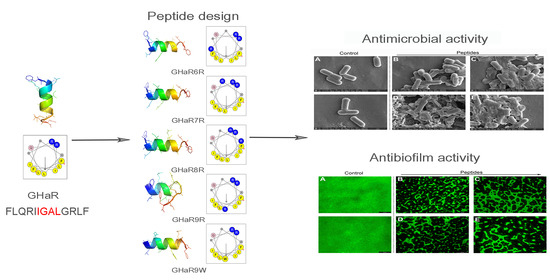
Temporin-Like Peptides Show Antimicrobial and Anti-Biofilm Activities against Streptococcus mutans with Reduced Hemolysis
Abstract
1. Introduction
2. Results
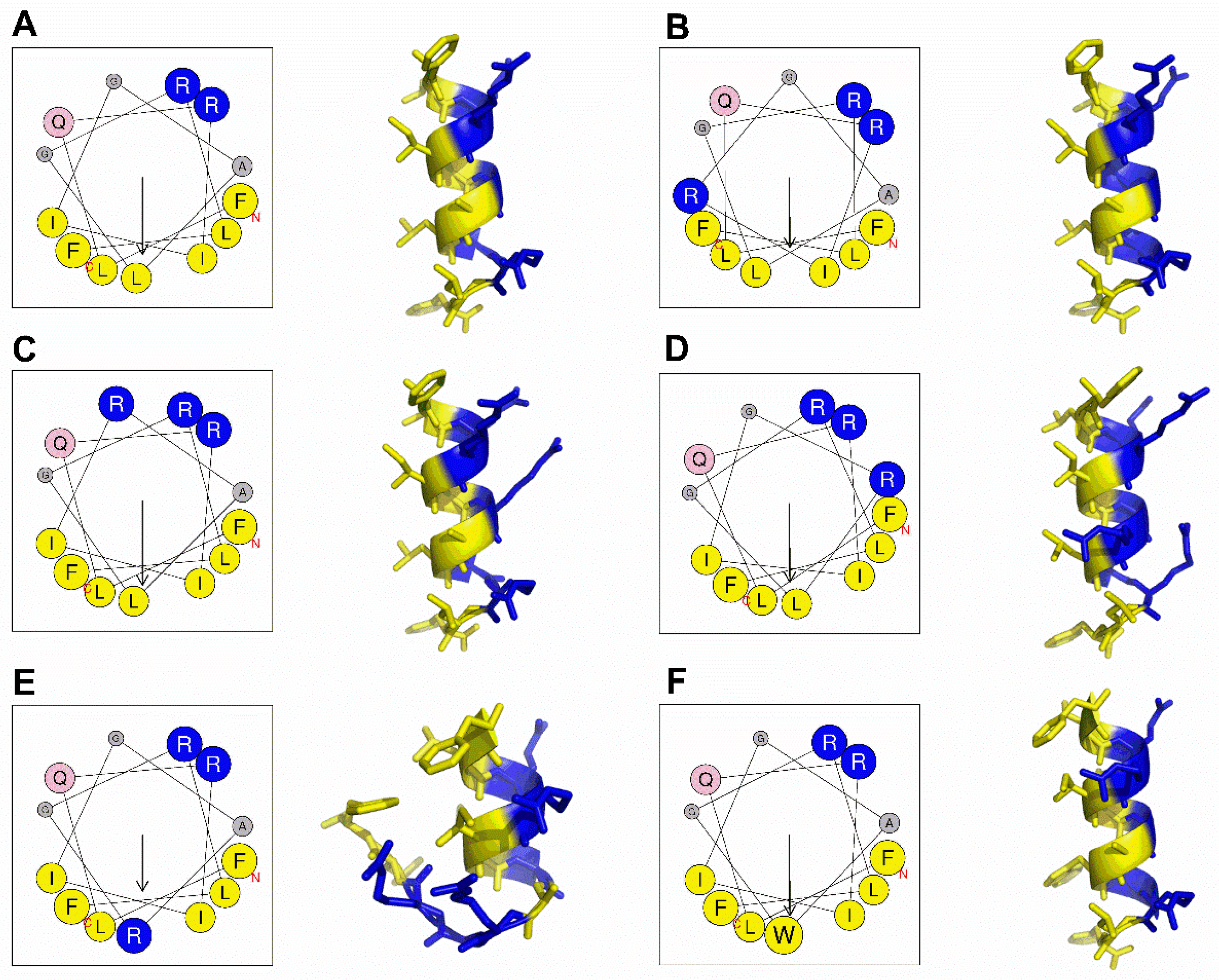
2.1. Peptide Design and Structure Prediction
2.2. Physicochemical Properties Analysis and Antimicrobial Activity Prediction
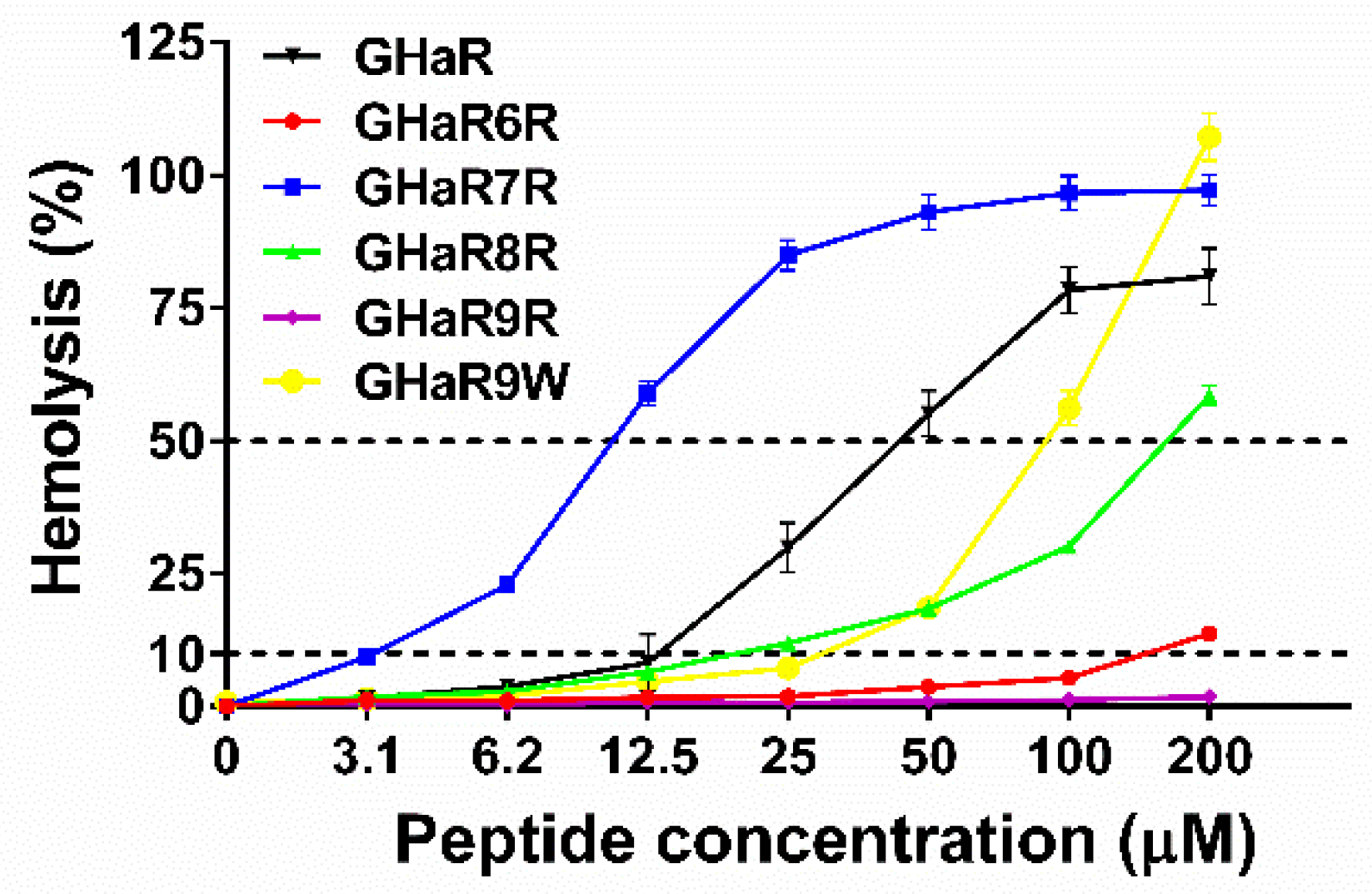
2.3. Hemolysis Assay
2.4. Bacterial Susceptibility Test
2.5. Growth Inhibition Kinetics
2.6. Killing Kinetics
2.7. Membrane Permeation Assay
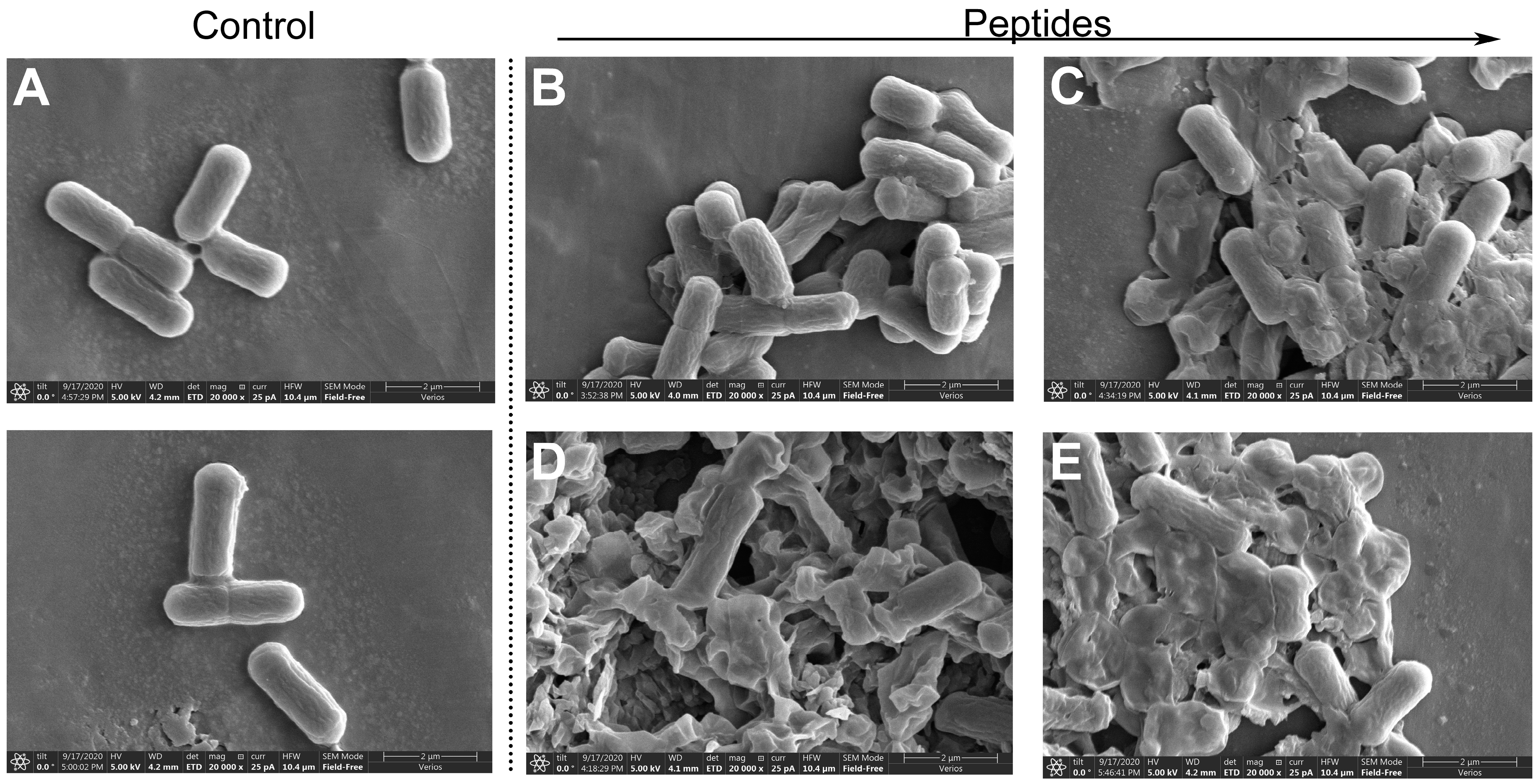
2.8. Scanning Electron Microscopy Analysis
2.9. Anti-Biofilm Activity
2.10. Fluorescence Microscope Analysis
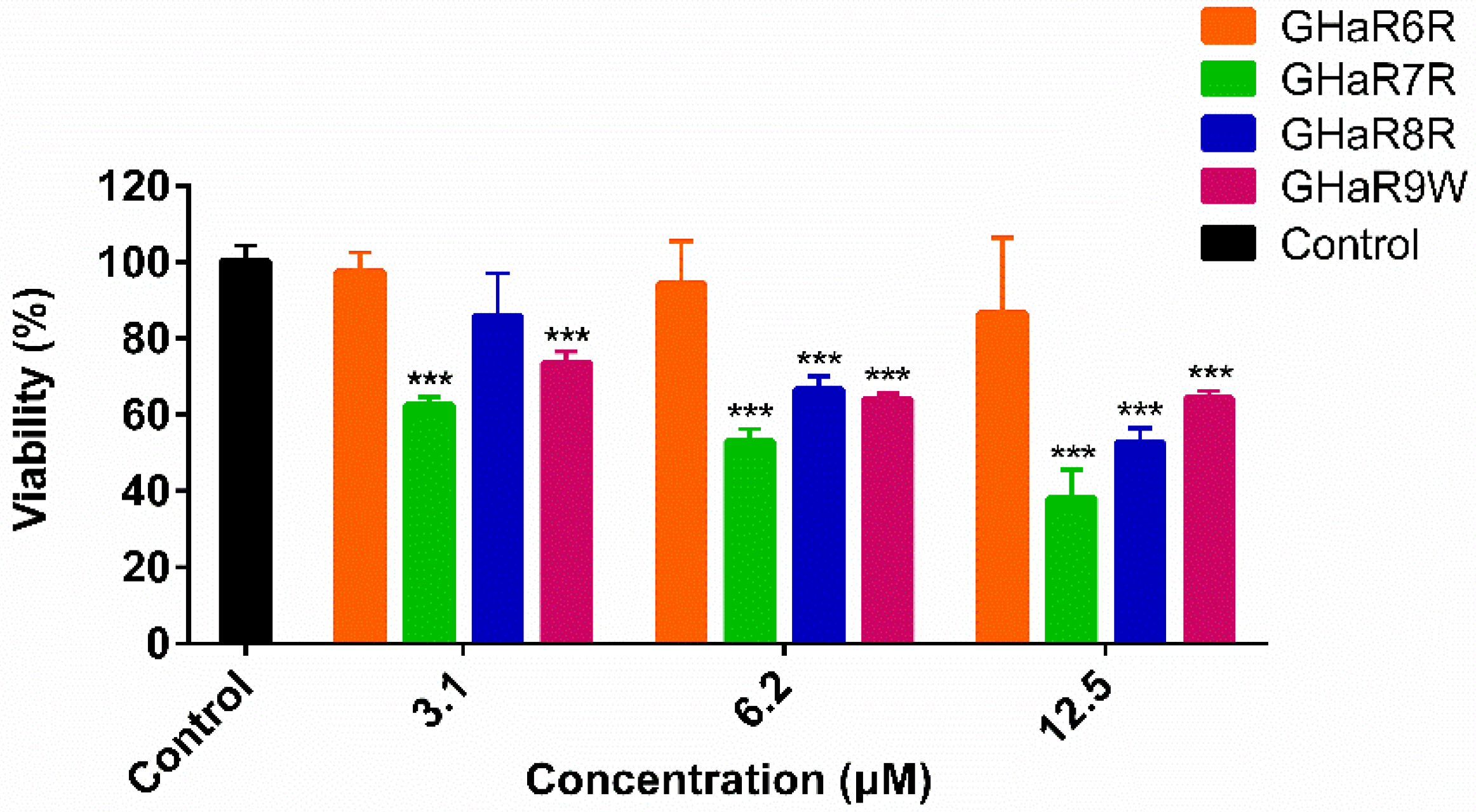
2.11. Viability of Human Oral Epithelial Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Peptide Design and Structure Prediction
4.3. Physicochemical Properties and Antimicrobial Activity Analysis
4.4. Synthesis and Storage of Peptides
4.5. Hemolysis Assay
4.6. Antimicrobial Activity Detection
4.7. Influence on Growth of S. mutans
4.8. Time-Killing Curves
4.9. Propidium Iodide Uptake Assay
4.10. Scanning Electron Microscopy
4.11. Effect of the Peptides on Biofilms
4.12. Biofilm Observation by Fluorescence Microscope
4.13. Cytotoxicity Assay
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kolenbrander, P.E.; Palmer, R.J.; Rickard, A.H.; Jakubovics, N.S.; Diaz, P.I. Bacterial interactions and successions during plaque development. Periodontol. 2000 2010, 42, 47–79. [Google Scholar] [CrossRef] [PubMed]
