Design, Synthesis, and Molecular Docking of Paracyclophanyl-Thiazole Hybrids as Novel CDK1 Inhibitors and Apoptosis Inducing Anti-Melanoma Agents
Abstract
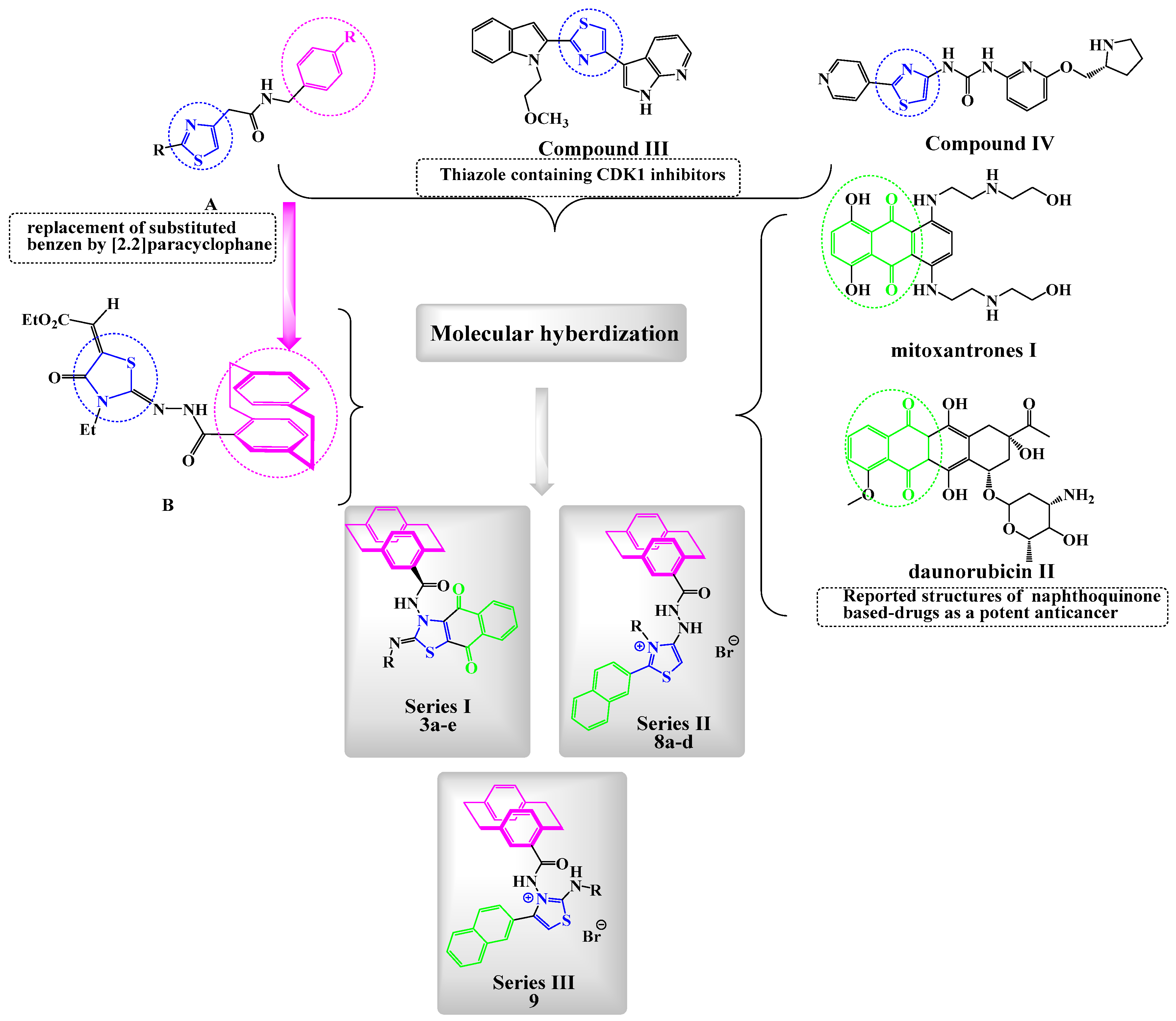
1. Introduction
Compound Design Rationale
2. Results and Discussion
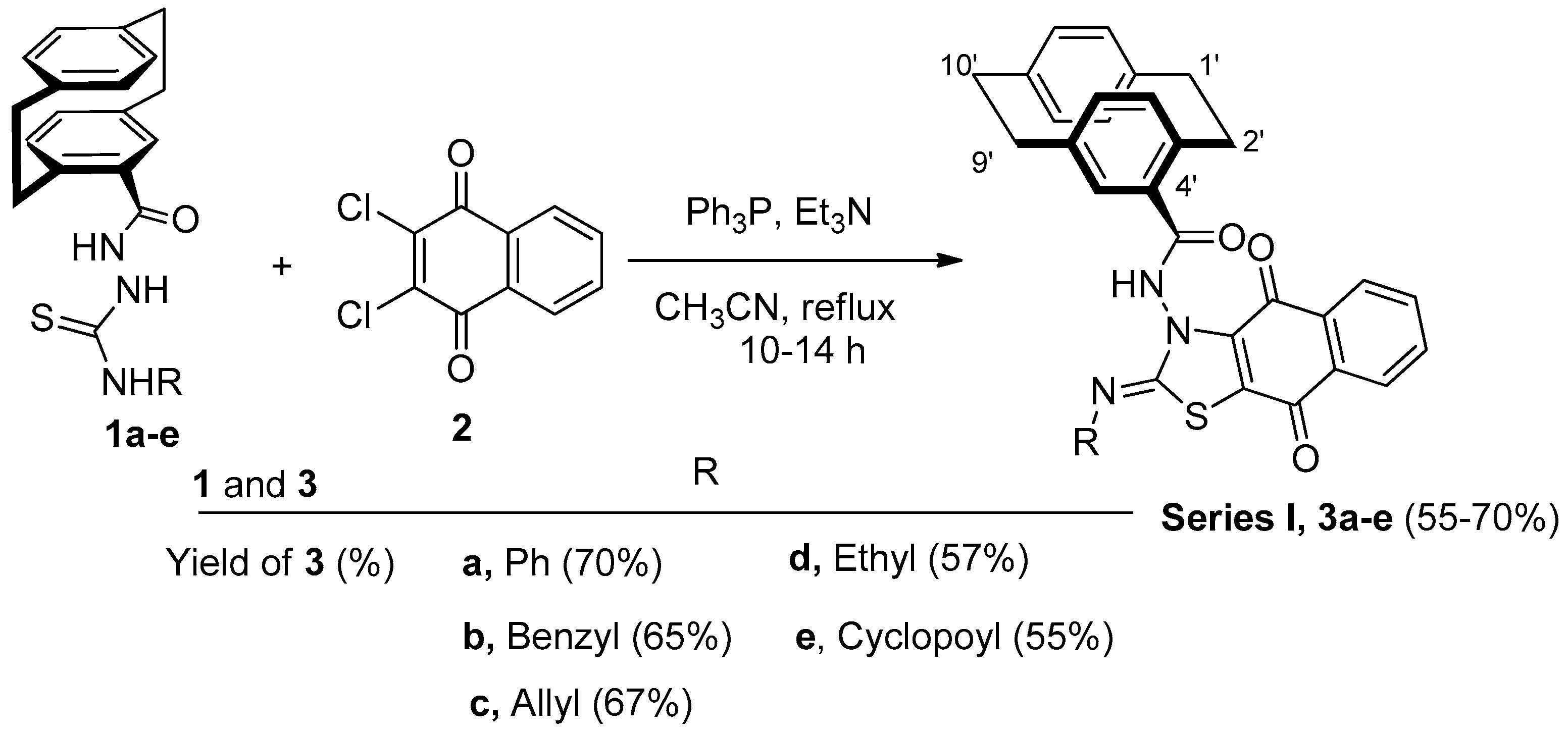
2.1. Chemistry
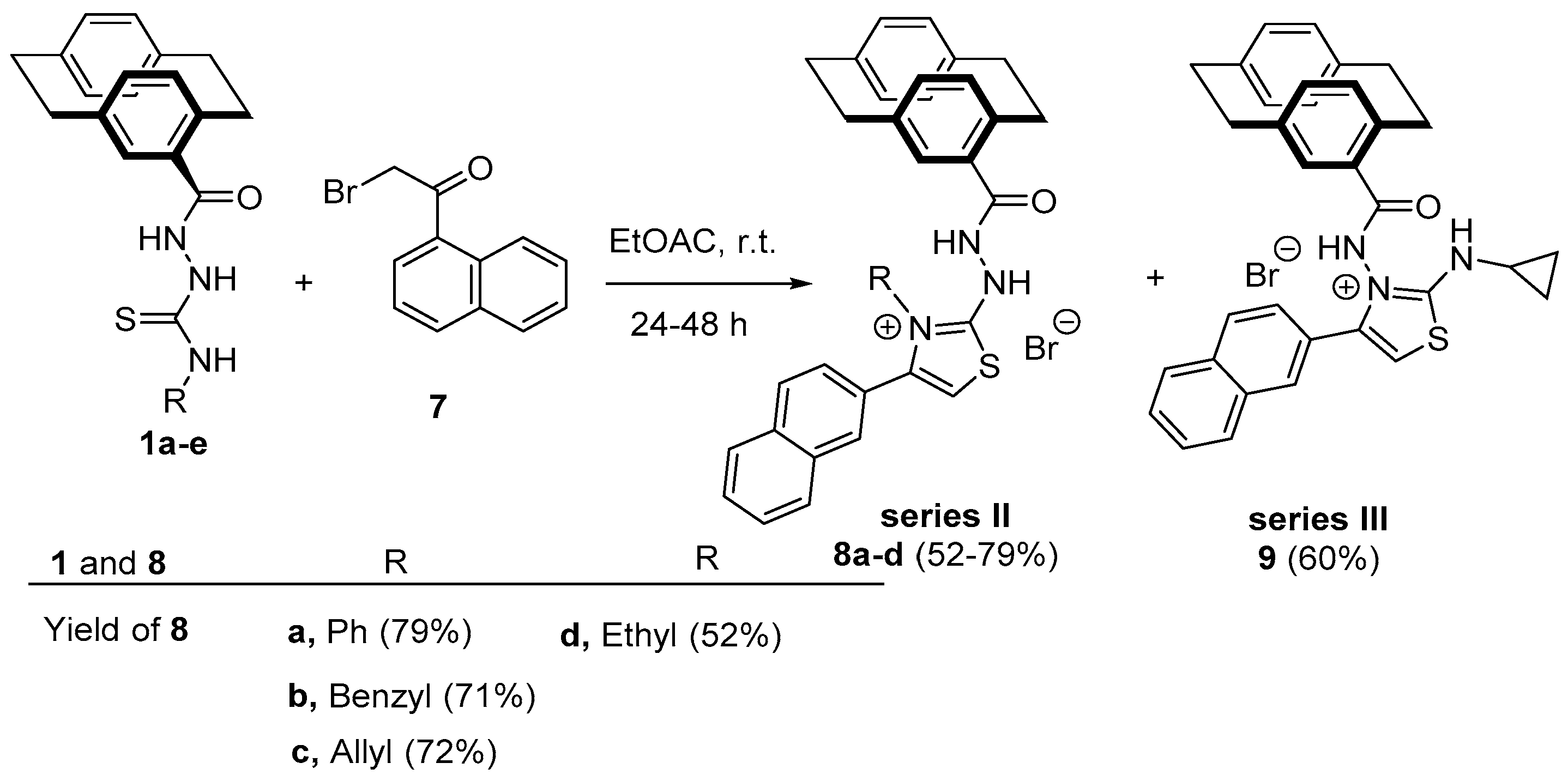
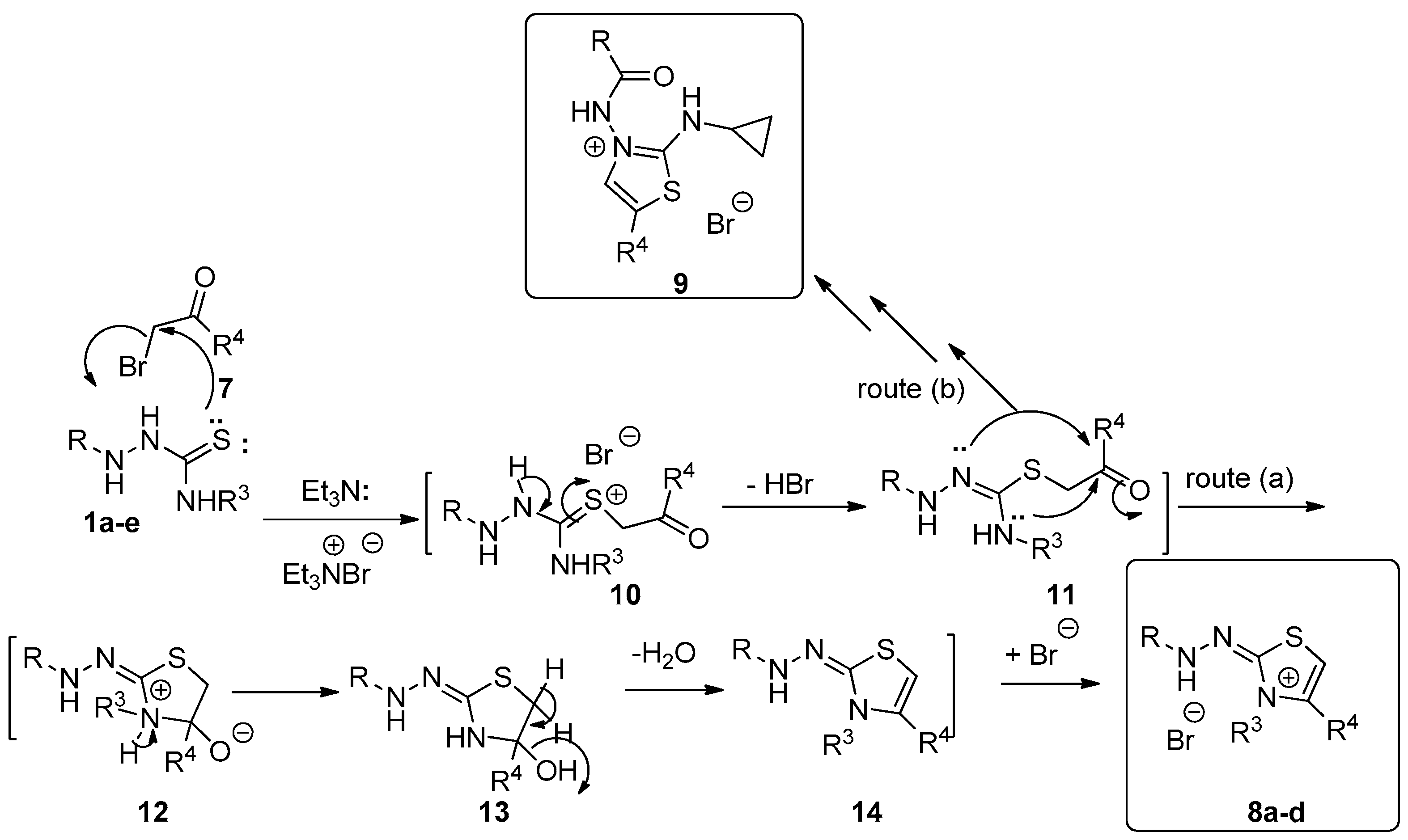
Synthesis of Substituted Thiazoles 8a–d and 9
2.2. Biological Activity Evaluation
2.2.1. In Vitro Screening of One-Dose Anticancer Activity on 60 Cancer Cell Lines
2.2.2. In Vitro Five-Dose Full NCI 60 Cell Panel Assay
