Abstract
The ability of Staphylococcus epidermidis to produce virulence factors, such as biofilm, added to its increased resistance to antimicrobials can cause infections that are difficult to treat. Many staphylococcal virulence factors are under the control of the accessory gene regulator (agr). The objective of this study was to establish the agr locus and susceptibility of biofilm-producing S. epidermidis specimens to antimicrobial agents, through PCR reactions, reverse transcription polymerase chain reaction (RT-PCR), and the determination of minimum inhibitory concentration (MIC), and to analyze the clonal profile of 300 strains isolated from blood culture specimens from inpatients at a University Hospital in Brazil, over a 20-year period by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) techniques. The ica operon expression was shown in 83.6% strains, bhp gene in 11.5%, and aap gene in 32.8%. Oxacillin resistance was detected in 90.1%, while 4.9% showed tigecycline resistance, and intermediate resistance to quinupristin/dalfopristin was identified in 0.4%. Clonal profile determination showed 11 clusters, with the ST2 type determined as the major cluster. The S. epidermidis biofilm producer demonstrated a predominance of agr I locus, oxacillin resistance, and SCCmec III as well as the potential dissemination of pathogenic clones in hospital settings over long periods.
1. Introduction
Coagulase-negative Staphylococci (CoNS) are members of the genus Staphylococcus and major colonizers of the human microbiota. Staphylococcus epidermidis is the most commonly isolated microorganism in clinical materials and being opportunistic, it can cause serious infections, mainly in immunosuppressed patients and those who use prosthetics [1]. The ability to produce virulence factors, such as biofilm, and increased resistance to antimicrobials may make it difficult to treat infections caused by these microorganisms [2].
Biofilm, considered the main virulence factor of CoNS, is defined as a complex interaction of microorganisms incorporated into an extracellular matrix of polysaccharide, proteins, and nucleic acid [3], which confers protection to the involved microorganisms and prevent them from the host’s immune system action during infection [4]. Although the exact mechanism of biofilm formation has not yet been clarified, it is known that this process involves four steps: adhesion, accumulation, maturation, and detachment. In the constitution of the biofilm of Staphylococcus spp., some genes are involved in the coding of the most important substances and proteins: the ica operon, which carries the icaA, icaC, icaD, and icaB genes, the accumulation-associated protein (Aap), and the biofilm-associated protein homologue (Bhp) [3].
The operon ica synthesizes a polysaccharide intercellular adhesion (PIA) which allows cell-cell bonding and multilayer formation, its main function being the contribution to biofilm formation [3]. Despite its importance, it was recognized that the synthesis of PIA is not essential for the formation of biofilm in strains of S. epidermidis [5], because the formation of biofilm independent of PIA is related to proteins that can replace this polysaccharide, such as accumulation-associated protein (Aap) and biofilm-associated protein (Bhp).
The Aap protein is important for the accumulation and growth of the polymers that form the biofilm [5]. Aap is a protein with an N-terminal of domain A and one of domain B, processed by proteases, and its active form is a fibrillar protein, released in “tufts”. Its accumulation is through the dimerization of Zn2 + -dependent domain B on neighboring cells, while domain A is responsible for adhering to corneocytes, aiding adherence to the skin [3]. The Bhp protein is important as it promotes primary fixation on abiotic surfaces, as well as intercellular adhesion during biofilm formation. The Bhp protein is homologous to the Bap of S. aureus, and its mechanism of contribution to Biofilm formation is still unclear [5].
Biofilm formation, as well as other virulence factors in Staphylococcus spp., can be regulated by a system with quorum sensing activity, which allows communication between bacterial cells, the detection of cell density, and a phenotypic reaction according to the growth where the culture is located [6,7]. The accessory gene regulator (agr) is the main system with quorum sensing activity, and three polymorphisms of the agr locus (agr I, II, and III) are described in S. epidermidis [8]. The agr locus contributes to the regulation of the virulence of the CoNS at different stages of the infection, with adherence-related proteins produced in the exponential phase of the bacterial growth curve, and some exoproteins secreted in the post-exponential phase [9].
In addition to the biofilm formation capacity of CoNS, increasing antimicrobial resistance makes it difficult to treat infections caused by these bacteria. The high rates of oxacillin resistance highlight the importance of CoNS capable of forming a biofilm, since this is related to the persistence of infections and a decrease in the effectiveness of antimicrobial activity [3,10]. It has been suggested that the matrix of biofilms can be responsible for the increased resistance to antibiotics by acting as a diffusion barrier [11].
Oxacillin is a semi-synthetic penicillin used in Brazil as a sensitivity test and treatment for infections caused by Staphylococcus and 66% to 95% of clinical CoNS isolates are resistant to this drug [12]. Oxacillin resistance is mediated by the mecA gene, which encodes a supplemental penicillin-binding protein (PBP2a) that has low affinity for semi-synthetic penicillins. The mecA gene is located in a mobile genetic element identified as the staphylococcal chromosome cassette mec (SCCmec), composed of the mec complex, which encompasses the mecA gene and its regulators mecI and mecRI, by the complex of the ccr gene, responsible for the integration and excision of SCCmec, and the J region, which is not essential for the formation of SCCmec, but can carry genes that encode resistance to other non-β-lactam antimicrobials [13]. To date, 13 types of SCCmec (www.sccmec.org) have been identified, which are defined by the combination of the types of the ccr gene complex and the class of the mec gene complex, with the subtypes being defined by polymorphisms of the J region in the same combination of the mec and ccr complexes [13].