- Ajdic, D.; McShan, W.M.; McLaughlin, R.E.; Savic, G.; Chang, J.; Carson, M.B.; Primeaux, C.; Tian, R.; Kenton, S.; Jia, H.; et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 2002, 99, 14434–14439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, C.; Chen, J.; Zhou, S.; Zhao, Y.; Xu, M.; Xu, H. Dual Mode of Anti-Biofilm Action of G3 against Streptococcus mutans. ACS Appl. Mater. Interfaces 2020, 12, 27866–27875. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]
- David, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar]
- Selwitz, R.H.; Ismail, A.I.; Pitts, N.B. Dental caries. Lancet 2007, 369, 51–59. [Google Scholar] [CrossRef]
- Chen, L.; Ren, Z.; Zhou, X.; Zeng, J.; Zou, J.; Li, Y. Inhibition of Streptococcus mutans biofilm formation, extracellular polysaccharide production, and virulence by an oxazole derivative. Appl. Microbiol. Biotechnol. 2016, 100, 857–867. [Google Scholar] [CrossRef]
- Figuero, E.; Nóbrega, D.F.; García-Gargallo, M.; Tenuta, L.M.A.; Herrera, D.; Carvalho, J.C. Mechanical and chemical plaque control in the simultaneous management of gingivitis and caries: A systematic review. J. Clin. Periodontol. 2017, 44, S116–S134. [Google Scholar] [CrossRef]
- Baehni, P.; Takeuchi, Y. Anti-plaque agents in the prevention of biofilm-associated oral diseases. Oral Dis. 2003, 9, 23–29. [Google Scholar] [CrossRef]
- Cieplik, F.; Jakubovics, N.S.; Buchalla, W.; Maisch, T.; Hellwig, E.; Al-Ahmad, A. Resistance Toward Chlorhexidine in Oral Bacteria–Is There Cause for Concern? Front. Microbiol. 2019, 10, 587. [Google Scholar] [CrossRef]
- Sitaram, N.; Nagaraj, R. Interaction of antimicrobial peptides with biological and model membranes: Structural and charge requirements for activity. Biochim. Biophys. Acta 1999, 1462, 29–54. [Google Scholar] [CrossRef]
