-Halder, C.V. Calix[6]arene bypasses human pancreatic cancer aggressiveness: Downregulation of receptor tyrosine kinases and induction of cell death by reticulum stress and autophagy. BBA-Mol. Cell. Res. 2013, 1833, 2856–2865. [Google Scholar] [CrossRef] [PubMed]
- Schrama, D.; Reisfeld, R.A.; Becker, J.C. Antibody targeted drugs as cancer therapeutics. Nat. Rev. Drug Discov. 2006, 5, 147–159. [Google Scholar] [CrossRef]
- Geraci, C.; Consoli, G.M.L.; Granata, G.; Galante, E.; Palmigiano, A.; Pappalardo, M.; Di Puma, S.D.; Spadaro, A. First Self-Adjuvant Multicomponent Potential Vaccine Candidates by Tethering of Four or Eight MUC1 Antigenic Immunodominant PDTRP Units on a Calixarene Platform: Synthesis and Biological Evaluation. Bioconjug. Chem. 2013, 24, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.; Plumb, J.A.; Johnston, B.F.; Wheate, N.J. Folding of dinuclear platinum anticancer complexes within the cavity of para-sulphonatocalix[4]arene. Inorg. Chim. Acta. 2012, 393, 182–186. [Google Scholar] [CrossRef]
- Gutsche, C.D.; Bauer, L.J. Calixarenes. 13. The conformational properties of calix[4]arenes, calix[6]arenes, calix[8]arenes, and oxacalixarenes. J. Am. Chem. Soc. 1985, 107, 6052–6059. [Google Scholar] [CrossRef]
- Guo, D.S.; Liu, Y.J. Supramolecular Chemistry of p-Sulfonatocalix[n]arenes and Its Biological Applications. J. Chem. Res. 2014, 47, 1925–1934. [Google Scholar] [CrossRef]