The objective of this study was to determine the agr and antimicrobial susceptibility of S. epidermidis isolated from blood culture capable of producing biofilm and to analyze the clonal profile of blood culture isolates from patients hospitalized over a period of 20 years.
2. Results
2.1. Identification of Strains
The CoNS identification by the biochemical method detected 223 (74.3%) Staphylococcus epidermidis, 27 (9.0%) Staphylococcus haemolyticus, 22 (7.3%) Staphylococcus hominis, 14 (4.7%) Staphylococcus warneri, nine (3.0%) Staphylococcus lugdunensis, and five (1.7%) Staphylococcus capitis. In contrast the genotypic technique of internal transcribed spacer by polymerase chain reaction (ITS-PCR) detected 223 (74.3%) S. epidermidis, 29 (9.7%) S. haemolyticus, 23 (7.7%) S. hominis, 11 (3.7%) S. warneri, nine (3.0%) S. lugdunensis, and five (1.7%) S. capitis. There was 98% agreement between the two methods used to identify the species of CoNS.
2.2. Detection of Biofilm Genes
We investigated the genes related to biofilm formation in the 300 isolates and the presence of ica operon genes and/or bhp and aap genes was detected in 163 (53.3%). The icaA gene was positive in 123 (41.3%) CoNS, the icaB in 165 (55.0%), the icaC in 222 (74.0%), and the icaD gene in 236 (78.7%). The presence of icaA, icaB, icaC, and icaD genes, concomitantly, was detected in 107 (35.7%) strains. We detected the bhp and aap genes in 37 (12.3%) and 104 (34.7%) of the CoNS, respectively. The distribution of these genes in relation to CoNS species can be visualized in Table 1.

Table 1.
Distribution of genes involved in the formation of Biofilm in Coagulase-negative Staphylococci (CoNS) species.
2.3. Detection of mRNA of Biofilm Genes in CoNS Samples
The 163 strains that were positive for the operon ica, bhp, and aap genes were submitted to the reverse transcription polymerase chain reaction (RT-PCR) technique for detection of mRNA. Plankton cells in which the mRNA of the operon ica or the bhp or aap genes were detected were considered capable of producing biofilm. The capacity to produce biofilm was verified in 61 (37.4%) S. epidermidis. Of these 61 samples, the complete detection of ica operon was confirmed in 51 (83.6%) S. epidermidis and the mRNA of the bhp and aap genes was detected in seven (11.5%) and 20 (32.8%), respectively. There was concomitant expression of the ica operon and bhp gene in three S. epidermidis and the operon ica and aap gene in 12 S. epidermidis. There was no expression of genes related to biofilm formation in the other species studied.
2.4. Investigation of Biofilm Production by Adherence to Polystyrene Plates
The 61 S. epidermidis that presented mRNA for icaA, icaD, bhp, and/or aap genes were submitted to the adhesion technique on polystyrene plates to verify the production of biofilm. Strong adherence was observed in 49.0% S. epidermidis, weak adherence in 33% and non-adherence in 18.0% (Figure 1).
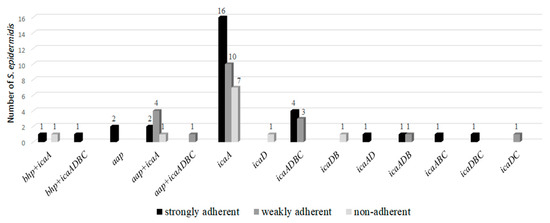
Figure 1.
Correlation between adherence and expression of the detected genes.
2.5. Determination of the agr Locus in Biofilm-Producing S. epidermidis
We investigated the agr locus (I, II, and III) in 61 S. epidermidis that expressed genes related to biofilm production. The agr I was detected in 47 (77.1%); 45.9% in those which expressed only the ica operon, 6.6% in those which expressed only the aap gene, 4.9% in those which expressed the bhp gene and ica operon, and 19.7% in those which expressed the aap gene and ica operon. The agr II was detected in 12 (19.6%) S. epidermidis, and also detected in all samples that expressed only the bhp gene. The agr locus was not found in two S. epidermidis that presented ica operon expression. These results are presented in Table 2.

Table 2.
Relationship of genes related to biofilm production and detection of accessory gene regulator (agr) locus in Staphylococcus epidermidis positive by the reverse transcription polymerase chain reaction (RT-PCR) technique.
2.6. Detection of mecA Gene and SCCmec in Biofilm-Producing S. epidermidis
We performed the mecA gene research on the 61 samples that confirmed the ability to produce biofilm by the RT-PCR technique and the mecA gene was detected in 55 (90.1%). The SCCmec typing determined five (9.0%) type I, 37 (60.6%) type III, and 14 (25.5%) type IV. The distribution of the mecA gene and the SCCmec types in the CoNS are shown in Table 3.

Table 3.
Distribution of the agr locus, mecA gene, and Staphylococcal chromosomal cassette mec (SCCmec) in S. epidermidis positive by the RT-PCR technique.