2.2.3. Evaluation of In Vitro Antiproliferative Activities against Melanoma SK-MEL-5 Cancer Cell Line and Nontumorigenic SK-MEL-5 Cell Line
Structure-Activity Relationship
2.3. Selectivity Profiling of Compound 3c
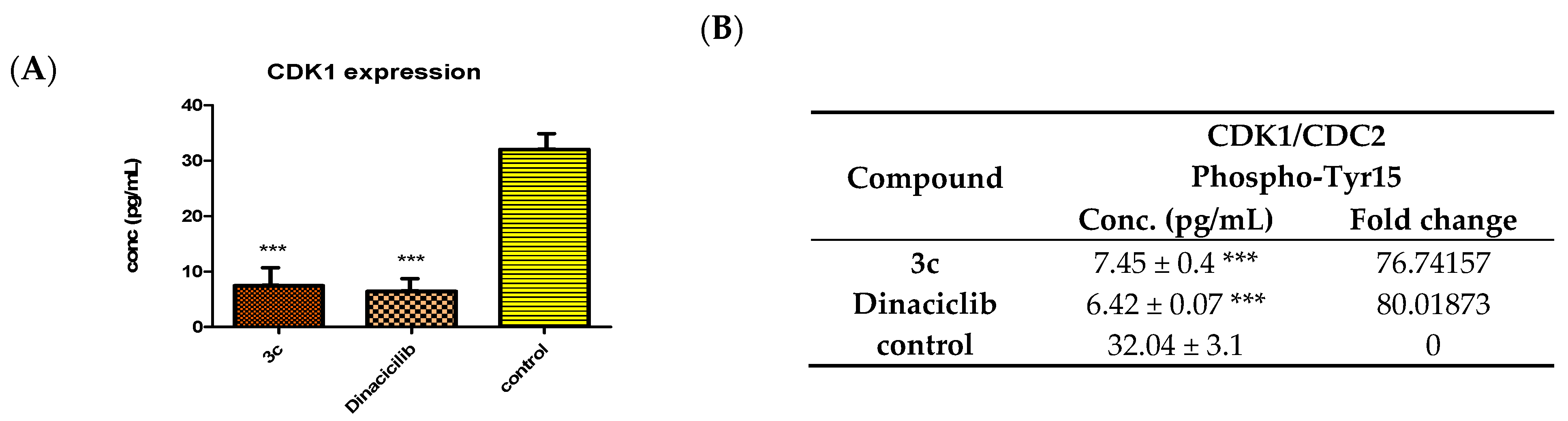
2.4. Inhibition of Phospho-CDK1/CDC2 Cell-Based Phosphorylation in SK-MEL-5 Cancer Cells
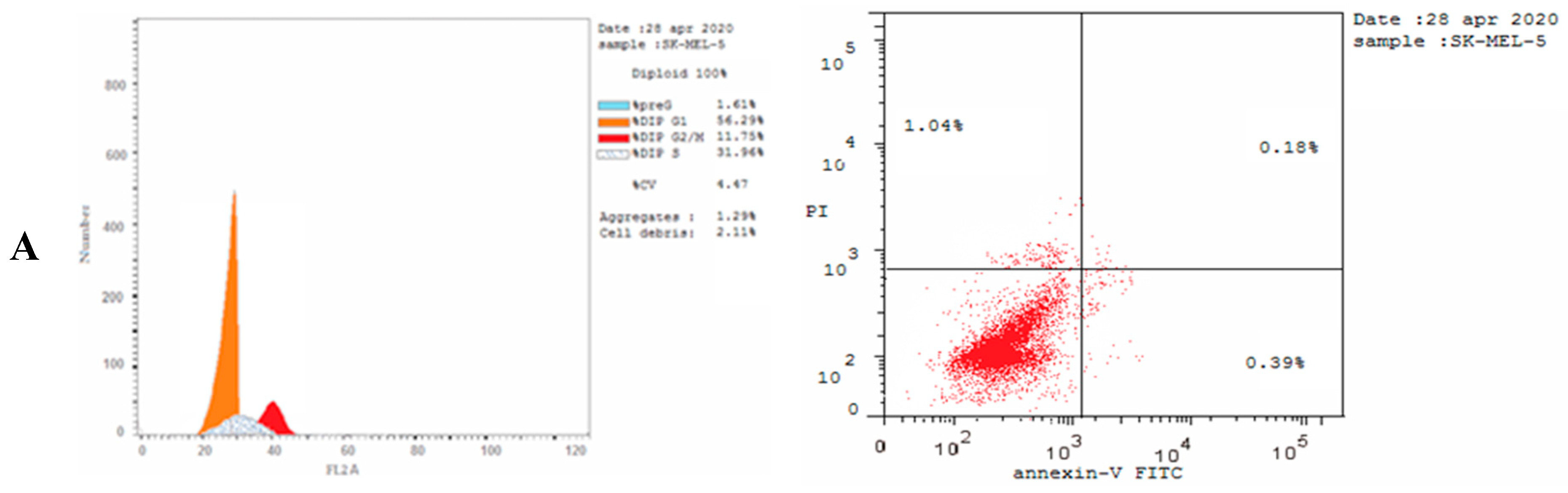
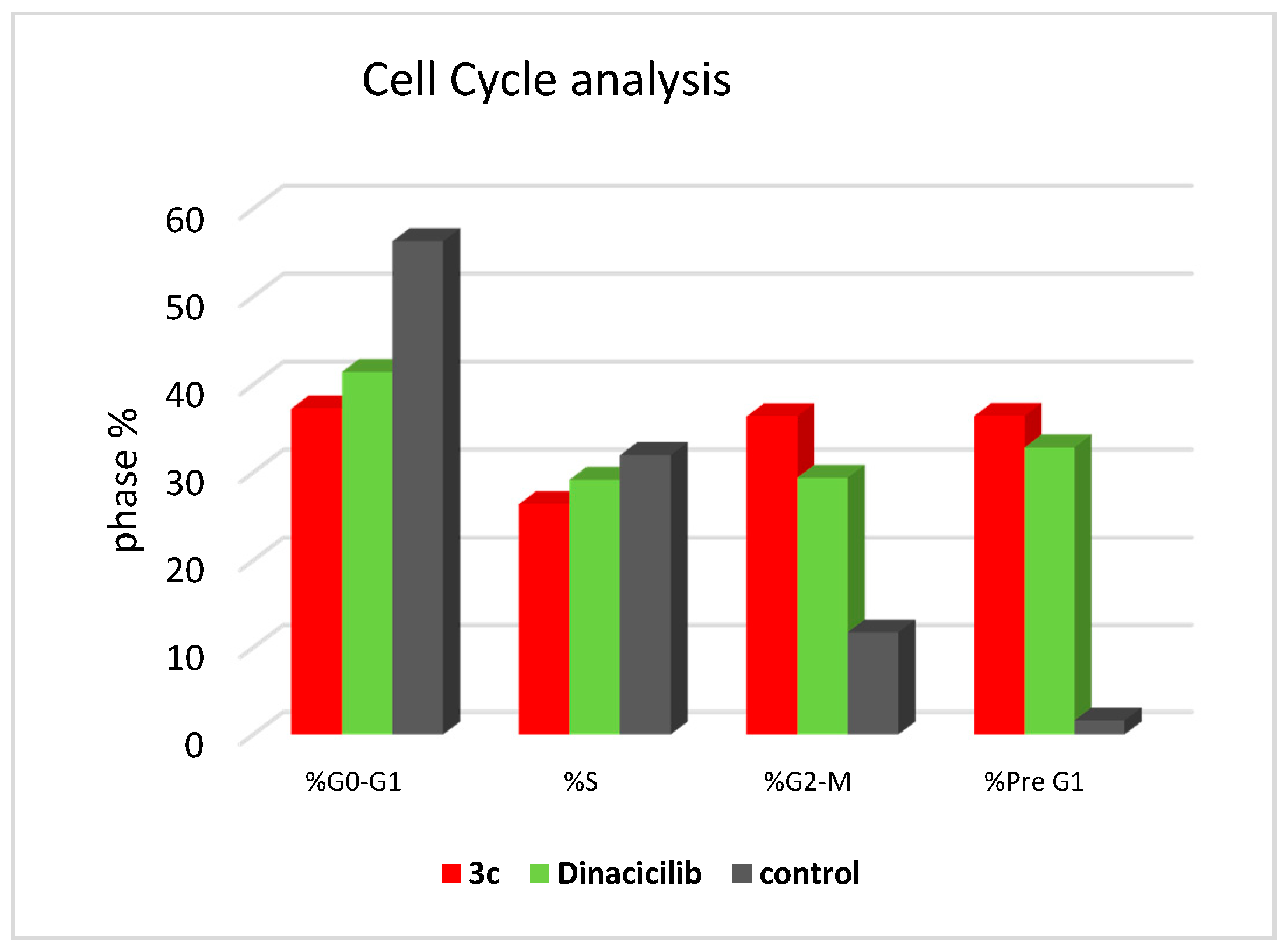
2.5. Cell Cycle Analysis
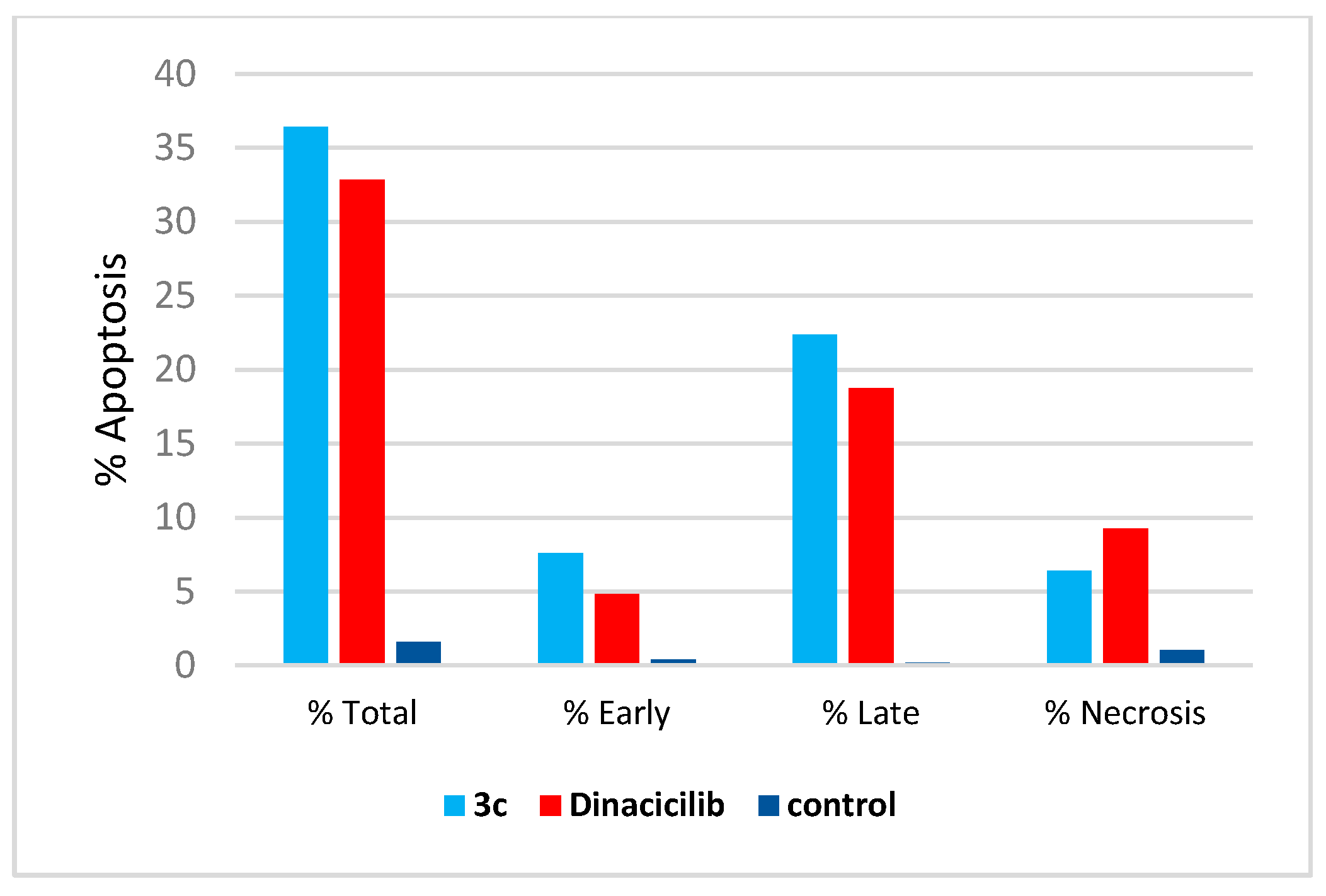
2.6. Caspase-3 Activation Assay
2.7. Molecular Modeling
3. Experimental
3.1. Chemistry
3.1.1. Starting Materials
3.1.2. Reactions of Hydrazinecarbothioamides 1a–e with DCHNQ (2); Preparation of Compounds 3a–e