- Wei, G.X.; Campagna, A.N.; Bobek, L.A. Effect of MUC7 peptides on the growth of bacteria and on Streptococcus mutans biofilm. J. Antimicrob. Chemother. 2006, 57, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Jia, L.; Zhang, Q.; Zhou, X.; Liu, Z.; Li, B.; Zhu, Z.; Wang, F.; Yu, C.; Zhang, Q.; et al. A novel antimicrobial peptide against dental-caries-associated bacteria. Anaerobe 2017, 47, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, W.; Fan, M.; Tong, Z.; Kuang, R.; Jiang, W.; Ni, L. Antimicrobial and anti-biofilm effect of Bac8c on major bacteria associated with dental caries and Streptococcus mutans biofilms. Peptides 2014, 52, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Luo, W.; Zhong, H.; Wang, M.; Song, Y.; Deng, S.; Zhang, Y. Molecular cloning and characterization of antimicrobial peptides from skin of Hylarana guentheri. Acta Biochim. Biophys. Sin. 2017, 49, 450–457. [Google Scholar] [CrossRef]
- Zelezetsky, I.; Tossi, A. Alpha-helical antimicrobial peptides-using a sequence template to guide structure-activity relationship studies. Biochim. Biophys. Acta 2006, 1758, 1436–1449. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Sugishita, K.I.; Harada, M.; Fujii, N.; Miyajima, K. Interactions of an antimicrobial peptide, magainin 2, with outer and inner membranes of Gram-negative bacteria. Biochim. Biophys. Acta Biomembr. 1997, 1327, 119–130. [Google Scholar] [CrossRef]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Rational Design of α-Helical Antimicrobial Peptides with Enhanced Activities and Specificity/Therapeutic Index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef]