2.7. Determination of Minimum Inhibitory Concentration (MIC) of Antimicrobials in Biofilm-Producing S. epidermidis
We determined the MICs of the antimicrobials in the biofilm-producer S. epidermidis strains. Resistance to oxacillin was found in 83.6% of S. epidermidis and to tigecycline in 4.9%. However, intermediate resistance to quinupristin/dalfopristin was detected in one S. epidermidis presenting a MIC of 2 mg/mL for this drug. MIC values capable of inhibiting 50% and 90% of bacterial growth (MIC50 and MIC90) were determined, these values being considered indicators of oxacillin resistance. With the exception of oxacillin, vancomycin was the drug with the highest values of MIC50 and MIC90, but vancomycin resistance in S. epidermidis was not detected. The other drugs presented MIC values in the susceptibility range according to CLSI recommendations (2013). The results are shown in Table 4.

Table 4.
Minimum inhibitory concentration (MIC) of antimicrobials (mg/mL), range of MIC (mg/mL), and percentage of resistant CoNS.
2.8. Determination of the Clonal Profile of the Biofilm-Producer S. epidermidis
Determination of the clonal profile of biofilm-producer S. epidermidis was performed by the pulsed-field gel electrophoresis (PFGE) technique. The macro-synthesis analysis of chromosomal DNA, using an established coefficient of 80% of similarity, allowed identification of the presence of 11 clusters, designated alphabetic letters from A to K. Four larger clusters were determined with 6–10 S. epidermidis isolates: A, G, H, and J.
Cluster A comprised six S. epidermidis, four of which were identical (100% similarity) and isolated over a period of 17 years, from 1993 to 2009, and all of them were positive for the ica operon, SCCmec III, and agrI. Cluster G grouped eight samples isolated from 2000 to 2009, all with SCCmec IV, agrI, and ica operon, and bhp in three isolates. Cluster H was the largest group, with 10 S. epidermidis isolated from 2001 to 2004, all of them positive for SCCmec III, agrI, and ica operon, and aap genes in three samples. Cluster J grouped six samples from the period 2000 to 2007, with ica operon, SCCmec III, and agrI.
The other groups, including a smaller number of clones, were important, presenting isolated samples over long periods. Cluster E presented a single sample in 1992 and again in 2000, while cluster F presented isolated samples over a range of 12 years, with isolation in 1996, 2004, and 2008 (Figure 2).
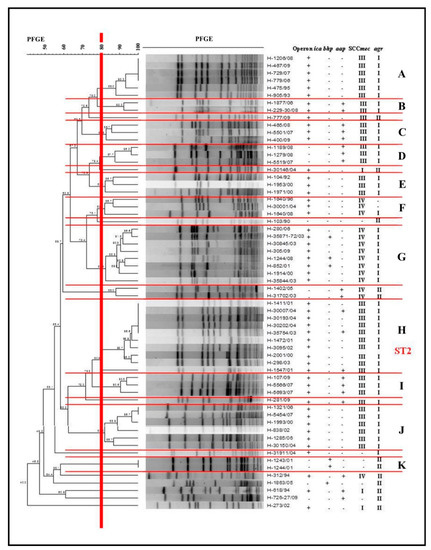
Figure 2.
Dendrogram generated by the Dice/UPGMA (Bionumerics Applied Maths) analysis of Staphylococcus epidermidis biofilm producers isolated from 1990 to 2009, indicating the presence (+) or absence of (−) operon ica expression, bhp and aap genes, and carrying of the SCCmec and accessory gene regulator (agr) locus.
We used the multilocus sequence typing (MLST) technique of Cluster H to characterize the largest number of isolates. A sample was selected and then submitted to the amplification and sequencing of the seven housekeeping genes, in order to obtain the type of sequence (ST). It was then characterized as ST2.
3. Discussion
Coagulase-negative staphylococci are opportunistic microorganisms capable of causing serious infections, mainly due to their ability of synthesizing biofilm, which protects the CoNS and increases the time spent in the hospital environment. In the current work, the capacity to produce biofilm was analyzed in 300 CoNS isolated from blood cultures, in addition to the possible mechanisms involved and susceptibility to antimicrobials used in the treatment of infections caused by these microorganisms.
The ability to adhere to surfaces and, consequently, to produce biofilm is the main virulence factors of CoNS, especially in S. epidermidis, which is the species most frequently isolated from clinical material [14]. The genes of ica operon were detected in all species studied, but the complete operon was detected in only 35.7% of the isolates, 98% of which belonged to the species S. epidermidis. The ability to produce polysaccharide intercellular adhesin (PIA), through the presence of mRNA, was detected only in the S. epidermidis strains, and the ica operon was expressed in 83.6%. Similar results were found in 2007 by Qin et al. [15], who detected ica genes in 21.8% of S. epidermidis from a group of 24 biofilm producing strains. These authors pointed out that the frequency of biofilm production and the presence of the ica operon are related to the different sources of lineages studied, such as sputum, blood cultures, catheters, and wounds. According to some studies, clinical isolates of S. epidermidis are those that most commonly present ica operon genes, as these genes are less frequent in CoNS isolated from healthy individuals, since the synthesis of PIA may require a great deal of adaptive cost, which is not advantageous for these bacteria [3].