3.1.3. (Z)-N-(4,9-Dioxo-2-(phenylimino)-4,9-dihydronaphtho[2,3-d]thiazol-3(2H)-yl)-4′-[2.2]paracyclophanylamide (3a)
3.1.4. (Z)-N-(2-(Benzylimino)-4,9-dioxo-4,9-dihydronaphtho[2,3-d]thiazol-3(2H)-yl)-4′-[2.2]paracyclophanylamide (3b)
3.1.5. (Z)-N-(2-(Allylimino)-4,9-dioxo-4,9-dihydronaphtho[2,3-d]thiazol-3(2H)-yl)-4′-[2.2]paracyclophanylamide (3c)
3.1.6. (Z)-N-(2-(Ethylimino)-4,9-dioxo-4,9-dihydronaphtho[2,3-d]thiazol-3(2H)-yl)4′-[2.2]paracyclophanylamide (3d)
3.1.7. (Z)-N-(2-(Cyclopropylimino)-4,9-dioxo-4,9-dihydronaphtho[2,3-d]thiazol-3(2H)-yl)-4′-[2.2]paracyclophanylamide (3e)
3.1.8. Reactions of Hydrazinecarbothioamide Derivatives 1a–f with 2-bromo-2′-acetonaphthone (4); Preparation of Compounds 8a–d and 9.