- Bi, X.; Wang, C.; Ma, L.; Sun, Y.; Shang, D. Investigation of the role of tryptophan residues in cationic antimicrobial peptides to determine the mechanism of antimicrobial action. J. Appl. Microbiol. 2013, 115, 663–672. [Google Scholar] [CrossRef]
- Saporito, P.; Vang Mouritzen, M.; Løbner-Olesen, A.; Jenssen, H. LL-37 fragments have antimicrobial activity against Staphylococcus epidermidis biofilms and wound healing potential in HaCaT cell line. J. Pept. Sci. 2018, 24, e3080. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Shi, Y.Q.; Pan, X.H.; Cao, P. Antibacterial effects of cinnamon (Cinnamomum zeylanicum) bark essential oil on Porphyromonas gingivalis. Microb. Pathog. 2018, 116, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Mishra, B.; Lushnikova, T.; Narayana, J.L.; Wang, G. Amino Acid Composition Determines Peptide Activity Spectrum and Hot-Spot-Based Design of Merecidin. Adv. Biosyst. 2018, 2, 1700259. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Wei, H.; Meng, J.; Cheng, T.; Song, Y.; Wang, M.; Zhang, Y. The Analogs of Temporin-GHa Exhibit a Broader Spectrum of Antimicrobial Activity and a Stronger Antibiofilm Potential against Staphylococcus aureus. Molecules 2019, 24, 4173. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta 2009, 1788, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Vasil, A.I.; Hale, J.D.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic α-helical cationic antimicrobial peptides. Pept. Sci. 2008, 90, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Dathe, M.; Nikolenko, H.; Meyer, J.; Beyermann, M.; Bienert, M. Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Lett. 2001, 501, 146–150. [Google Scholar] [CrossRef]