It has recently been recognized that PIA is not essential for the formation of biofilm in S. epidermidis, since strains that did not have ica genes were isolated from biofilm-related infections. In some strains, biofilm formation may be additionally or exclusively mediated by proteins such as Bhp or Aap [16]. The presence of bhp and aap genes was also detected in our study, in 12.3% and 34.7% of the isolates, respectively. Although these percentages are lower than those presented by the ica operon, the genes were positive by the RT-PCR technique and were isolated or in concomitance with the ica operon, and the expression capacity was detected in 11.5% for the bhp gene and 32.8% for the aap gene. Similar results were found by Bowden et al. [17], who investigated these genes in clinical and nonclinical samples, determining the bhp gene in 9% of bacteremia isolates and 13% of skin colonizers, and the aap gene in 27% of bacteremias, 50% of contaminants in blood cultures, and 47% of S. epidermidis colonizers of the skin. The presence of the bhp and aap genes, concomitantly with the ica operon genes, suggests an association of the Bhp and Aap proteins with PIA, playing a role in intercellular adhesion and in the accumulation phase [17].
S. epidermidis that presented mRNA for icaA, icaD, bhp and/or aap genes were submitted to the adhesion technique on polystyrene plates to verify the production of biofilm. Strong adherence was observed in 49.0% S. epidermidis, weak adherence in 33% and non-adherent in 18.0%. The samples that expressed icaA (90.1%) were the ones that presented the strongest and weakest adherence. The expression of the bhp and aap genes were also important for and strong and weak adherence. The expression of the icaD gene not associated with icaA was less frequent (6.5%) and two samples were non-adherent. The environmental conditions or mechanism of gene regulation may have influenced the production of the protein and studies highlight the importance of the icaA gene in the production of staphylococcal biofilm and also of other genes, such as bhp and aap, in independent PIA strains [18,19].
The agr locus was investigated in S. epidermidis capable of producing biofilm, this being the most commonly detected agrI associated with the ica operon and aap gene. Strains that were positive for bhp gene expression showed agrII. As the regulation of the agr system is associated with cell density, these loci may be related to the release phases of biofilm cells, with the agrI and agrII loci being involved in this process [6,20]. S. epidermidis produces a series of exoproteases strictly regulated by the agr system, such as δ-toxin and other phenol-soluble modulins. These exoproteases act as surfactants, inhibiting the non-covalent bonds of the bacteria with the surface of the biofilm, resulting in detachment of the cells [3]. Studies have shown that in an S. epidermidis agr deletion mutant, increased biofilm development and intensified colonization in rabbits was due to non-regulation of exoproteins [21].
The mecA gene was positive in 90.1% of the biofilm-producer S. epidermidis, being present mainly in the samples that expressed the ica operon and aap gene and absent in those that expressed only the bhp gene. In the studies performed by Cabrera-Contreras et al. [22], the mecA gene was detected in 95% of S. epidermidis biofilm producers, and according to these authors, the presence of the mecA gene is enhanced in these samples, conferring resistance to the other β-lactam antibiotics. The SCCmec typing made possible to characterize SCCmec I, III, and IV in these samples, with SCCmec III being the most frequent. SCCmec III is the most commonly found element in multiresistant staphylococci related to nosocomial infections, since this element can carry other genes that confer resistance to other antimicrobial agents [23].
In addition to the mecA gene, the minimal inhibitory concentration of some antimicrobials used in the treatment of infections caused by CoNS was determined. Oxacillin presented 83.6% resistance, with MIC50 values above 256 mg/mL. The results also revealed 4.3% of tigecycline resistant samples and a sample with intermediate resistance to quinupristin/dalfopristin. Low MIC values were detected for linezolid and daptomycin, but higher MIC50 and MIC90 values for vancomycin were found. According to some authors, non-biofilm-producing bacteria are more susceptible to the action of antimicrobials and the host immune system. Therefore, the selective pressure imposed by the use of antimicrobials in the hospital environment favors the most adapted strains capable of producing biofilm [15,21]. Although there is an adaptive cost in the transport of genes related to biofilm formation and resistance to antimicrobials, a compensatory effect may occur through additional mutations favoring these microorganisms [24].
It is noteworthy that antimicrobial concentration values below the minimum inhibitory concentration (sub-MIC) can trigger an increase in the production of staphylococcus virulence factors, such as biofilm production. A study by Lázaro-Díez et al. [11] investigated the effects of subinhibitory concentrations of ceftaroline on the formation of biofilm by methicillin-resistant strains of S. aureus (MRSA) and found that sub-MIC of ceftaroline increased biofilm production by some strains of MRSA. Szczuka et al. [25] evaluated the sub-MICs of tigecycline and ciprofloxacin in strains of S. epidermidis and found that ciprofloxacin decreased biofilm formation, while tigecycline stimulated this process. The expression of icaA was increased by one-fold to 52-fold when the isolates were grown in the presence of 0.5 MIC tigecycline and 0.25 MIC resulted in an increase in icaA mRNA levels (by 2.6–12.6-fold) in three of the five isolates tested. These studies emphasize the importance of maintaining effective bactericidal concentrations of antimicrobials to combat infections related to Staphylococcus biofilm [11,25].