- Coleman, A.W.; Jebors, S.; Cecillon, S.; Perret, P.; Garin, D.; Marti-Battle, D.; Moulin, M. Toxicity and biodistribution of para-sulfonato-calix[4]arene in mice. New J. Chem. 2008, 32, 780–782. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistiga, D.; Warren, T.J.; Bokesch, H.; Kenney, S.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Allam, R.M.; Al-Abd, A.M.; Khedr, A.; Sharaf, O.A.; Nofal, S.M.; Khalifa, A.E.; Mosli, H.A.; Abdel-Naim, A.B. Fingolimod interrupts the cross talk between estrogen metabolism and sphingolipid metabolism within prostate cancer cells. Toxicol. Lett. 2018, 291, 77–85. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K.S. ulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A. Gaussian 09, Revision A.02, Revision D.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Andrae, D.; Häusermann, U.; Dolg, M.; Stoll, H.; Preuss, H. Energy-adjusted ab initio pseudopotentials for the second and third row transition elements. Theor. Chim. Acta 1990, 77, 123. [Google Scholar] [CrossRef]
- El-Rahman, M.K.; Mazzone, G.; Mahmoud, A.M.; Sicilia, E.; Shoeib, T. Spectrophotometric determination of choline in pharmaceutical formulations via host-guest complextaion with a biomimetic calixarene receptor. Microchem. J. 2019, 146, 735–741. [Google Scholar] [CrossRef]
- Cossi, M.; Barone, V. Solvent effect on vertical electronic transitions by the polarizable continuum model. J. Chem. Phys. 2000, 112, 2427–2435. [Google Scholar] [CrossRef]
- Boys, S.F.; Bernardi, F. The calculation of small molecular interactions by the differences of separate total energies. Mol. Phys. 1970, 19, 553. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723, Ver. 4.1.0. Available online: http://www.expasy.org/spdbv/ (accessed on 28 November 2020). [CrossRef]
- Wheate, N.J. Improving platinum(II)-based anticancer drug delivery using cucurbit[n]urils. J. Inorg. Biochem. 2008, 102, 2060–2066. [Google Scholar] [CrossRef]
