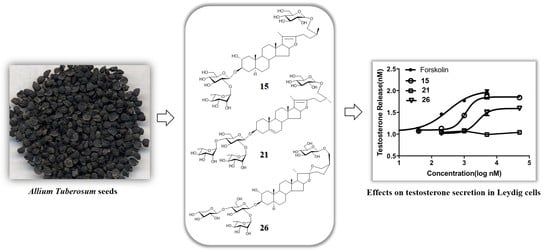
Steroidal Glycosides from Allium tuberosum Seeds and Their Roles in Promoting Testosterone Production of Rat Leydig Cells
Abstract
1. Introduction
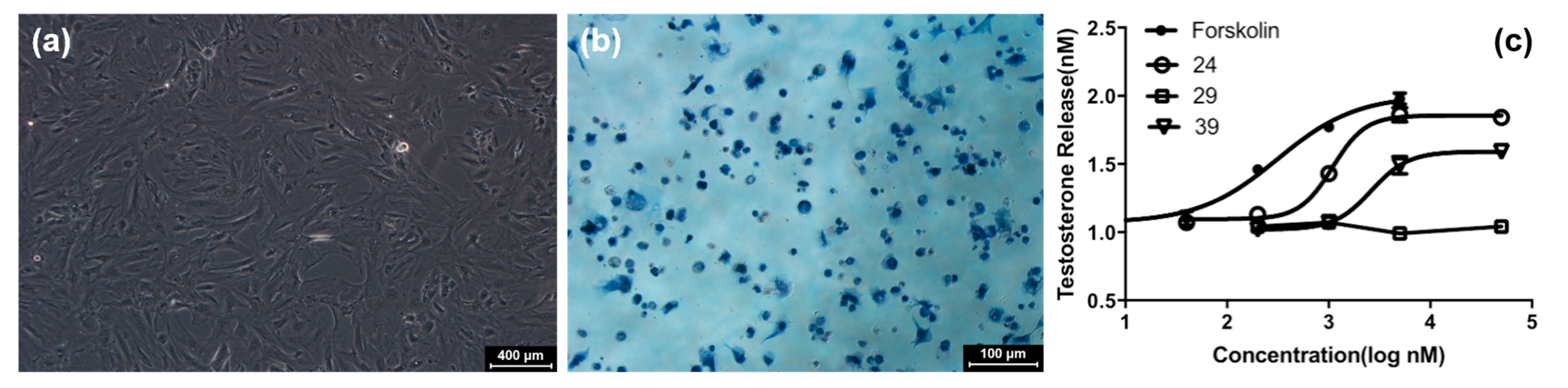
2. Results and Discussion
3. Materials and Methods
3.1. Experimental Procedures for Phytochmistry Study
3.1.1. General Experimental Procedures
3.1.2. Plant Material
3.1.3. Extraction and Isolation
3.1.4. Acid Hydrolysis and Absolute Configuration Determination
3.2. Bioactivity Assay
3.2.1. Preparation of Rat Leydig Cells and Primary Culture
3.2.2. Cellular Viability and Testosterone Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Williams, J.R.; Gong, H. Biological activities and syntheses of steroidal saponins: the shark-repelling pavoninins. Lipids 2007, 42, 77–86. [Google Scholar] [CrossRef]
- Qin, Y.; Wu, X.; Huang, W.; Gong, G.; Li, D.; He, Y.; Zhao, Y. Acute toxicity and sub-chronic toxicity of steroidal saponins from Dioscorea zingiberensis C.H. Wright in rodents. J. Ethnopharmacol. 2009, 126, 543–550. [Google Scholar] [CrossRef]
- Chen, M.H.; Chen, X.J.; Wang, M.; Lin, L.G.; Wang, Y.T. Ophiopogon japonicus—A phytochemical, ethnomedicinal and pharmacological review. J. Ethnopharmacol. 2016, 181, 193–213. [Google Scholar] [CrossRef]
- Wang, Y.H.; Niu, H.M.; Zhang, Z.Y.; Hu, X.Y.; Li, H. Medicinal values and their chemical bases of Paris. Zhongguo Zhongyao Zazhi 2015, 40, 833–839. (In Chinese) [Google Scholar] [PubMed]
- Wang, Y.; Xu, L.; Lou, L.L.; Song, S.J.; Yao, G.D.; Ge, M.Y.; Hayashi, T.; Tashiro, S.I.; Onodera, S.; Ikejima, T. Timosaponin AIII induces apoptosis and autophagy in human melanoma A375-S2 cells. Arch. Pharm. Res. 2016, 40, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Sobolewska, D.; Michalska, K.; Podolak, I.; Grabowska, K. Steroidal saponins from the genus Allium. Phytochem. Rev. 2016, 15, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.H.; Lu, Y.H.; Mao, R.G.; Wei, D.Z.; Ma, Z.Z.; Zhang, H. Aphrodisiac properties of Allium tuberosum seeds extract. J. Ethnopharmacol. 2009, 122, 579–582. [Google Scholar]