The clonal profile of the biofilm-producing S. epidermidis samples was established, with 11 clusters, including 4 larger clusters with a greater number of samples (6–10). The expression of the ica operon, agrI locus, and SCCmec III was found in clusters A, H, and J, and that of SCCmec IV in G. The potential to form biofilm favors these strains, which can colonize catheters, prostheses, and other materials for clinical use. This can cause invasive infections, and the proven multidrug resistance of these samples, as well as their ability to escape from the host’s immune system and the action of other antimicrobials, making it difficult to treat these infections. In addition, in the clusters mentioned above, as well as in the smaller clusters (E, F) the presence of these bacteria for long periods was verified, as it was possible to observe the same clone (A) over an interval of 17 years. The MLST analysis characterized the major cluster (H) as ST2. According to Otto [16], strains of S. epidermidis present a high level of diversity, and up to 74 types of sequences (STs) have been identified, the majority belonging to the clonal complex 2 (CC2), which includes the most frequently isolated ST2. The success of ST2 propagation may be due to the fact that all ST2s contain the IS256 insertion sequence and ica genes, factors that are correlated to invasive S. epidermidis isolates. In our study, ST2 isolates demonstrated biofilm formation capacity in vitro.
It is well established that biofilm-producing S. epidermidis are more pathogenic than non-biofilm producers, considered as commensal microorganisms. With the aim of better characterizing pathogenic strains of S. epidermidis isolated from clinical materials, studies suggest that the detection of biofilm expression capacity could be an excellent marker for the determination of invasive strains of S. epidermidis [18]. Biofilms can form on abiotic surfaces, such as surgical implants and catheters, and result in persistent infections that are difficult to treat, aggravating infections caused by S. epidermidis and increasing the length of stay of hospitalized patients.
4. Materials and Methods
4.1. Strains
A total of 300 strains isolated from blood cultures of patients hospitalized in the University Hospital of the Botucatu School of Medicine (HC-FMB) between 1990 and 2009 were studied. The strains were stored in the bacterial cultures collection of the Department of Microbiology and Immunology of the Institute of Biosciences, UNESP.
The strains were isolated as described by Koneman et al. [26] and coagulase- negative staphylococci were submitted to biochemical tests proposed in 2004 by Cunha et al. [27] for phenotypic identification of species.
Genotypic identification was performed using conserved sequence primers adjacent to the 16S and 23S genes for amplification of the ITS-PCR with the primers G1 and L1, as described by Barry et al. [28] and Couto et al. [29].
4.2. DNA Extraction
Total nucleic acid was extracted from S. aureus isolates cultured on blood agar, inoculated individually into brain-heart infusion broth, and incubated at 37 °C for 24 h. DNA extraction was performed using the Illustra kit (IllustraTM blood genomic Prep Mini Spin Kit), following the protocol described by Pereira et al. [30].
4.3. Detection of the Biofilm and mecA Gene
PCR reactions for the detection of genes related to biofilm formation (icaA, icaB, icaC, icaD, aap, and bhp) and the mecA gene were performed in microcentrifuge tubes of 0.5 mL in total volumes of 25 µL containing 10pmol of each primer (Table 5), 2.5 U of Taq DNA polymerase, 200 µM of deoxyribonucleotide triphosphate, 20 mM of Tris-HCl (pH 8.4), 0.75 mM of MgCl2, and 3 µL of DNA. The incubation was performed in a PTC-100 MJ research thermocycler, employing different parameters for each gene [31,32]. The efficiency of the amplifications was monitored by electrophoresis of the reaction in 2% agarose gel stained with SaberTM Safe.

Table 5.
Oligonucleotides used in PCR techniques for genotypic identification, mecA gene, biofilm and SCCmec type.
4.4. RNA Extraction and RT-PCR
Positive strains for genes related to biofilm formation were subjected to RNA extraction with the Illustra RNAspin Mini kit (GE Healthcare), following the manufacturer’s standards and the protocol described by Pereira et al. [30]. Total RNA was extracted from stationary phase planktonic cultures using the Illustra RNA spin Mini kit according to the manufacturer’s recommendations. After treatment with DNAse, the mRNA samples were converted into cDNA. For this purpose, 12 μL mRNA treated with DNAse was added to 1μL of random primer (75 ng/µL), 6 μL nuclease-free water, and 1 μL dNTP (200 μM). The mixture was heated for 5 min at 65 °C for RNA denaturation and primer binding and 4 µL reverse transcription buffer, 1 µL dithiothreitol and 1 µL SuperScriptTM III (200 U/μL) were added. cDNA was synthesized in a PTC-100 thermocycler using one cycle at 25 °C for 5 min, 50 °C for 60 min, and 70 °C for 15 min, followed by cooling at 4 °C. As internal control, the expression of 16S rRNA using 16S1 and 16S2 primers (Table 5) was analyzed, which corresponds to gene regions that are conserved in staphylococci and specific to the genus. The cDNA obtained was amplified by PCR and the resulting products were visualized by electrophoresis.