- Wheate, N.J.; Abbott, G.M.; Tate, R.J.; Clements, C.J.; Edrada-Ebel, R.; Johnston, B.F. Side-on binding of p-sulphonatocalix[4]arene to the dinuclear platinum complex trans-[{PtCl(NH3)2}2mu-dpzm]2+ and its implications for anticancer drug delivery. J. Inorg. Biochem. 2009, 103, 448–454. [Google Scholar] [CrossRef]
- Guo, D.-S.; Uzunova, V.D.; Su, X.; Liu, Y.; Nau, W.M. Operational calixarene-based fluorescent sensing systems for choline and acetylcholine and their application to enzymatic reactions. Chem. Sci. 2011, 2, 1722. [Google Scholar] [CrossRef]
- Shinkai, S.; Araki, K.; Matsuda, T.; Manabe, O. NMR Determination of Association Constants for Aqueous Calixarene Complexes and Guest Template Effects on the Conformational Freedom. Bull. Chem. Soc. Jpn. 1989, 62, 3856–3862. [Google Scholar] [CrossRef]
- Shinkai, K.; Araki, O.; Manabe, S. NMR determination of association constants for calixarene complexes. Evidence for the formation of a 1:2 complex with calix[8]arene. J. Am. Chem. Soc. 1998, 110, 7214–7215. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Ponte, F.; Sicilia, E.; Azzazy, H.M.E.-S. Experimental and Computational Investigations of Carboplatin Supramolecular Complexes. ACS Omega 2020, 5, 31456–31466. [Google Scholar] [CrossRef]
- Olson, E.J.; Buhlmann, P. Getting More out of a Job Plot: Determination of Reactant to Product Stoichiometry in Cases of Displacement Reactions and n:n Complex Formation. J. Org. Chem. 2011, 76, 8406. [Google Scholar] [CrossRef]