3.1.9. 2-(2-(4′-[2.2]Paracyclophonyl)hydrazinyl)-4-(naphth-2-yl)-3-phenylthiazol-3-ium Bromide (8a)
3.1.10. 2-(2-(4′-[2.2]Paracyclophonyl)hydrazineyl)-3-benzyl-4-(naphth-2-yl)-thiazol-3-ium Bromide (8b)
3.1.11. 3-Allyl-2-(2-(4′-[2.2]Paracyclophonyl)hydrazineyl)-4-(naphth-2-yl)-thiazol-3-ium Bromide (8c)
3.1.12. 2-(2-(4′-[2.2]Paracyclophonyl)hydrazineyl)-3-ethyl-4-(naphth-2-yl)-thiazol-3-ium Bromide (8d)
3.1.13. 3-(4′-[2.2]Paracyclophan)amido-2-(cyclopropylamino)-4-(naphth-2-yl)thiazol-3-ium Bromide (9)
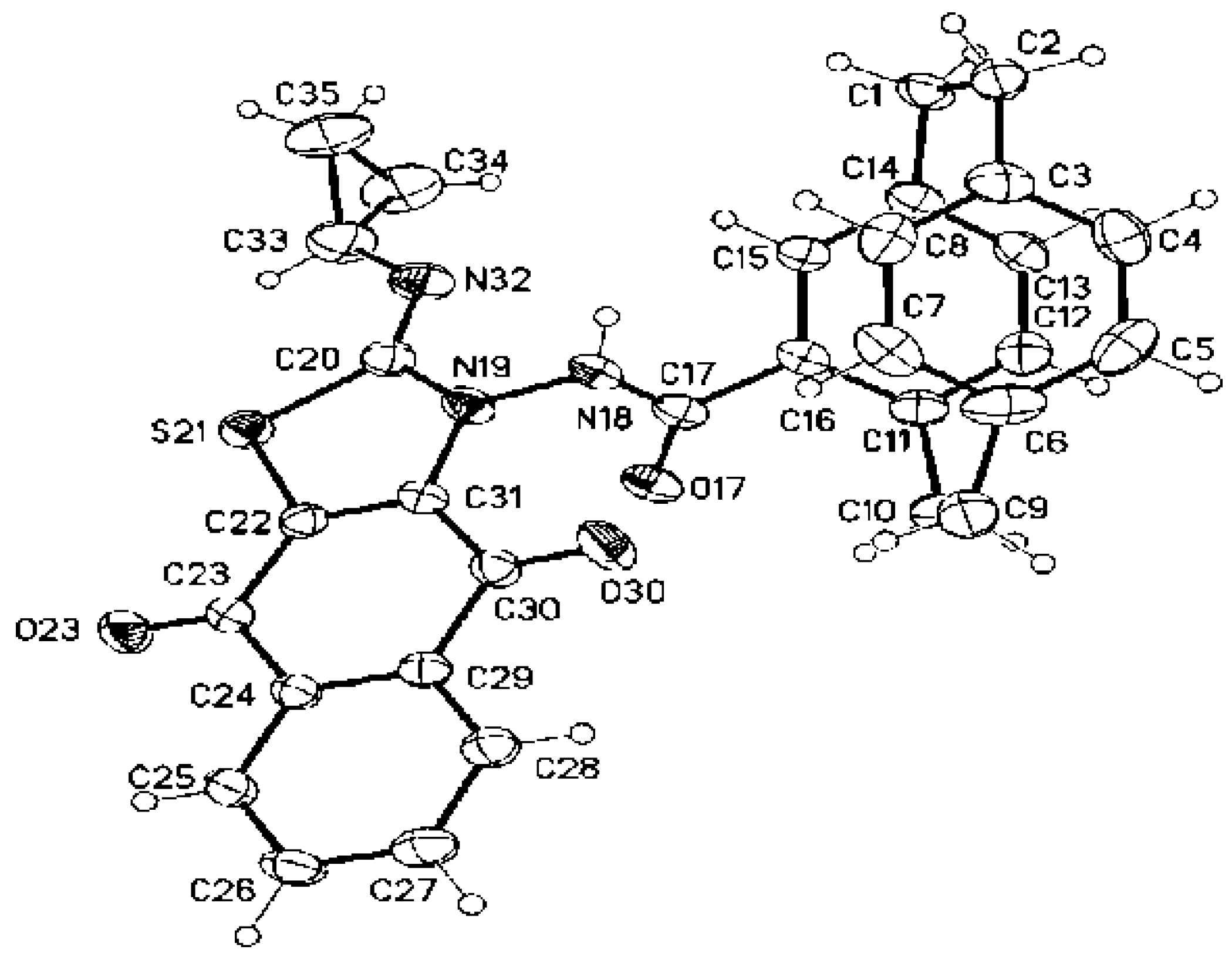
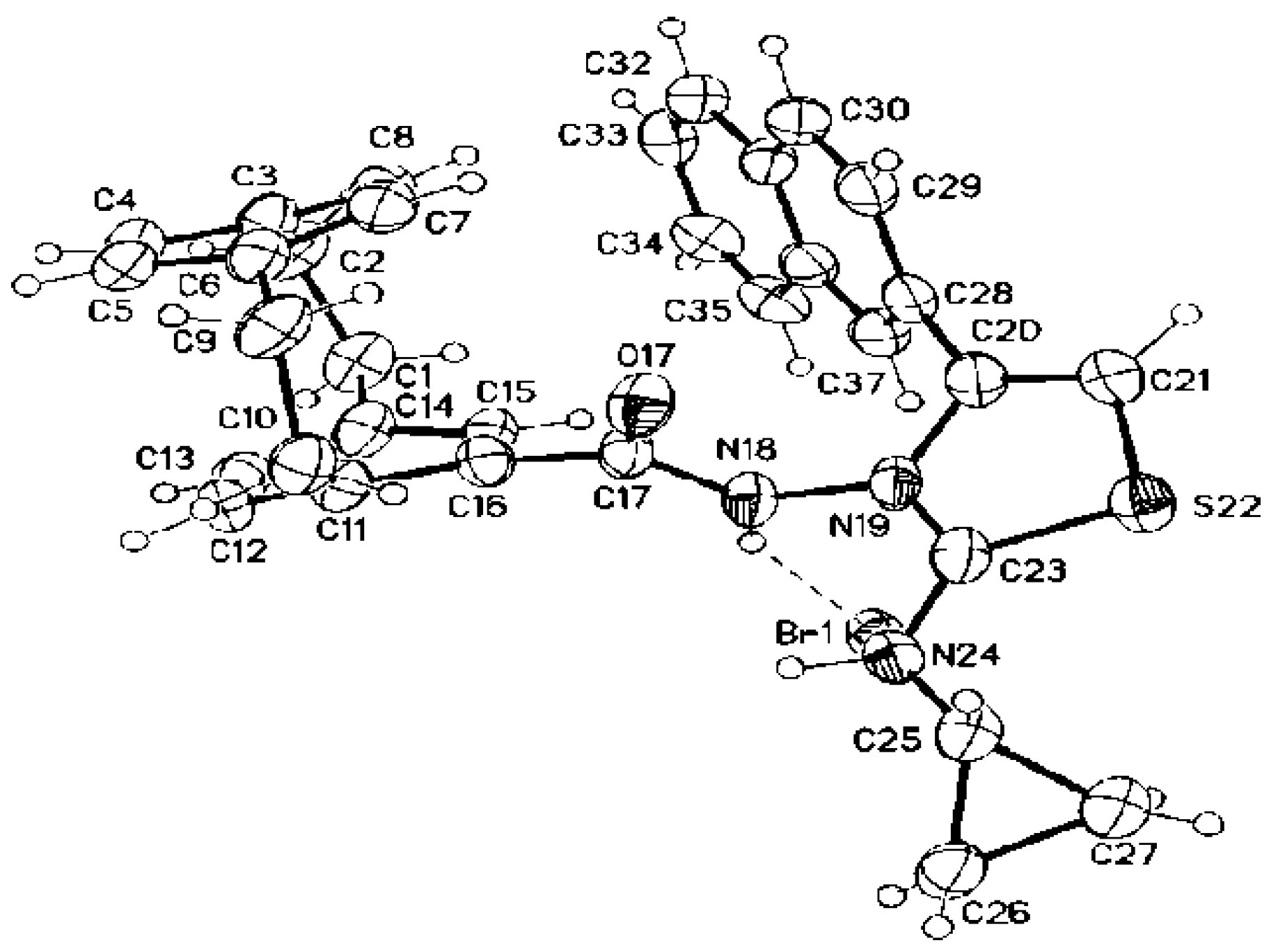
3.1.14. Crystal Structure Determinations of 3e, 8c, and 9
3.2. Biological Evaluation
3.2.1. Sixty Cancer Cell Lines Screening at the NCI
3.2.2. Cytotoxic Activity Using the MTT Assay and Evaluation of IC50
3.2.3. CDK Inhibitory Assay
3.2.4. Cell Cycle and Annexin-V FITC Apoptotic Study
3.2.5. Inhibition of Phospho-CDK1/CDC2 Cell-Based Phosphorylation in SK-MEL-5 Cancer Cells
3.2.6. Caspase-3 Activation Assay
3.2.7. Docking Study
3.2.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Higa, M.; Noha, N.; Yokaryo, H.; Ogihara, K.; Yogi, S. Three New Naphthoquinone Derivatives from Diospyros maritima B LUME. Chem. Pharm. Bull. 2002, 50, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Schüffler, A.; Liermann, J.C.; Kolshorn, H.; Opatz, T.; Anke, H. New naphthoquinone derivatives from the ascomycete IBWF79B-90A. Z. Nat. C 2009, 64, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Lin, Z.; Wang, D.; Feng, Y.; Sun, H. Plumbasides A—C three naphthoquinone derivatives from Ceratostigma minus. Phytochemistry 1994, 35, 1023–1025. [Google Scholar] [CrossRef]
- Elliott, M.C.; Long, M.S. Studies towards the total synthesis of batzelladine A. Org. Biomol. Chem. 2004, 2, 2003–2011. [Google Scholar] [CrossRef] [PubMed]
- Lednicer, D. Strategies for Organic Drug Synthesis and Design; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Neto, B.A.; Lapis, A.A.; Bernd, A.B.; Russowsky, D. Studies on the Eschenmoser coupling reaction and insights on its mechanism. Application in the synthesis of Norallosedamine and other alkaloids. Tetrahedron 2009, 65, 2484–2496. [Google Scholar] [CrossRef]
- Zhang, C.; Ondeyka, J.G.; Zink, D.L.; Basilio, A.; Vicente, F.; Collado, J.; Platas, G.; Bills, G.; Huber, J.; Dorso, K. Isolation, structure, and antibacterial activity of phaeosphenone from a Phaeosphaeria sp. discovered by antisense strategy. J. Nat. Prod. 2008, 71, 1304–1307. [Google Scholar] [CrossRef]
- Meazza, G.; Dayan, F.E.; Wedge, D.E. Activity of quinones on Colletotrichum species. J. Agric. Food Chem. 2003, 51, 3824–3828. [Google Scholar] [CrossRef]
- Fan, E.; Shi, W.; Lowary, T.L. Synthesis of daunorubicin analogues containing truncated aromatic cores and unnatural monosaccharide residues. J. Org. Chem. 2007, 72, 2917–2928. [Google Scholar] [CrossRef]
- Verma, R.P. Anti-cancer activities of 1,4-naphthoquinones: A QSAR study. Anticancer Agents Med. Chem. 2006, 6, 489–499. [Google Scholar] [CrossRef]
- Spyroudis, S. Hydroxyquinones: Synthesis and reactivity. Molecules 2000, 5, 1291–1330. [Google Scholar] [CrossRef]
- Liu, H.B.; Cai, B.; Cui, C.B.; Gu, Q.Q.; Zhao, Q.C.; Guan, H.S. Pterocaryquinone, a novel naphthoquinone derivative from Pterocarya tonkinesis. Chin. J. Chem. 2006, 24, 1683–1686. [Google Scholar] [CrossRef]
- Gu, C.; Zhai, J.; Jiang, J.; Liu, H.; Wang, L.; Zhu, D.; Ji, Y. An Efficient One-pot Synthesis of Aryl-substituted 1-(Thiazol-2-yl)-1H-pyrazole-3-carboxylates via a Hantzsch Synthesis-Knorr Reaction Sequence. Chin. J. Chem. 2014, 32, 179–190. [Google Scholar] [CrossRef]
- Aly, A.A.; El-Sheref, E.M.; Brown, A.B.; Bräse, S.; Nieger, M.; Abdelhafez, E.-S.M. New one-pot synthesis of 2-ylidenehydrazono-thiazoles. J. Sulfur Chem. 2019, 40, 641–647. [Google Scholar] [CrossRef]
- Payton, M.; Chung, G.; Yakowec, P.; Wong, A.; Powers, D.; Xiong, L.; Zhang, N.; Leal, J.; Bush, T.L.; Santora, V. Discovery and evaluation of dual CDK1 and CDK2 inhibitors. Cancer Res. 2006, 66, 4299–4308. [Google Scholar] [CrossRef]
- Parrino, B.; Attanzio, A.; Spano, V.; Cascioferro, S.; Montalbano, A.; Barraja, P.; Tesoriere, L.; Diana, P.; Cirrincione, G.; Carbone, A. Synthesis, antitumor activity and CDK1 inhibition of new thiazole nortopsentin analogues. Eur. J. Med. Chem. 2017, 138, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Spruck, C.H.; Won, K.-A.; Reed, S.I. Deregulated cyclin E induces chromosome instability. Nature 1999, 401, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wu, J.; Wu, Y.; Chen, C.; Zou, F.; Wang, A.; Wu, H.; Hu, Z.; Jiang, Z.; Liu, Q. Discovery of 4-(((4-(5-chloro-2-(((1s,4s)-4-((2-methoxyethyl)amino)cyclohexyl)amino)pyridin-4-yl)thiazol-2-yl) amino)methyl) tetrahydro-2H-pyran-4-carbonitrile (JSH-150) as a novel highly selective and potent CDK9 kinase inhibitor. Eur. J. Med. Chem. 2018, 158, 896–916. [Google Scholar] [CrossRef] [PubMed]