- Hawrani, A.; Howe, R.A.; Walsh, T.R.; Dempsey, C.E. Origin of low mammalian cell toxicity in a class of highly active antimicrobial amphipathic helical peptides. J. Biol. Chem. 2008, 283, 18636–18645. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Adv. Exp. Med. Biol. 2019, 1117, 3–6. [Google Scholar] [CrossRef]
- WILSON, M. Susceptibility of oral bacterial biofilms to antimicrobial agents. J. Med. Microbiol. 1996, 44, 79–87. [Google Scholar] [CrossRef]
- Dong, N.; Ma, Q.; Shan, A.; Lv, Y.; Hu, W.; Gu, Y.; Li, Y. Strand length-dependent antimicrobial activity and membrane-active mechanism of arginine- and valine-rich β-hairpin-like antimicrobial peptides. Antimicrob. Agents Chemother. 2012, 56, 2994–3003. [Google Scholar] [CrossRef]
- Mishra, B.; Wang, X.; Lushnikova, T.; Zhang, Y.; Golla, R.M.; Narayana, J.L.; Wang, C.; McGuire, T.R.; Wang, G. Antibacterial, antifungal, anticancer activities and structural bioinformatics analysis of six naturally occurring temporins. Peptides 2018, 106, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Joycharat, N.; Thammavong, S.; Limsuwan, S.; Homlaead, S.; Voravuthikunchai, S.P.; Yingyongnarongkul, B.E.; Dej-adisai, S.; Subhadhirasakul, S. Antibacterial substances from Albizia myriophylla wood against cariogenic Streptococcus mutans. Arch. Pharm. Res. 2013, 36, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, Y.; Song, Q.; Wang, F.; Sun, L.; Liu, L.; Yang, X.; Yi, J.; Bao, Y.; Ma, H.; et al. Anti-biofilm activities from bergenia crassifolia leaves against Streptococcus mutans. Front. Microbiol. 2017, 8, 1738. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Jiang, W.; Wang, K.; Luo, J.; Li, W.; Zhou, X.; Zhang, L. Antimicrobial peptide GH12 suppresses cariogenic virulence factors of Streptococcus mutans. J. Oral Microbiol. 2018, 10, 1442089. [Google Scholar] [CrossRef]