- Salem, M.Y.; El-Kosasy, A.M.; El-Bardicy, M.G.; Abd El-Rahman, M.K. Spectrophotometric and spectrodensitometric methods for the determination of rivastigmine hydrogen tartrate in presence of its degradation product. Drug Test. Anal. 2010, 2, 225. [Google Scholar] [CrossRef]
- Darwish, H.W.; Hassan, S.A.; Salem, M.Y.; El-Zeany, A.B. Comparative study between derivative spectrophotometry and multivariate calibration as analytical tools applied for the simultaneous quantitation of Amlodipine, Valsartan and Hydrochlorothiazide. Spectrochim. Acta A Mol.Biomol. Spectrosc. 2013, 113, 215. [Google Scholar] [CrossRef]
- Nebsen, M.; Abd El-Rahman, M.K.; Salem, M.Y.; El-Kosasy, A.M.; El-Bardicy, M.G. Stability-indicating spectrophotometric and spectrodensitometric methods for the determination of diacerein in the presence of its degradation product. Drug Test. Anal. 2011, 3, 221. [Google Scholar] [CrossRef]
- Bosque-Sendra, J.M.; Almansa-López, E.; García-Campaña, A.M.; Cuadros- Rodríguez, L. Data Analysis in the Determination of Stoichiometries and Stability Constants of Complexes. Anal. Sci. 2003, 19, 1431. [Google Scholar] [CrossRef][Green Version]
- Carvalho, C.P.; Uzunova, V.D.; Da Silva, J.P.; Nau, W.M.; Pischel, U. A photoinduced pH jump applied to drug release from cucurbit[7]uril. Chem. Commun. 2011, 47, 8793–8795. [Google Scholar] [CrossRef]
- Marquez, C.; Nau, W.M. Two Mechanisms of Slow Host-Guest Complexation between Cucurbit[6]uril and Cyclohexylmethylamine: pH-Responsive Supramolecular Kinetics. Angew. Chem. Int. Ed. 2001, 40, 3155–3160. [Google Scholar] [CrossRef]