- Kim, S.Y.; Park, K.W.; Kim, J.Y.; Jeong, I.Y.; Byun, M.W.; Park, J.E.; Yee, S.T.; Kim, K.H.; Rhim, J.S.; Yamada, K.; et al. Thiosulfinates from Allium tuberosum L. induce apoptosis via caspase-dependent and -independent pathways in PC-3 human prostate cancer cells. Bioorg. Med. Chem. Lett. 2008, 18, 199–204. [Google Scholar] [CrossRef]
- Park, K.W.; Kim, S.Y.; Jeong, I.Y.; Byun, M.W.; Park, K.H.; Yamada, K.; Seo, K.I. Cytotoxic and antitumor activities of thiosulfinates from Allium tuberosum L. J. Agric. Food Chem. 2007, 55, 7957–7961. [Google Scholar] [CrossRef]
- Lee, J.H.; Yang, H.S.; Park, K.W.; Kim, J.Y.; Lee, M.K.; Jeong, I.Y.; Shim, K.H.; Kim, Y.S.; Yamada, K.; Seo, K.I. Mechanisms of thiosulfinates from Allium tuberosum L.-induced apoptosis in HT-29 human colon cancer cells. Toxicol. Lett. 2009, 188, 142–147. [Google Scholar] [CrossRef]
- Sang, S.M.; Lao, A.N.; Wang, H.C.; Chen, Z.L. Furostanol saponins from Allium tuberosum. Phytochemistry 1999, 52, 1611–1615. [Google Scholar] [CrossRef]
- Sang, S.M.; Lao, A.N.; Wang, H.C.; Chen, Z.L. Two new spirostanol saponins from Allium tuberosum. J. Nat. Prod. 1999, 62, 1028–1029. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.M.; Zou, M.L.; Xia, Z.H.; Lao, A.N.; Chen, Z.L.; Ho, C.T. New spirostanol saponins from Chinese chives (Allium tuberosum). J. Agric. Food Chem. 2001, 49, 4780–4783. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.M.; Mao, S.L.; Lao, A.N.; Chen, Z.L.; Ho, C.T. Four new steroidal saponins from the seeds of Allium tuberosum. J. Agric. Food Chem. 2001, 49, 1475–1478. [Google Scholar] [CrossRef]
- Zou, Z.M.; Yu, D.Q.; Cong, P.Z. A steroidal saponin from the seeds of Allium tuberosum. Phytochemistry 2001, 57, 1219–1222. [Google Scholar] [CrossRef]
- Fang, Y.S.; Cai, L.; Li, Y.; Wang, J.P.; Xiao, H.; Ding, Z.T. Spirostanol steroids from the roots of Allium tuberosum. Steroids 2015, 100, 1–4. [Google Scholar] [CrossRef]