- Abraham, P.; George, S.; Kumar, K.S. Novel antibacterial peptides from the skin secretion of the Indian bicoloured frog Clinotarsus curtipes. Biochimie 2014, 97, 144–151. [Google Scholar] [CrossRef]
- Zhong, H.; Xie, Z.; Wei, H.; Zhang, S.; Song, Y.; Wang, M.; Zhang, Y. Antibacterial and Antibiofilm Activity of Temporin-GHc and Temporin-GHd Against Cariogenic Bacteria, Streptococcus mutans. Front. Microbiol. 2019, 10, 2854. [Google Scholar] [CrossRef]
- Ko, S.J.; Kang, N.H.; Kim, M.K.; Park, J.; Park, E.; Park, G.H.; Kang, T.W.; Na, D.E.; Park, J.B.; Yi, Y.E.; et al. Antibacterial and anti-biofilm activity, and mechanism of action of pleurocidin against drug resistant Staphylococcus aureus. Microb. Pathog. 2019, 127, 70–78. [Google Scholar] [CrossRef]
- Li, H.; Cheng, J.W.; Yu, H.Y.; Xin, Y.; Tang, L.; Ma, Y. Effect of the antimicrobial peptide D-Nal-Pac-525 on the growth of Streptococcus mutans and its biofilm formation. J. Microbiol. Biotechnol. 2013, 23, 1070–1075. [Google Scholar] [CrossRef]
- Nilsson, M.; Rybtke, M.; Givskov, M.; Hoiby, N.; Twetman, S.; Tolker-Nielsen, T. The dlt genes play a role in antimicrobial tolerance of Streptococcus mutans biofilms. Int. J. Antimicrob. Agents 2016, 48, 298–304. [Google Scholar] [CrossRef]
- Huang, L.; Chen, D.; Wang, L.; Lin, C.; Ma, C.; Xi, X.; Chen, T.; Shaw, C.; Zhou, M. Dermaseptin-PH: A Novel Peptide with Antimicrobial and Anticancer Activities from the Skin Secretion of the South American Orange-Legged Leaf Frog, Pithecopus (Phyllomedusa) hypochondrialis. Molecules 2017, 22, 1805. [Google Scholar] [CrossRef]