- Miskolczy, Z.; Biczok, L. Photochromism in Cucurbit[8]uril Cavity: Inhibition of Hydrolysis and Modification of the Rate of Merocyanine–Spiropyran Transformations. J. Phys. Chem. B 2011, 115, 12577–12583. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Isaacs, L. Cucurbit[7Cao]uril Complexation Drives Thermal trans–cis-Azobenzene Isomerization and Enables Colorimetric Amine Detection. Chem. Eur. J. 2009, 15, 11675–11680. [Google Scholar] [CrossRef] [PubMed]
- Pischel, U.; Uzunova, V.D.; Remon, P.; Nau, W.M. Supramolecular logic with macrocyclic input and competitive reset. Chem. Commun. 2010, 46, 2635–2637. [Google Scholar] [CrossRef]
- Praetorius, A.; Bailey, D.M.; Schwarzlose, T.; Nau, W.M. Design of a Fluorescent Dye for Indicator Displacement from Cucurbiturils: A Macrocycle-Responsive Fluorescent Switch Operating through a pKa Shift. Org. Lett. 2008, 10, 4089–4092. [Google Scholar] [CrossRef]
- Koner, A.L.; Nau, W.M. Cucurbituril Encapsulation of Fluorescent Dyes. Supramol. Chem. 2007, 19, 55–66. [Google Scholar] [CrossRef]
- Arantes, L.M.; Varejao, E.V.; Pelizzaro-Rocha, K.J.; Cereda, C.M.; de Paula, E.; Lourenco, M.P.; Duarte, H.A.; Fernandes, S.A. Benzocaine Complexation with p-Sulfonic Acid Calix[n]arene: Experimental (1H-NMR) and Theoretical Approaches. Chem. Biol. Drug Des. 2014, 83, 550–559. [Google Scholar] [CrossRef]
- Shaikh, M.; Mohanty, P.K.J.; Singh, W.M.; Nau, P.H. Complexation of acridine orange by cucurbit[7]uril and β-cyclodextrin: Photophysical effects and pKa shifts. Photobiol. Sci. 2008, 7, 408–414. [Google Scholar] [CrossRef]