- Matsushime, H.; Quelle, D.; Shurtleff, S.; Shibuya, M.; Sherr, C.; Kato, J. D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol. 1994, 14, 2066–2076. [Google Scholar] [CrossRef]
- Abdel-Hamid, O.M.; Abeer, A.N.; Emam, M.A.; Elshimaa, M.A. The ameliorative effect of Vitamin C in experimentally induced colon cancer in rats. Benha Vet. Med. J. 2018, 34, 329–343. [Google Scholar] [CrossRef]
- Gibson, S.E.; Knight, J.D. [2.2]Paracyclophane derivatives in asymmetric catalysis. Org. Biomol. Chem. 2003, 1, 1256–1269. [Google Scholar] [CrossRef]
- Gulder, T.; Baran, P.S. Strained cyclophane natural products: Macrocyclization at its limits. Nat. Prod. Rep. 2012, 29, 899–934. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Z.; Spuling, E.; Knoll, D.M.; Bräse, S. Regioselective functionalization of [2.2]paracyclophanes: Recent synthetic progress and perspectives. Angew. Chem. Int. Ed. 2020, 59, 2156–2170. [Google Scholar] [CrossRef] [PubMed]
- Aly, A.A.; Brown, A.B. Asymmetric and fused heterocycles based on [2.2]paracyclophane. Tetrahedron 2009, 65, 8055–8089. [Google Scholar] [CrossRef]
- Aly, A.A.; Hopf, H.; Ernst, L.; Dix, I.; Jones, P.G. New Cycloadditions of (E)-N,α-Dimethyl-α-(4-[2.2] paracylophanyl)nitrone. Eur. J. Org. Chem. 2006, 2006, 3001–3006. [Google Scholar] [CrossRef]
- Aly, A.A. Cycloaddition of (E)-N-[2-([2.2]paracyclophan-4-yl) ethylidene] methylamine-N-oxide with 2,3-diphenylcyclopropenones and dibenzoyl acetylene; synthesis of new paracyclophanylpyrroles. J. Chem. Res. 2007, 2007, 451–454. [Google Scholar] [CrossRef]
- Aly, A.A.; Hopf, H.; Jones, P.G.; Dix, I. Cycloadditions of α-(4-[2.2] paracyclophanyl)-N-methyl nitrone. Tetrahedron 2006, 62, 4498–4505. [Google Scholar] [CrossRef]
- Hopf, H.; Aly, A.A.; Swaminathan, V.N.; Ernst, L.; Dix, I.; Jones, P.G. A simple route to a pyridinyl[2.2] paracyclophane. Eur. J. Org. Chem. 2005, 2005, 68–71. [Google Scholar] [CrossRef]
- Aly, A.A.; Bräse, S.; Weis, P. Tridentate and bidentate copper complexes of [2.2]paracyclophanyl-substituted thiosemicarbazones, thiocarbazones, hydrazones and thioureas. J. Mol. Struct. 2019, 1178, 311–326. [Google Scholar] [CrossRef]
- Hammam, A.; Bayoumy, B. Reaction of thioamides with 2,3-dichloro-1,4-naphthoquinone. A novel synthesis of naphtho [2, 3-d] thiazole-4, 9-diones. Collect. Czechoslov. Chem. Commun. 1985, 50, 71–79. [Google Scholar] [CrossRef]
- Aly, A.A.; Ahmed, E.K.; El-Mokadem, K.M. A convenient and efficient method for the synthesis of benzo-and naphthothiazolediones. J. Sulfur Chem. 2006, 27, 419–426. [Google Scholar] [CrossRef]
- Li, Z.; Yang, Q.; Qian, X. Novel heterocyclic family of phenyl naphthothiazole carboxamides derived from naphthalimides: Synthesis, antitumor evaluation, and DNA photocleavage. Bioorg. Med. Chem. 2005, 13, 3149–3155. [Google Scholar] [CrossRef]
- Konstantinova, L.S.; Lysov, K.A.; Souvorova, L.I.; Rakitin, O.A. Synthesis of 2,3-dihydronaphtho [2,3-d][1,3]thiazole-4,9-diones and 2,3-dihydroanthra[2,3-d][1,3]thiazole-4,11-diones and novel ring contraction and fusion reaction of 3H-spiro[1,3-thiazole-2,1′-cyclohexanes] into 2,3,4,5-tetrahydro-1H-carbazole-6,11-diones. Beilstein J. Org. Chem. 2013, 9, 577–584. [Google Scholar]
- Ding, A.; Guo, H. Comprehensive Organic Synthesis; Elsevier: Oxford, UK, 2014. [Google Scholar]
- Hussaini, S.R.; Chamala, R.R.; Wang, Z. The Eschenmoser sulfide contraction method and its application in the synthesis of natural products. Tetrahedron 2015, 36, 6017–6086. [Google Scholar] [CrossRef]
- Olson, E. Combination Therapies in Advanced, Hormone Receptor–Positive Breast Cancer. J. Adv. Pract. Oncol. 2018, 9, 43. [Google Scholar]
- Kumar, S. Caspase function in programmed cell death. Cell Death Differ. 2007, 14, 32–43. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT-integrated space-group and crystal-structure determination. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Aly, A.A.; Bräse, S.; Hassan, A.A.; Mohamed, N.K.; El-Haleem, L.E.A.; Nieger, M.; Morsy, N.M.; Abdelhafez, E.M.N. New Paracyclophanylthiazoles with Anti-Leukemia Activity: Design, Synthesis, Molecular Docking, and Mechanistic Studies. Molecules 2020, 25, 3089. [Google Scholar] [CrossRef]
- Parsons, S.; Flack, H.D.; Wagner, T. Use of intensity quotients and differences in absolute structure refinement. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2013, 69, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, M.; Abdelhamid, D.; Abdelhafez, E.M.; Ibrahim, M.A.; Gamal-Eldeen, A.M.; Aly, O.M. Synthesis, antiproliferative, anti-tubulin activity, and docking study of new 1,2,4-triazoles as potential combretastatin analogues. Eur. J. Med. Chem. 2017, 141, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Maklad, R.M.; AbdelHafez, E.-S.M.; Abdelhamid, D.; Aly, O.M. Tubulin Inhibitors: Discovery of a New Scaffold Targeting Extra-binding Residues within the Colchicine Site through Anchoring Substituents Properly Adapted to their Pocket by a Semi-flexible Linker. Bioorg. Chem. 2020, 103767. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.P.; Poon, R.Y.; Ma, H.T. Inhibitory phosphorylation of cyclin-dependent kinase 1 as a compensatory mechanism for mitosis exit. Mol. Cell. Biol. 2011, 31, 1478–1491. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lenardo, M.J. Roles of caspases in apoptosis, development, and cytokine maturation revealed by homozygous gene deficiencies. J. Cell Sci. 2000, 113, 753–757. [Google Scholar] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Hawkins, P.C.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef]