4.5. Investigation of Biofilm Production by Adherence to Polystyrene Plates
The quantitative method of adherence to polystyrene plates (TCP) proposed by Christensen et al. [36] was used in the present study, with modifications proposed by Oliveira and Cunha [14]. Trypticase soy broth (TSB) cultures were used, incubated for 24 h and then diluted 1:1 with TSB, prepared with 2% glucose. Sterile plates composed of 96 wells with a flat bottom (Costar, model 3599 manufactured by Corning Incorporated) were used. The wells were filled in quadruplicate with 200 μL of the diluted culture, using a multichannel pipette. International reference strains used as positive (S. epidermidis ATCC 35983) and negative controls (S. epidermidis ATCC 12228) and sterile TSB were included in all tests. The plates were incubated for 24 h at 37 °C and then the contents of each well were carefully aspirated using a multichannel pipette and then washed four times with 200 μL of phosphate buffered saline (PBS), pH 7.2. The plate was dried at room temperature for one hour. Then, the wells were stained with 2% violet crystal for one minute, and then the volume was aspirated, and the excess dye removed by washing the plates with distilled water using a multichannel pipette and subsequently dried at room temperature for 60 min, and the optical density reading was carried out in an ELISA reader (Labsystems, model Multiskan EX) using a 540-nm filter.
The isolates were classified into three categories: non-adherent, optical density equal to or lower than 0.111; weakly adherent, optical density higher than 0.111 or equal to or lower than 0.222; and strongly adherent, optical density higher than 0.222. When the cut-off corresponded to non-adherent, the isolates were classified as negative, and as positive when the cut-off corresponded to weakly or strongly adherent.
4.6. Determination of SCCmec
The SCCmec type was determined in mecA positive strains using multiplex PCR. The primers (Table 5) and parameters described by Oliveira et al. [34] and Machado et al. [37] were used for amplification.
4.7. Determination of Minimum Inhibitory Concentration by the E-Test
The susceptibility of the CoNS strains to the following antimicrobials were tested in vitro: oxacillin, vancomycin, daptomycin, linezolid, quinupristin/dalfopristin, and tigecycline. The MIC of these drugs was determined through the Etest® (BioMérieux, Marcy l’Étoile, France). The procedure uses an inert plastic strip, which incorporates a stabilized concentration gradient of the antimicrobial to be investigated. First an inoculum of the sample was seeded following the 0.5 McFarland scale on a Mueller-Hinton plate and then the plastic strips containing each of the antimicrobials were applied to the inoculum and incubated at 35 °C for 24 h.
4.8. Determination of agr Group
All positive isolates from the RT-PCR technique for the detection of mRNA of genes related to biofilm were subjected to typing of the agr locus. The primers and parameters described by Li et al. [8] were used for amplification. The efficiency of the amplifications was monitored by electrophoresis of the reaction in 2% agarose gel stained with SaberTM Safe.
4.9. Pulsed-Field Gel Electrophoresis
The positive isolates from the RT-PCR technique were submitted to clonal profile analysis through the PFGE technique. The clonal profile of the Staphylococcus spp. isolates was determined using the modified protocol of McDougal et al. [38], described by Pereira et al. [30].
BioNumerics software, version 6.1 (Applied Maths, Belgium), was used for analysis of similarity, calculation of the Dice correlation coefficient, and construction of the dendrogram by the UPGMA method (unweighted pair group method using arithmetic averages). Band position tolerance and optimization were set at 1.25% and 0.5%, respectively. A similarity coefficient of 80% was chosen for the definition of clusters.
4.10. Determination of Multilocus Sequence Typing (MLST)
MLST was performed according to standards described in 2000 by Enright et al. [35]. Each primer pair amplifies an internal fragment of the housekeeping gene (about 500 bp) (Table 5): carbamate kinase dehydrogenase (arcC), chiquimate dehydrogenase (aroE), glycerol kinase (glpF), guanylatoquinase (gmk_), phosphate acetyltransferase (pta_), triosephosphate isomerase (tpi_), and acetyl coenzyme A acetyltransferase (yqiL).
Purification was performed by the HiYield ™ Gel/PCR Fragments Extraction Kit and reactions were performed on an ABI3500 8 capillary (50 cm) sequencer using POP7 (Applied Biosystems) as the polymer. Sequence visualization (electrophoresis) was performed by Mega, Laser Gene and Bionumerics (version 7.1, Applied Maths, Belgium). The analysis and comparison of the sequences were performed on an internet database (http://www.mlst.net).
5. Conclusions
For the increased knowledge of the clones that persist in the Hospital das Clínicas of Botucatu, SP, Brazil over a period of 20 years, a characterization of multiresistance, the role ofthe agrI locus, and ability of biofilm formation in S. epidermidis strains isolated from blood culture is of great importance. Characterization of the major clone as ST2 highlights the relevance of these invasive samples in the hospital environment, emphasizing their potential to cause serious infections in hospitalized patients, and a high dissemination potential of these clones within the hospital environment.
Summary points:
- In some strains, biofilm formation may be additionally or exclusively mediated by proteins, such as Bhp or Aap.
- The agrI was associated with the ica operon and aap gene and strains that were positive for bhp gene expression showed agrII.
- The mecA gene was positive in 90.1% of the S. epidermidis capable of producing biofilm.
- The SCCmec typing made possible the characterization of SCCmec I, III, and IV in these samples, with SCCmec III being the most frequent.
- The expression of the ica operon, agrI locus, and SCCmec III was found in clusters A, H, and J, and that of SCCmec IV in G.
- The MLST analysis characterized the major cluster (H) as ST2.