- Ikeda, T.; Tsumagari, H.; Nohara, T. Steroidal oligoglycosides from the seeds of Allium tuberosum. Chem. Pharm. Bull. 2000, 48, 362–365. [Google Scholar] [CrossRef]
- Gupta, R.K.; Jain, D.C.; Thakur, R.S. Two furostanol saponins from Trigonella foenum-graecum. Phytochemistry 1986, 25, 2205–2207. [Google Scholar] [CrossRef]
- Murakami, T.; Kishi, A.; Matsuda, H.; Yoshikawa, M. Medicinal foodstuffs. XVII. Fenugreek seed. (3): Structures of new furostanol-type steroid saponins, trigoneosides Xa, Xb, XIb, XIIa, XIIb, and XIIIa, from the seeds of Egyptian trigonella foenum-graecum L. Chem. Pharm. Bull. 2000, 48, 994–1000. [Google Scholar] [CrossRef]
- Shvets, S.A.; Gutsu, O.N.; Kintia, P.K. Steroidal glycosides from Nicotiana tabacum L. seeds and their biological activity. Adv. Exp. Med. Biol. 1996, 405, 247–257. [Google Scholar]
- Zuo, Y.M.; Xu, Y.L.; Zhang, Z.L.; Liu, D.H.; Cai, M.T. Chemical components in roots and rhizome of Trillium tschonoskii (V). Shizhen Guoyi Guoyao 2016, 27, 260–263. (In Chinese) [Google Scholar]
- Hayes, P.Y.; Jahidin, A.H.; Lehmann, R.; Penman, K.; Kitching, W.; De Voss, J.J. Structural revision of shatavarins I and IV, the major components from the roots of Asparagus racemosus. Tetrahedron Lett. 2006, 47, 6965–6969. [Google Scholar] [CrossRef]
- Hu, K.; Dong, A.J.; Yao, X.S.; Kobayashi, H.; Iwasaki, S. Antineoplastic agents. Part 2. Four furostanol glycosides from rhizomes of Dioscorea collettii var. hypoglauca. Planta Med. 1997, 63, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.L.; Liu, R.H.; Shao, F. Structural determination of two new steroidal saponins from Smilax china. Magn. Reson. Chem. 2009, 47, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Jian, R.; Zeng, K.W.; Li, J.; Li, N.; Jiang, Y.; Tu, P.F. Anti-neuroinflammatory constituents from Asparagus cochinchinensis. Fitoterapia 2013, 84, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.K. Dependence of 1H-NMR chemical shifts of geminal protons of glycosyloxy methylene (H2-26) on the orientation of the 27-methyl group of furostane-type steroidal