- Norbury, C.J.; Hickson, I.D. Cellular responses to DNA damage. Annu. Rev. Pharmacol. Toxicol. 2001, 41, 367–401. [Google Scholar] [CrossRef]
- Aly, A.A.; Sayed, S.M.; Abdelhafez, E.-S.M.; Abdelhafez, S.M.N.; Abdelzaher, W.Y.; Raslan, M.A.; Ahmed, A.E.; Thabet, K.; El-Reedy, A.A.; Brown, A.B. New quinoline-2-one/pyrazole derivatives; design, synthesis, molecular docking, anti-apoptotic evaluation, and caspase-3 inhibition assay. Bioorg. Chem. 2020, 94, 103348. [Google Scholar] [CrossRef]
- Gasteiger, J.; Marsili, M. Iterative Partial Equalization of Orbital Electronegativity—A Rapid Access to Atomic Charges. Tetrahcdron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Wood, D.J.; Korolchuk, S.; Tatum, N.J.; Wang, L.-Z.; Endicott, J.A.; Noble, M.E.M.; Martin, M.P. Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition. Cell Chem. Biol. 2019, 26, 121–130.e5. [Google Scholar] [CrossRef]
- Gordon, J.C.; Myers, J.B.; Folta, T.; Shoja, V.; Heath, L.S.; Onufriev, A. H++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005, 33, W368–W371. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed]







| Panel/Cell Line | 3a | 3b | 3c | 3d | 3e | 8a | 8b | 8c | 8d | |
|---|---|---|---|---|---|---|---|---|---|---|
| Leukemia | CCRF-CEM | 20.15 | 59.23 | 103.65 | 116.03 | 77.39 | 7.77 | 49.02 | 58.83 | 48.46 |
| HL-60(TB) | 8.41 | 72.97 | 120.12 | 118.34 | 97.61 | 3.67 | 17.11 | 50.76 | 46.34 | |
| K-562 | 22.24 | 51.17 | 112.34 | 128.49 | 74.12 | 14.31 | 54.53 | 65.03 | 58.59 | |
| MOLT-4 | 16.68 | 78.95 | 115.46 | 132.80 | 91.47 | 11.35 | 32.30 | 54.75 | 43.07 | |
| RPMI-8226 | 24.80 | 41.03 | 127.65 | 133.56 | 80.48 | 9.83 | 37.81 | 74.17 | 62.22 | |
| SR | 38.87 | 51.66 | 108.54 | 103.66 | 70.59 | 11.99 | 53.27 | 60.88 | 45.35 | |
| Non-Small Cell Lung Cancer | A549/ATCC | 35.96 | 20.00 | 76.74 | 138.85 | 32.30 | 2.64 | 12.65 | 43.90 | 28.88 |
| EKVX | 29.59 | 11.57 | 120.17 | 161.32 | 32.49 | 7.32 | 21.69 | 41.82 | 31.03 | |
| HOP-62 | 33.63 | 22.46 | 80.49 | 158.60 | 45.90 | 6.03 | 22.55 | 24.84 | 19.99 | |
| HOP-92 | 0 | 0 | 78.49 | 109.79 | 4.51 | 3.02 | 11.28 | 40.36 | 28.25 | |
| NCI-H226 | 48.81 | 33.73 | 51.50 | 26.96 | 35.15 | 5.88 | 13.23 | 24.77 | 21.36 | |
| NCI-H23 | 69.39 | 68.04 | 134.07 | 132.12 | 90.37 | 2.21 | 21.82 | 44.16 | 39.27 | |
| NCI-H322M | 0.21 | 0 | 32.21 | 19.52 | 3.95 | 1.65 | 5.69 | 2.44 | 4.81 | |
| NCI-H460 | 28.14 | 25.07 | 87.28 | 90.38 | 46.08 | 0 | 39.19 | 47.70 | 29.96 | |
| NCI-H522 | 31.97 | 47.13 | 120.20 | 134.81 | 62.67 | 7.64 | 18.21 | 23.62 | 22.73 | |
| Colon Cancer | COLO 205 | 0 | 0 | 80.85 | 57.45 | 3.67 | 0 | 15.44 | 55.51 | 39.55 |
| HCC-2998 | 13.21 | 0 | 133.50 | 137.87 | 38.63 | 0 | 2.86 | 10.89 | 0 | |
| HCT-116 | 39.13 | 44.14 | 132.42 | 150.92 | 109.10 | 8.46 | 63.36 | 72.90 | 59.89 | |
| HCT-15 | 48.69 | 44.66 | 168.35 | 168.11 | 97.21 | 0 | 57.16 | 50.52 | 33.91 | |
| HT29 | 8.23 | 0.35 | 73.95 | 50.97 | 0 | 5.99 | 26.49 | 69.86 | 50.14 | |
| KM12 | 21.19 | 25.15 | 93.96 | 129.88 | 64.80 | 0 | 18.15 | 43.43 | 17.79 | |
| SW-620 | 24.10 | 22.49 | 159.75 | 156.56 | 81.66 | 4.29 | 20.05 | 27.07 | 18.93 | |
| CNS Cancer | SF-268 | 32.26 | 41.37 | 91.21 | 110.65 | 54.82 | 15.04 | 19.26 | 37.67 | 29.00 |
| SF-295 | 12.21 | 5.24 | 30.80 | 69.03 | 20.44 | 9.26 | 14.92 | 54.00 | 36.89 | |
| SF-539 | 7.45 | 12.52 | 198.29 | 194.41 | 94.83 | 0 | 4.73 | 22.07 | 18.69 | |
| SNB-19 | 39.25 | 25.88 | 95.41 | 174.30 | 66.30 | 1.20 | 9.35 | 33.22 | 17.91 | |
| SNB-75 | 63.66 | 66.13 | 194.63 | 198.23 | 110.40 | 20.65 | 27.60 | 46.73 | 41.79 | |
| U251 | 17.35 | 29.29 | 99.78 | 181.45 | 67.40 | 8.65 | 42.13 | 46.10 | 26.06 | |
| Melanoma | LOX IMVI | 37.84 | 64.25 | 180.28 | 183.86 | 99.49 | 4.55 | 39.20 | 40.06 | 22.40 |
| MALME-3M | 169.26 | 94.48 | 188.59 | 187.38 | 164.33 | 0 | 0.23 | 23.30 | 14.64 | |
| M14 | 95.86 | 80.70 | 193.88 | 193.47 | 121.83 | 0 | 13.94 | 24.45 | 14.96 | |
| MDA-MB-435 | 172.49 | 185.53 | 193.49 | 197.05 | 193.24 | 0 | 7.06 | 32.55 | 24.57 | |
| SK-MEL-2 | 24.77 | 22.73 | 117.29 | 140.56 | 50.50 | 0 | 0.55 | 14.41 | 21.90 | |
| SK-MEL-28 | 7.59 | 8.64 | 135.36 | 181.24 | 59.83 | 0 | 8.15 | 27.78 | 18.01 | |
| SK-MEL-5 | 60.33 | 48.69 | 198.49 | 196.93 | 177.60 | 3.12 | 44.92 | 110.14 | 83.18 | |
| UACC-257 | 37.58 | 47.42 | 172.55 | 190.50 | 128.61 | 0 | 5.58 | 51.31 | 33.52 | |
| UACC-62 | 29.18 | 33.59 | 136.03 | 154.04 | 112.39 | 5.12 | 34.09 | 47.31 | 46.24 | |