Author Contributions
V.C.P. and M.d.L.R.d.S.d.C. designed the study. V.C.P., L.C.R. and M.d.L.R.d.S.d.C. wrote the manuscript. V.C.P., L.P.-H., A.d.O., D.F.M.R., K.B.M. and L.C.R. performed the experiments. All authors wrote and agreed with the final version. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the State funding agency São Paulo Research Foundation (FAPESP—Grant 2011/23742-2 and the National Council for Scientific and Technological Development (CNPq; grant 304051/2017-9).
Acknowledgments
We thank the State funding agency São Paulo Research Foundation (FAPESP—Grant 2011/23742-2 and Grant 2020/15256-4), the National Council for Scientific and Technological Development (CNPq; grant 304051/2017-9) and the Coordination for the Improvement of Higher Education Personnel (CAPES/PROEX 0708/2018) for the financial support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kloos, W.E. Staphylococcus and Micrococcus. In Manual of Clinical Microbiology; Murray, P.R., Baron, E.J., Pfaller, M.A., Tenover, F.C., Yolken, R.H., Eds.; American Society Microbiology: Washington, DC, USA, 1999; pp. 264–282. [Google Scholar]
- Rowlinson, M.C.; LeBourgeois, P.; Ward, K.; Song, Y.; Finegold, S.M.; Bruckner, D.A. Isolation of a strictly anaerobic strain of Staphylococcus epidermidis. J. Clin. Microbiol. 2006, 44, 857–860. [Google Scholar] [CrossRef]
- Fey, P.D.; Olson, M.E. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010, 5, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Virulence factors of the coagulase-negative staphylococci. Front. Biosci. 2004, 9, 841. [Google Scholar] [CrossRef] [PubMed]
- Cucarella, C.; Tormo, M.Á.; Úbeda, C.; Trotonda, M.P.; Monzón, M.; Peris, C.; Amorena, B.; Lasa, Í.; Penadés, J.R. Role of Biofilm-Associated Protein Bap in the Pathogenesis of Bovine Staphylococcus aureus. Infect. Immun. 2004, 72, 2177–2185. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Geisinger, E. Quorum sensing in staphylococci. Annu. Rev. Genet. 2008, 42, 541–564. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, J.A. Avaliação da Funcionalidade do Locus Acessory Gene Regulator (Agr) em Cepas de Staphylococcus aureus Brasileiras com Suscetibilidade Reduzida aos Glicopeptídeos. Ph.D. Thesis, Universidade de São Paulo, Sao Paulo, Brazil, 2006. [Google Scholar]
- Li, M.; Guan, M.; Jiang, X.F.; Yuan, F.Y.; Xu, M.; Zhang, W.Z.; Lu, Y. Genetic polymorphism of the accessory gene regulator (agr) locus in Staphylococcus epidermidis and its association with pathogenicity. J. Med. Microbiol. 2004, 53, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Huseby, M.; Shi, K.; Kent Brown, C.; Digre, J.; Mengistu, F.; Keun, S.S.; Bohach, G.A.; Schlievert, P.M.; Ohlendorf, D.H.; Earhart, C.A. Structure and biological activities of beta toxin from Staphylococcus aureus. J. Bacteriol. 2007, 189, 8719–8726. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar] [CrossRef]
- Lázaro-Díez, M.; Remuzgo-Martínez, S.; Rodríguez-Mirones, C.; Acosta, F.; Icardo, J.M.; Martínez-Martínez, L.; Ramos-Vivas, J. Effects of Subinhibitory Concentrations of Ceftaroline on Methicillin-Resistant Staphylococcus aureus (MRSA) Biofilms. PLoS ONE 2016, 11, e0147569. [Google Scholar] [CrossRef]
- Bouchami, O.; Achour, W.; Mekni, M.A.; Rolo, J.; Hassen, A. Ben Antibiotic resistance and molecular characterization of clinical isolates of methicillin-resistant coagulase-negative staphylococci isolated from bacteremic patients in oncohematology. Folia Microbiol. (Praha) 2011, 56, 122–130. [Google Scholar] [CrossRef]
- International Working Group on the Staphylococcal Cassette Chromosome Elements SCCmec about IWG-SCC EN. Available online: http://www.sccmec.org/Pages/SCC_aboutIWG-SCCEN.html (accessed on 17 August 2018).