- Kaur, G.; Balamurugan, P.; Uma Maheswari, C.; Anitha, A.; Princy, S.A. Combinatorial Effects of Aromatic 1,3-Disubstituted Ureas and Fluoride on In vitro Inhibition of Streptococcus mutans Biofilm Formation. Front. Microbiol. 2016, 7, 861. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tao, R.; Tong, Z.; Ding, Y.; Kuang, R.; Zhai, S.; Liu, J.; Ni, L. Effect of a novel antimicrobial peptide chrysophsin-1 on oral pathogens and Streptococcus mutans biofilms. Peptides 2012, 33, 212–219. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |





| Peptides | Sequence | MW a | μH b | Charge a | PI c | BI a | GRAVY a |
|---|---|---|---|---|---|---|---|
| GHaR | FLQRIIGALGRLF | 1503.86 | 0.762 | 2 | 12.0 | 0.08 | 1.115 |
| GHaR6R | FLQRIRGALGRLF | 1546.89 | 0.693 | 3 | 12.3 | 1.61 | 0.423 |
| GHaR7R | FLQRIIRALGRLF | 1603.00 | 0.837 | 3 | 12.3 | 1.30 | 0.800 |
| GHaR8R | FLQRIIGRLGRLF | 1588.97 | 0.780 | 3 | 12.3 | 1.37 | 0.631 |
| GHaR9R | FLQRIIGARGRLF | 1545.89 | 0.554 | 3 | 12.3 | 1.61 | 0.477 |
| GHaR9W | FLQRIIGAWGRLF | 1575.91 | 0.804 | 2 | 12.0 | 0.28 | 0.754 |
| Species | Strains | MIC/MBC (μM) | |||||
|---|---|---|---|---|---|---|---|
| GHaR | GHaR6R | GHaR7R | GHaR8R | GHaR9R | GHaR9W | ||
| Gram+ | S. aureus | 1.6/1.6 | 3.1/12.5 | 3.1/3.1 | 1.6–3.1/3.1 | 6.2/12.5 | 3.1/6.2 |
| B. subtilis | 12.5/25 | 25/50 | >50/>50 | 12.5/12.5 | >50/>50 | 12.5/50 | |
| S. mutans | 3.1/6.2 | 12.5/12.5 | 6.2/12.5 | 6.2/6.2 | >50/>50 | 6.2/6.2 | |
| MRSA | 3.1/3.1 | 6.2/12.5 | 3.1/6.2 | 3.1/3.1 | >50/>50 | 6.2/12.5 | |
| MRSA-1 | 3.1/25 | 12.5/25 | 6.2/25 | 6.2/6.2 | >50/>50 | 6.2/12.5 | |
| MRSA-2 | 6.2/12.5 | 12.5/25 | 6.2/50 | 3.1/12.5 | >50/>50 | 6.2/12.5 | |