| Ovarian Cancer | IGROV1 | 51.23 | 46.96 | 120.49 | 139.81 | 65.42 | 0 | 12.89 | 18.70 | 19.70 |
| OVCAR-3 | 35.24 | 48.92 | 126.39 | 118.83 | 98.89 | 3.35 | 30.37 | 46.79 | 39.78 | |
| OVCAR-4 | 30.63 | 52.34 | 196.31 | 198.41 | 59.51 | 10.84 | 29.10 | 66.51 | 59.08 | |
| OVCAR-5 | 0 | 0 | 165.70 | 177.37 | 0 | 0 | 0 | 0 | 0 | |
| OVCAR-8 | 37.77 | 53.68 | 94.40 | 182.60 | 96.21 | 2.18 | 11.63 | 34.51 | 22.47 | |
| NCI/ADR-RES | 29.79 | 35.70 | 112.75 | 122.75 | 82.21 | 0.82 | 8.21 | 32.84 | 28.33 | |
| SK-OV-3 | 23.68 | 27.13 | 44.32 | 38.03 | 40.91 | 4.34 | 19.80 | 17.91 | 21.63 | |
| Renal Cancer | 786-0 | 19.04 | 17.33 | 121.83 | 190.37 | 31.40 | 0.60 | 12.69 | 29.94 | 22.26 |
| A498 | 6.32 | 13.32 | 44.03 | 48.42 | 10.55 | 26.71 | 13.08 | 37.26 | 27.25 | |
| ACHN | 57.35 | 57.98 | 195.25 | 190.19 | 74.36 | 0 | 11.65 | 27.66 | 15.91 | |
| CAKI-1 | 67.87 | 56.64 | 79.67 | 91.15 | 69.16 | 9.75 | 42.98 | 39.11 | 32.21 | |
| RXF 393 | 29.32 | 39.72 | 126.32 | 187.24 | 46.32 | 0 | 13.75 | 25.92 | 26.73 | |
| SN12C | 29.81 | 38.18 | 196.64 | 191.17 | 69.12 | 7.24 | 19.77 | 25.74 | 18.58 | |
| TK-10 | −8.43 | 0 | 41.72 | 11.39 | 0 | 0 | 8.81 | 20.32 | 20.52 | |
| UO-31 | 66.37 | 61.82 | 135.24 | 173.08 | 78.47 | 32.71 | 42.83 | 57.68 | 49.11 | |
| Prostate Cancer | PC-3 | 33.67 | 42.21 | 99.63 | 102.79 | 69.33 | 14.03 | 54.49 | 62.23 | 51.20 |
| DU-145 | 28.33 | 11.20 | 190.66 | 191.42 | 66.79 | 0 | 7.12 | 25.02 | 12.24 | |
| Breast Cancer | MCF7 | 54.39 | 59.97 | 162.80 | 163.07 | 88.50 | 16.30 | 35.02 | 64.10 | 47.06 |
| MDA-MB-231/ATCC | 60.00 | 59.61 | 106.76 | 107.49 | 84.61 | 0 | 10.63 | 7.06 | 4.91 | |
| HS 578T | 21.13 | 27.78 | 109.43 | 108.98 | 53.83 | 15.85 | 21.79 | 41.31 | 38.71 | |
| BT-549 | 32.10 | 22.75 | 199.09 | 197.40 | 62.82 | 7.56 | 17.13 | 46.06 | 39.07 | |
| T-47D | 28.91 | 64.88 | 103.21 | 119.84 | 79.61 | 11.05 | 46.29 | 79.21 | 75.60 | |
| MDA-MB-468 | 0 | 20.05 | 130.08 | 133.70 | 67.59 | 0 | 32.25 | 60.86 | 56.41 | |
| Panel | Cell Line | 3c | 3d | 3e | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GI50 | TGI | LC50 | GI50 | TGI | LC50 | GI50 | TGI | LC50 | ||||||||
| Conc. Per Cell Line | Subpanel MID b | Selectivity Ratio (MID a: MID b) | Conc. Per Cell Line | Subpanel MID b | Selectivity Ratio (MID a: MID b) | Conc. Per Cell Line | Subpanel MID b | Selectivity Ratio (MID a: MID b) | ||||||||
| leukemia | CCRF-CEM | 2.39 | 2.21 | 0.99 | >100 | >100 | 9.98 | 7.87 | 0.73 | 3.33 | >3.33 | 2.53 | 2.14 | 1.16 | >100 | >100 |
| HL-60(TB) | 3.95 | 3.26 | >100 | 7.70 | 2.05 | >3.33 | 2.18 | 6.26 | >100 | |||||||
| K-562 | 1.69 | -- | >100 | 8.79 | 3.33 | >3.33 | 2.43 | >100 | >100 | |||||||
| MOLT-4 | 1.22 | 5.76 | >100 | 5.99 | 1.72 | >3.33 | 2.22 | 8.89 | >100 | |||||||
| RPMI-8226 | 2.21 | -- | >100 | 7.17 | 2.01 | >3.33 | 1.86 | 6.47 | >100 | |||||||
| SR | 1.78 | >100 | >100 | 7.57 | 2.95 | >3.33 | 1.64 | -- | >100 | |||||||
| Non-Small Cell Lung Cancer | A549/ATCC | 2.26 | 1.69 | 1.03 | 5.61 | >100 | 8.35 | 5.07 | 1.14 | 2.07 | >3.33 | 2.42 | 2.31 | 1.07 | 6.07 | >100 |
| EKVX | 1.39 | 3.46 | 8.60 | 5.01 | 1.15 | 2.64 | 1.87 | 1.25 | ||||||||
| HOP-62 | 1.96 | 4.85 | 1.65 | 1.85 | 6.48 | 1.53 | 4.28 | 1.72 | ||||||||
| HOP-92 | 1.56 | 4.19 | >100 | 7.41 | 2.60 | 1.56 | 2.26 | 1.44 | ||||||||
| NCI-H226 | 1.61 | 3.35 | 6.99 | 9.36 | 4.09 | 1.37 | 1.81 | 5.83 | ||||||||
| NCI-H23 | 1.46 | 3.76 | 9.72 | 5.50 | 1.50 | >3.33 | 1.31 | 4.32 | >100 | |||||||
| NCI-H322M | 1.55 | 2.92 | 5.50 | 1.61 | 5.90 | 1.43 | 1.93 | 5.13 | ||||||||
| NCI-H460 | 1.94 | 3.97 | 8.11 | 1.06 | 3.60 | 1.50 | 3.35 | 1.22 | ||||||||
| NCI-H522 | 1.49 | 4.19 | >100 | 5.51 | 1.37 | 4.64 | 1.60 | 4.69 | >100 | |||||||
| Colon Cancer | COLO 205 | 1.94 | 2.61 | 0.84 | 4.24 | 9.28 | 3.63 | 5.99 | 0.96 | 9.06 | 2.26 | 3.36 | 2.50 | 0.99 | 1.27 | 6.62 |
| HCC-2998 | 1.62 | 3.14 | 6.06 | 9.48 | 2.91 | 1.06 | 2.34 | 6.67 | 2.40 | |||||||
| HCT-116 | 5.77 | 2.58 | 9.31 | 6.67 | 1.70 | 6.89 | 1.94 | 6.45 | 2.73 | |||||||
| HCT-15 | 1.52 | 3.50 | 8.07 | 5.49 | 1.18 | 2.54 | 1.51 | 3.24 | 6.96 | |||||||
| HT29 | 3.09 | >100 | >100 | 1.15 | 3.18 | >3.33 | 3.32 | 2.87 | >100 | |||||||
| KM12 | 2.68 | 8.80 | 9.05 | 9.99 | 3.56 | >3.33 | 3.12 | 1.37 | 8.18 | |||||||
| SW-620 | 1.63 | 3.45 | 7.30 | 5.51 | 1.20 | 2.62 | 1.92 | 4.32 | 9.71 | |||||||
| CNS Cancer | SF-268 | 1.68 | 2.79 | 0.78 | 4.48 | >100 | 6.15 | 5.12 | 1.13 | 1.65 | >3.33 | 2.54 | 2.43 | 1.02 | 1.10 | 5.91 |
| SF-295 | 1.68 | 3.08 | 5.66 | 6.15 | 1.27 | 2.60 | 3.55 | 1.27 | 3.57 | |||||||
| SF-539 | 1.69 | 3.12 | 5.79 | 5.54 | 1.02 | 1.89 | 1.67 | 3.06 | 5.58 | |||||||
| SNB-19 | 1.67 | 3.44 | 7.05 | 6.45 | 1.64 | 5.31 | 3.09 | 1.17 | 3.42 | |||||||
| SNB-75 | 8.43 | 2.76 | 7.79 | 1.32 | 7.46 | 1.74 | 1.32 | 6.04 | 2.51 | |||||||
| U251 | 1.60 | 4.28 | >100 | 5.12 | 1.12 | 2.43 | 2.41 | 1.11 | >100 | |||||||
| Melanoma | LOX IMVI | 5.32 | 2.32 | 0.95 | 2.10 | 5.86 | 4.06 | 5.77 | 1.00 | 9.04 | 2.01 | 9.66 | 3.12 | 0.79 | 2.62 | 6.98 |