- Oliveira, A.; Cunha, M.D.L.R. Comparison of methods for the detection of biofilm production in coagulase-negative staphylococci. BMC Res. Notes 2010, 3, 260. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Yang, X.; Yang, L.; Jiang, J.; Ou, Y.; Molin, S.; Qu, D. Formation and properties of in vitro biofilms of ica-negative Staphylococcus epidermidis clinical isolates. J. Med. Microbiol. 2007, 56, 83–93. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis—The “accidental” pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Bowden, M.G.; Chen, W.; Singvall, J.; Xu, Y.; Peacock, S.J.; Valtulina, V.; Speziale, P. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 2005, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Rohde, H.; Frankenberger, S.; Zähringer, U.; Mack, D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur. J. Cell Biol. 2010, 89, 103–111. [Google Scholar] [CrossRef]
- Pinheiro, L.; Brito, C.I.; Oliveira, A.; Pereira, V.C.; Cunha, M.L.R.S. Staphylococcus epidermidis and Staphylococcus haemolyticus: Detection of biofilm genes and biofilm formation in blood culture isolates from patients in a Brazilian teaching hospital. Diagn. Microbiol. Infect. Dis. 2016, 86, 11–14. [Google Scholar] [CrossRef]
- Schoenfelder, S.M.K.; Lange, C.; Eckart, M.; Hennig, S.; Kozytska, S.; Ziebuhr, W. Success through diversit—How Staphylococcus epidermidis establishes as a nosocomial pathogen. Int. J. Med. Microbiol. 2010, 300, 380–386. [Google Scholar] [CrossRef]
- Vuong, C.; Kocianova, S.; Yao, Y.; Carmody, A.B.; Otto, M. Increased Colonization of Indwelling Medical Devices by Quorum-Sensing Mutants of Staphylococcus epidermidis In Vivo. J. Infect. Dis. 2018, 190, 1498–1505. [Google Scholar] [CrossRef]
- Cabrera-Contreras, R.; Morelos-Ramírez, R.; Galicia-Camacho, A.N.; Meléndez-Herrada, E. Antibiotic Resistance and Biofilm production in Staphylococcus epidermidis Strains, Isolated from a Tertiary Care Hospital in Mexico City. Int. Sch. Res. Not. 2013, 2013, 1–5. [Google Scholar] [CrossRef]
- Ito, T.; Katayama, Y.; Hiramatsu, K. Cloning and Nucleotide Sequence Determination of the Entire mec DNA of Pre-Methicillin-Resistant Staphylococcus aureus N315 Cloning and Nucleotide Sequence Determination of the Entire mec DNA of Pre-Methicillin-Resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 1999, 43, 1449–1458. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef]
- Szczuka, E.; Jabłońska, L.; Kaznowski, A. Effect of subinhibitory concentrations of tigecycline and ciprofloxacin on the expression of biofilm-associated genes and biofilm structure of Staphylococcus epidermidis. Microbiology 2017, 163, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Koneman, E.W.; Allen, S.D.; Janda, W.M.; Schreckenberger, P.C.; Winn, W.C., Jr. Color Atlas and Textbook of Diagnostic Microbiology, 5th ed.; Lippincott: Philadelphia, PA, USA, 1997. [Google Scholar]
- Cunha, M.D.L.R.S.; Sinzato, Y.K.; Silveira, L.V. Comparison of methods for the identification of coagulase-negative staphylococci. Mem. Inst. Oswaldo Cruz 2004, 99, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Barry, T.; Colleran, G.; Glennon, M.; Dunican, L.K.; Gannon, F. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991, 1, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Couto, I.; Pereira, S.; Miragaia, M.; Santos Sanches, I.; De Lencastre, H. Identification of clinical staphylococcal isolates from humans by internal transcribed spacer PCR. J. Clin. Microbiol. 2001, 39, 3099–3103. [Google Scholar] [CrossRef] [PubMed]
- Pereira, V.C.; Pinheiro, L.; Oliveira, A.; Martins, K.B.; Riboli, D.F.; de Souza, M.D. Expression of superantigens and the agr system in Staphylococcus epidermidis. Microb. Pathogenes. 2018, 115, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Baldassarri, L.; Montanaro, L. Presence of icaA and icaD Genes and Slime Production in a Collection of Staphylococcal Strains from Catheter-Associated Infections. J. Clin. Microbiol. 2001, 39, 2151–2156. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Minamide, W.; Wada, K.; Nakamura, E.; Teraoka, H.; Watanabe, S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 1991, 29, 2240–2244. [Google Scholar] [CrossRef]
- Mason, W.J.; Blevins, J.S.; Beenken, K.; Wibowo, N.; Ojha, N.; Smeltzer, M.S. Multiplex PCR Protocol for the Diagnosis of Staphylococcal Infection. J. Clin. Microbiol. 2001, 39, 3332–3338. [Google Scholar] [CrossRef]
- Oliveira, D.C.; de Lencastre, H. Multiplex PCR Strategy for Rapid Identification of Structural Types and Variants of the mec Element in Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2155–2161. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.J.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus Sequence Typing for Characterization of Methicillin-Resistant and Methicillin-Susceptible Clones of Staphylococcus aureus Multilocus Sequence Typing for Characterization of Methicillin- Resistant and Methicillin-Susceptible Clones of Staphylococcus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [PubMed]
- Christensen, G.D.; Simpson, W.A.; Bisno, A.L.; Beachey, E.H. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 1982, 37, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.B.M.P.; Reiter, K.C.; Paiva, R.M.; Barth, A.L. Distribution of staphylococcal cassette chromosome mec (SCCmec) types I, II, III and IV in coagulase-negative staphylococci from patients attending a tertiary hospital in southern Brazil. J. Med. Microbiol. 2007, 56, 1328–1333. [Google Scholar] [CrossRef] [PubMed]
- McDougal, L.K.; Steward, C.D.; Killgore, G.E.; Chaitram, J.M.; McAllister, S.K.; Tenover, F.C. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: Establishing a national database. J. Clin. Microbiol. 2003, 41, 5113–5120. [Google Scholar] [CrossRef]
Sample Availability: The samples of compounds are not available from authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).