- Bakirci, H.; Koner, A.; Schwarzlose, T.; Nau, M.W. Analysis of host-assisted guest protonation exemplified for p-sulfonatocalix[4]arene—Towards enzyme-mimetic pKa shifts. Chem. Eur. J. 2006, 12, 4799–4807. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, H.; Ding, F.; Liu, Y. Preparation and characterization of inclusion complexes of topotecan with sulfonatocalixarene. Macrocycl. Chem. 2011, 69, 85–89. [Google Scholar] [CrossRef]
- Yang, W.; de Villiers, M.; Yang, W.; De Villiers, M. Effect of 4-Sulphonato-Calix[n]Arenes and Cyclodextrins on the Solubilizationof Niclosamide, a Poorly Water Soluble Anthelmintic. AAPS J. 2005, 7, 23. Available online: http://www.aapsj.org (accessed on 28 November 2020). [CrossRef] [PubMed]
- Zhang, D.; Zhang, J.; Jiang, K.; Li, K.; Cong, Y.; Pu, S.; Jin, Y.; Lin, J. Preparation, characterisation and antitumour activity of β-, γ-and HP-β-cyclodextrin inclusion complexes of oxaliplatin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 152, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, S.A.; Brüßler, J.; Ponte, F.; Abd El-Rahman, M.K.; Russo, N.; Sicilia, E.; Bakowsky, U.; Shoeib, T. A study on the physicochemical properties and cytotoxic activity of p-sulfocalix[4]arene-nedaplatin complex. J. Phys. Conf. Ser. 2019, 1310. [Google Scholar] [CrossRef]
- Yousaf, A.; Hamid, S.A.; Bunnori, N.M.; Ishola, A.A. Applications of calixarenes in cancer chemotherapy: Facts and perspectives. Drug Des. Devel. Ther. 2015, 9, 2831–2838. [Google Scholar] [PubMed]
- Dennington, R.; Keith, T.; Millam, J. (Eds.) GaussView; Version 6.1.1; Semichem Inc.: Shawnee Mission, KS, USA, 2019. [Google Scholar]
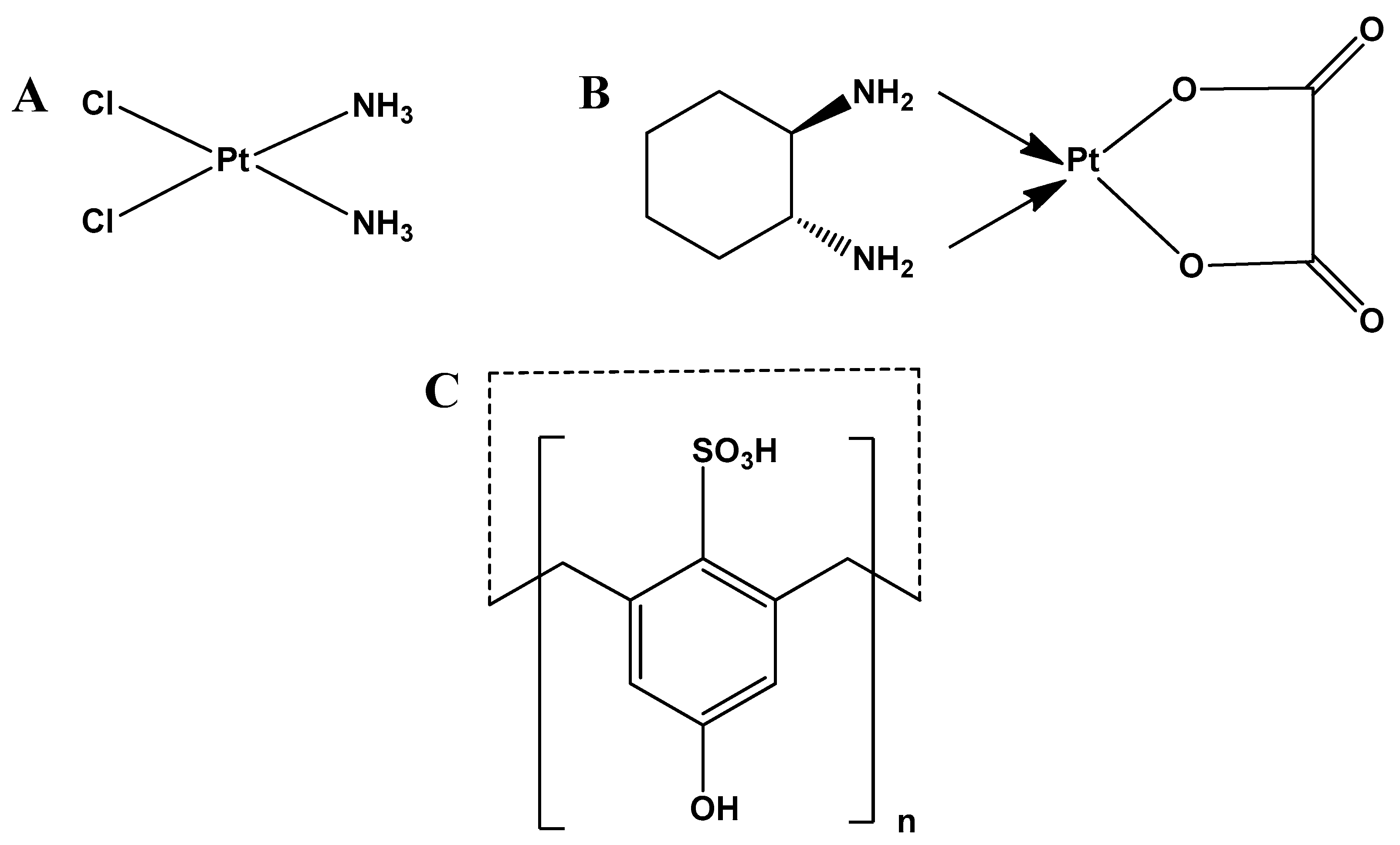
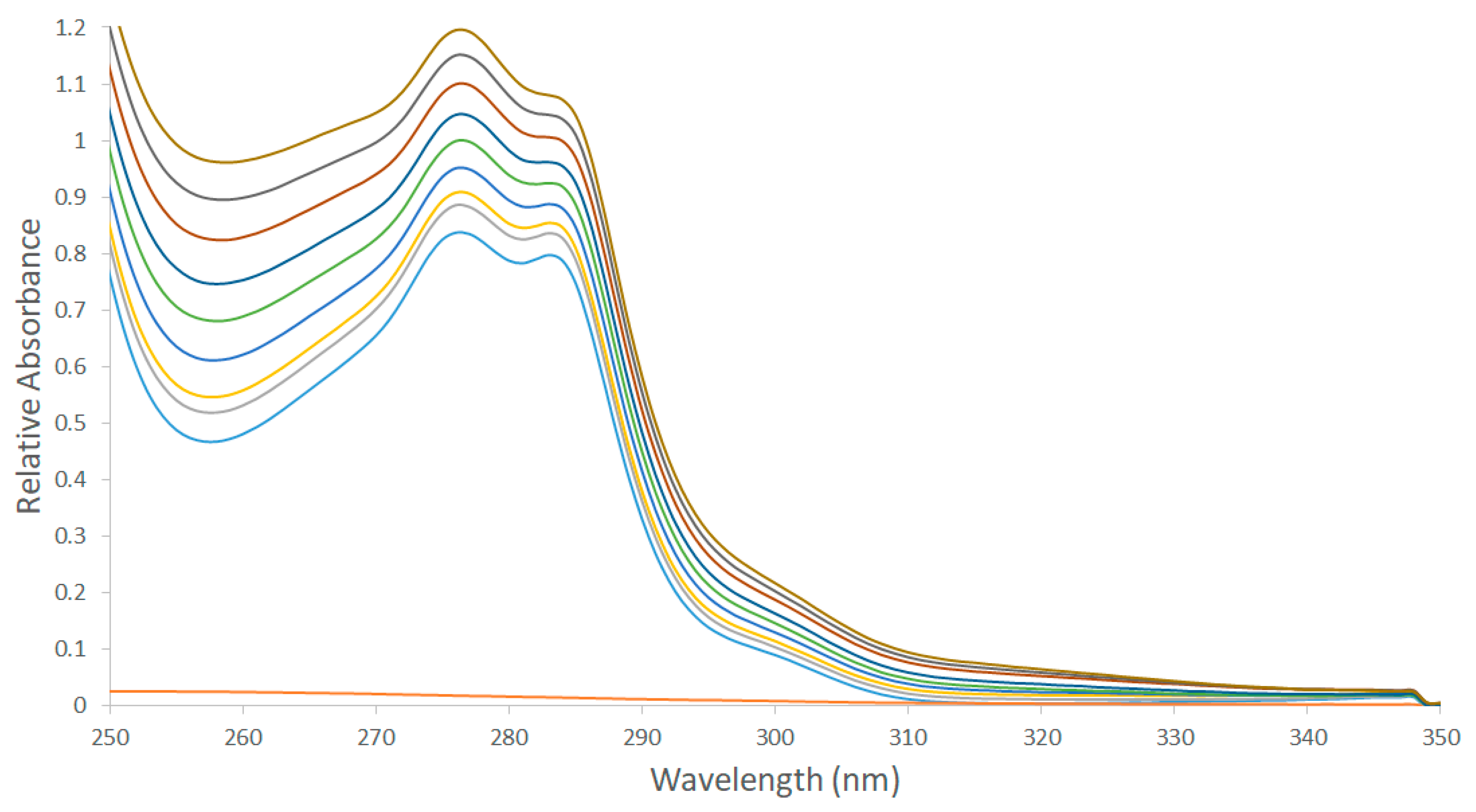
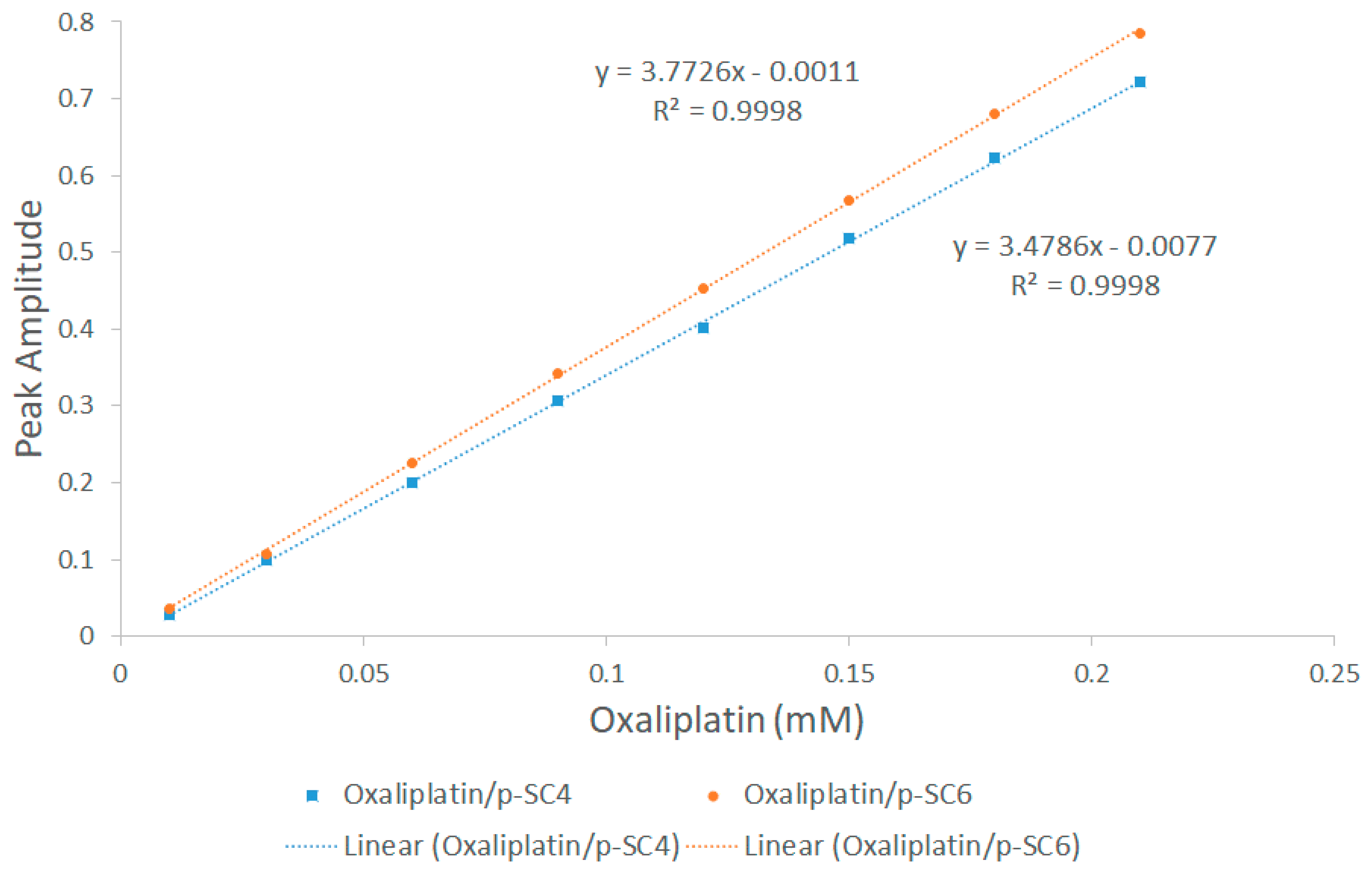
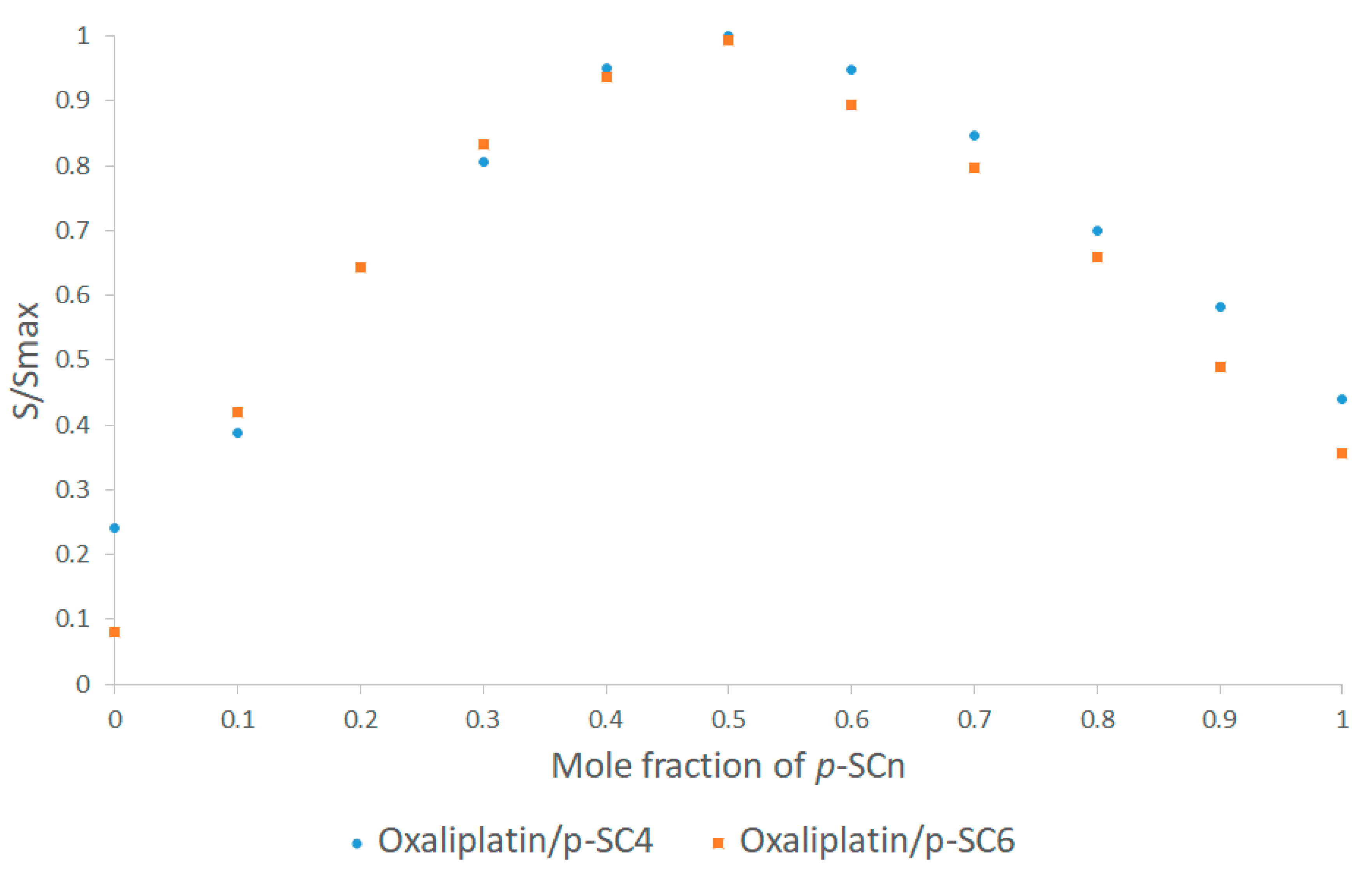