| MALME-3M | 3.83 | 1.58 | 4.00 | 4.42 | 8.70 | 1.71 | 5.70 | 1.75 | 4.19 | |||||||
| M14 | 1.63 | 3.30 | 6.65 | 5.54 | 1.06 | 2.01 | 1.33 | 2.83 | 6.03 | |||||||
| MDA-MB-435 | 1.94 | 3.63 | 6.82 | 5.98 | 1.09 | 1.97 | 1.76 | 3.23 | 5.93 | |||||||
| SK-MEL-2 | 1.74 | 4.32 | 7.27 | 7.39 | 1.58 | 9.60 | 2.47 | 7.25 | 9.29 | |||||||
| SK-MEL-28 | 1.87 | 3.35 | 6.00 | 6.87 | 1.21 | 2.15 | 2.40 | 5.22 | 1.39 | |||||||
| SK-MEL-5 | 1.41 | 2.75 | 5.37 | 5.51 | 1.02 | 1.89 | 1.43 | 2.76 | 5.32 | |||||||
| UACC-257 | 1.55 | 3.15 | 6.39 | 5.93 | 1.14 | 2.20 | 1.69 | 3.41 | 6.88 | |||||||
| UACC-62 | 1.63 | 3.91 | 9.38 | 6.23 | 1.33 | 2.84 | 1.62 | 3.52 | 7.66 | |||||||
| Ovarian Cancer | IGROV1 | 1.13 | 2.25 | 0.98 | 3.21 | 9.08 | 4.46 | 5.38 | 1.07 | 1.31 | 5.78 | 1.16 | 2.53 | 0.98 | 4.74 | 2.58 |
| OVCAR-3 | 3.28 | 1.43 | 5.03 | 5.22 | 1.28 | 3.16 | 1.53 | 4.42 | 1.59 | |||||||
| OVCAR-4 | 1.26 | 2.56 | 5.18 | 4.54 | 8.87 | 1.73 | 1.61 | 4.58 | 1.59 | |||||||
| OVCAR-5 | 2.03 | 4.37 | 9.42 | 5.86 | 1.15 | 2.26 | 1.83 | 3.53 | 6.79 | |||||||
| OVCAR-8 | 2.13 | 1.12 | >100 | 5.81 | 1.24 | 2.63 | 2.62 | 1.16 | 4.10 | |||||||
| NCI/ADR-RES | 2.24 | -- | >100 | 7.43 | 2.17 | >3.33 | 1.93 | 6.77 | >100 | |||||||
| SK-OV-3 | 3.70 | 2.34 | >100 | 4.35 | 8.75 | 1.76 | 7.03 | 2.14 | 5.23 | |||||||
| Renal Cancer | 786-0 | 2.30 | 2.25 | 0.98 | 5.70 | >100 | 7.48 | 4.82 | 1.20 | 1.70 | >3.33 | 2.75 | 2.53 | 0.98 | 7.93 | 9.61 |
| A498 | 1.05 | 4.70 | >100 | 6.93 | 2.90 | >3.33 | 2.07 | 6.34 | >100 | |||||||
| ACHN | 1.42 | 2.91 | 5.96 | 4.92 | 9.65 | 1.89 | 1.39 | 3.03 | 6.60 | |||||||
| CAKI-1 | 1.18 | 2.86 | 6.92 | 4.80 | 2.14 | 8.98 | 2.48 | 1.49 | 3.87 | |||||||
| RXF393 | 1.35 | 2.71 | 5.43 | 5.02 | 9.86 | 1.94 | 1.85 | 4.75 | 1.50 | |||||||
| SN12C | 1.22 | 2.79 | 6.42 | 5.06 | 1.07 | 2.24 | 1.33 | 2.61 | 5.11 | |||||||
| TK-10 | 4.48 | 8.14 | 4.13 | 1.03 | 1.71 | 2.85 | 1.84 | 3.47 | 6.54 | |||||||
| UO-31 | 4.96 | 2.09 | 4.60 | 3.34 | 8.60 | 2.21 | 8.50 | 2.77 | 7.85 | |||||||
| Prostate Cancer | PC-3 | 1.49 | 1.67 | 1.32 | 8.81 | >100 | 6.20 | 6.29 | 0.92 | 2.69 | >3.33 | 1.62 | 1.74 | 1.43 | 1.07 | >100 |
| DU-145 | 1.85 | 3.45 | 6.46 | 6.37 | 1.19 | 2.22 | 1.85 | 3.43 | 6.36 | |||||||
| Breast Cancer | MCF7 | 1.23 | 1.73 | 1.27 | 3.36 | 9.17 | 5.83 | 6.67 | 0.87 | 1.28 | 2.82 | 1.42 | 1.90 | 1.31 | 3.29 | 7.62 |
| MDA-MB-231/ATCC | 1.27 | 6.69 | >100 | 6.52 | 2.69 | >3.33 | 1.37 | 9.82 | >100 | |||||||
| HS 578T | 2.04 | 6.93 | >100 | 7.82 | 2.05 | >3.33 | 2.09 | 6.03 | >100 | |||||||
| BT-549 | 1.53 | 2.91 | 5.54 | 5.22 | 9.83 | 1.85 | 1.50 | 2.91 | 5.64 | |||||||
| T-47D | 2.03 | 7.29 | >100 | 6.98 | 2.30 | >3.33 | 2.49 | 1.44 | >100 | |||||||
| MDA-MB-468 | 2.28 | 5.72 | >100 | 7.66 | 2.04 | >3.33 | 2.55 | 7.32 | >100 | |||||||
| MID a | 2.20 | 5.77 | 2.48 | |||||||||||||
| Compound | Cytotoxicity IC50 (µM) a ± SEM |
|---|---|
| 3b | 9.11 ± 0.39 *** |
| 3c | 0.81 ± 0.03 *** |
| 3e | 4.18 ± 0.18 *** |
| 8a | 26.5 ± 1.13 ** |
| 8b | 21.5 ± 0.92 ** |
| 8c | 30.8 ± 1.32 ** |
| 8d | 87.3 ± 3.73 * |
| 9 | 12.1 ± 0.52 *** |
| Dinaciclib | 5.97 ± 0.25 *** |
| Control | 0 |
| Compound | Cytotoxicity IC50 ± SEM (µM) WI38 |
|---|---|
| 32.59 ± 1.44 | |
| 3e | 39.86 ± 1.76 |
| Dinaciclib | 22.01 ± 0.97 |
| Compound | Caspase 3 | |
|---|---|---|
| Conc. pg/mL | Fold Change | |
| 3c | 519.4 ± 5.8 *** | 8.66 |
| Dinaciclib | 476.7 ± 8.4 *** | 7.95 |
| Control | 59.95 ± 2.1 | 1 |
Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aly, A.A.; Bräse, S.; Hassan, A.A.; Mohamed, N.K.; El-Haleem, L.E.A.; Nieger, M.; Morsy, N.M.; Alshammari, M.B.; Ibrahim, M.A.A.; Abdelhafez, E.M.N. Design, Synthesis, and Molecular Docking of Paracyclophanyl-Thiazole Hybrids as Novel CDK1 Inhibitors and Apoptosis Inducing Anti-Melanoma Agents. Molecules 2020, 25, 5569. https://doi.org/10.3390/molecules25235569
Aly AA, Bräse S, Hassan AA, Mohamed NK, El-Haleem LEA, Nieger M, Morsy NM, Alshammari MB, Ibrahim MAA, Abdelhafez EMN. Design, Synthesis, and Molecular Docking of Paracyclophanyl-Thiazole Hybrids as Novel CDK1 Inhibitors and Apoptosis Inducing Anti-Melanoma Agents. Molecules. 2020; 25(23):5569. https://doi.org/10.3390/molecules25235569
Chicago/Turabian StyleAly, Ashraf A., Stefan Bräse, Alaa A. Hassan, Nasr K. Mohamed, Lamiaa E. Abd El-Haleem, Martin Nieger, Nesrin M. Morsy, Mohammed B. Alshammari, Mahmoud A. A. Ibrahim, and Elshimaa M. N. Abdelhafez. 2020. "Design, Synthesis, and Molecular Docking of Paracyclophanyl-Thiazole Hybrids as Novel CDK1 Inhibitors and Apoptosis Inducing Anti-Melanoma Agents" Molecules 25, no. 23: 5569. https://doi.org/10.3390/molecules25235569
APA StyleAly, A. A., Bräse, S., Hassan, A. A., Mohamed, N. K., El-Haleem, L. E. A., Nieger, M., Morsy, N. M., Alshammari, M. B., Ibrahim, M. A. A., & Abdelhafez, E. M. N. (2020). Design, Synthesis, and Molecular Docking of Paracyclophanyl-Thiazole Hybrids as Novel CDK1 Inhibitors and Apoptosis Inducing Anti-Melanoma Agents. Molecules, 25(23), 5569. https://doi.org/10.3390/molecules25235569