| MRSA-3 | 3.1/6.2 | 6.2/25 | 6.2/25 | 6.2/12.5 | >50/>50 | 6.2/12.5 | |
| Gram- | E. coli | 6.2/6.2 | 12.5/12.5 | 3.1/12.5 | 3.1/3.1 | >50/>50 | 12.5/12.5 |
| D31 | 12.5/25 | 25/50 | 12.5/50 | 12.5/25 | >50/>50 | 12.5/50 | |
| P. aeruginosa | 6.2/6.2 | >50/>50 | >50/>50 | 50/>50 | >50/>50 | >50/>50 | |
| PAO1 | 25/50 | 3.1/25 | >50/>50 | 6.2/12.5 | >50/>50 | 25/50 | |
| Fungi | C. albicans | 12.5/50 | 50/50 | 25/25 | 12.5/12.5 | >50/>50 | 25/25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, H.; Xie, Z.; Tan, X.; Guo, R.; Song, Y.; Xie, X.; Wang, R.; Li, L.; Wang, M.; Zhang, Y. Temporin-Like Peptides Show Antimicrobial and Anti-Biofilm Activities against Streptococcus mutans with Reduced Hemolysis. Molecules 2020, 25, 5724. https://doi.org/10.3390/molecules25235724
Wei H, Xie Z, Tan X, Guo R, Song Y, Xie X, Wang R, Li L, Wang M, Zhang Y. Temporin-Like Peptides Show Antimicrobial and Anti-Biofilm Activities against Streptococcus mutans with Reduced Hemolysis. Molecules. 2020; 25(23):5724. https://doi.org/10.3390/molecules25235724
Chicago/Turabian StyleWei, Hanqi, Zhipeng Xie, Xiuchuan Tan, Ran Guo, Yanting Song, Xi Xie, Rong Wang, Lushuang Li, Manchuriga Wang, and Yingxia Zhang. 2020. "Temporin-Like Peptides Show Antimicrobial and Anti-Biofilm Activities against Streptococcus mutans with Reduced Hemolysis" Molecules 25, no. 23: 5724. https://doi.org/10.3390/molecules25235724
APA StyleWei, H., Xie, Z., Tan, X., Guo, R., Song, Y., Xie, X., Wang, R., Li, L., Wang, M., & Zhang, Y. (2020). Temporin-Like Peptides Show Antimicrobial and Anti-Biofilm Activities against Streptococcus mutans with Reduced Hemolysis. Molecules, 25(23), 5724. https://doi.org/10.3390/molecules25235724