| Proton Signals | Individual Molecules | 1:1 Molar Ratio |
|---|---|---|
| 3ax | 0.92–1.00 | 0.19–0.25 |
| 2ax | 1.16–1.25 | 0.38–0.44 |
| 3eq | 1.39–1.46 | 0.98–1.04 |
| 2eq | 1.81–1.88 | 1.16–1.21 |
| 1 | 2.03–2.04 | 1.63–1.66 |
| Hy | 3.67 | 3.83 |
| Cells | In Vitro Anticancer Activity (IC50 in µg/mL) | ||||
|---|---|---|---|---|---|
| p-SC4 | p-SC6 | Oxaliplatin | Oxaliplatin/p-SC4 | Oxaliplatin/p-SC6 | |
| HT-29 | N/A | 10 | 3.9 | 2 | 1.3 |
| MCF-7 | N/A | 220 | 5 | 1.56 | 0.59 |
Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fahmy, S.A.; Ponte, F.; Fawzy, I.M.; Sicilia, E.; Bakowsky, U.; Azzazy, H.M.E.-S. Host-Guest Complexation of Oxaliplatin and Para-Sulfonatocalix[n]Arenes for Potential Use in Cancer Therapy. Molecules 2020, 25, 5926. https://doi.org/10.3390/molecules25245926
Fahmy SA, Ponte F, Fawzy IM, Sicilia E, Bakowsky U, Azzazy HME-S. Host-Guest Complexation of Oxaliplatin and Para-Sulfonatocalix[n]Arenes for Potential Use in Cancer Therapy. Molecules. 2020; 25(24):5926. https://doi.org/10.3390/molecules25245926
Chicago/Turabian StyleFahmy, Sherif Ashraf, Fortuna Ponte, Iten M. Fawzy, Emilia Sicilia, Udo Bakowsky, and Hassan Mohamed El-Said Azzazy. 2020. "Host-Guest Complexation of Oxaliplatin and Para-Sulfonatocalix[n]Arenes for Potential Use in Cancer Therapy" Molecules 25, no. 24: 5926. https://doi.org/10.3390/molecules25245926
APA StyleFahmy, S. A., Ponte, F., Fawzy, I. M., Sicilia, E., Bakowsky, U., & Azzazy, H. M. E.-S. (2020). Host-Guest Complexation of Oxaliplatin and Para-Sulfonatocalix[n]Arenes for Potential Use in Cancer Therapy. Molecules, 25(24), 5926. https://doi.org/10.3390/molecules25245926