saponins. Magn. Reson. Chem. 2004, 42, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Takamura, C.; Sugita, F.; Masuoka, C.; Yoshimitsu, H.; Ikeda, T.; Nohara, T. Two new steroid glycosides and a new sesquiterpenoid glycoside from the underground parts of Trillium kamtschaticum. Chem. Pharm. Bull. 2007, 55, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.S.; Pal, P.C.; Rajalakshmi, M. Isolation and culture of Leydig cells from adult rats. Indian J. Clin. Biochem. 2006, 21, 27–33. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
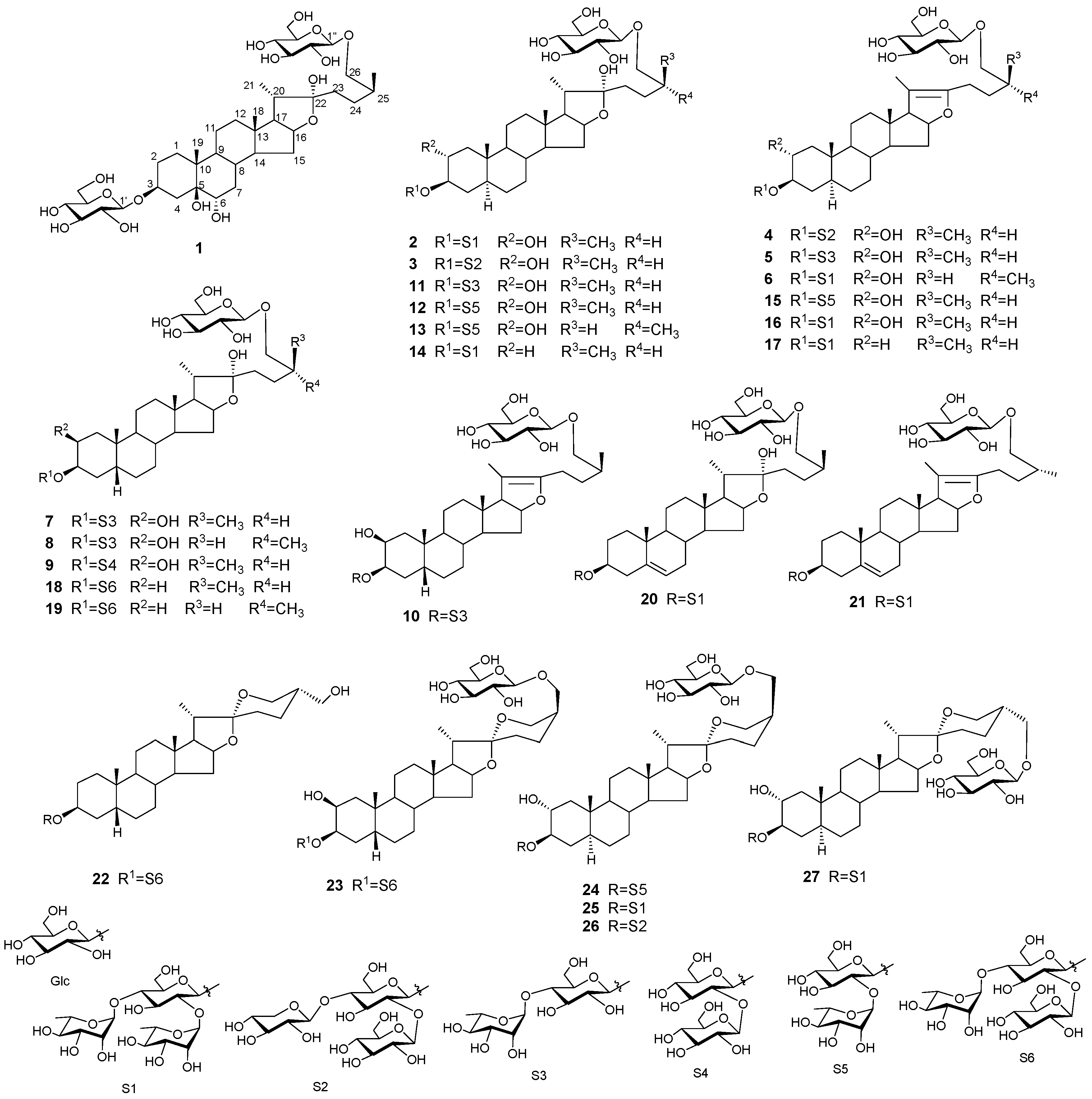
| POS. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| 1 | 36.0 | 45.9 | 45.9 | 46.0 | 45.8 | 45.9 | 40.3 | 40.3 |
| 2 | 29.2 | 70.6 | 70.6 | 70.5 | 70.4 | 70.6 | 66.9 | 66.9 |
| 3 | 79.2 | 85.1 | 85.1 | 85.1 | 84.9 | 85.1 | 80.2 | 80.2 |
| 4 | 35.2 | 33.5 | 33.5 | 33.5 | 33.9 | 33.5 | 31.7 | 31.7 |
| 5 | 73.1 | 44.6 | 44.7 | 44.7 | 44.6 | 44.7 | 36.4 | 36.4 |
| 6 | 66.2 | 28.2 | 28.2 | 28.2 | 28.1 | 28.2 | 26.2 | 26.2 |
| 7 | 35.6 | 32.3 | 32.3 | 32.5 | 32.4 | 32.4 | 26.7 | 26.7 |
| 8 | 34.6 | 34.6 | 34.6 | 34.4 | 34.4 | 34.4 | 35.5 | 35.5 |
| 9 | 44.7 | 54.5 | 54.5 | 54.4 | 54.4 | 54.4 | 41.4 | 41.4 |
| 10 | 43.1 | 36.9 | 36.9 | 36.9 | 36.9 | 36.9 | 36.9 | 36.9 |
| 11 | 21.9 | 21.5 | 21.5 | 21.6 | 21.6 | 21.6 | 21.3 | 21.4 |
| 12 | 40.1 | 40.1 | 40.2 | 39.8 | 39.8 | 39.8 | 40.5 | 40.5 |
| 13 | 41.0 | 41.1 | 41.1 | 43.7 | 43.7 | 43.8 | 41.2 | 41.2 |
| 14 | 56.3 | 56.3 | 56.3 | 54.7 | 54.6 | 54.6 | 56.3 | 56.3 |
| 15 | 32.4 | 32.4 | 32.4 | 34.4 | 34.3 | 34.4 | 32.4 | 32.4 |
| 16 | 81.1 | 81.1 | 81.2 | 84.5 | 84.5 | 84.5 | 81.2 | 81.2 |
| 17 | 63.8 | 63.9 | 63.9 | 64.6 | 64.6 | 64.6 | 64.0 | 64.0 |
| 18 | 16.7 | 16.7 | 16.7 | 14.4 | 14.4 | 14.4 | 16.7 | 16.7 |
| 19 | 17.6 | 13.6 | 13.6 | 13.6 | 13.4 | 13.6 | 23.8 | 23.8 |
| 20 | 40.7 | 40.7 | 40.7 | 103.6 | 103.6 | 103.6 | 40.7 | 40.7 |
| 21 | 16.5 | 16.5 | 16.5 | 11.8 | 11.8 | 11.8 | 16.5 | 16.5 |
| 22 | 110.7 | 110.6 | 110.6 | 152.4 | 152.4 | 152.4 | 110.6 | 110.6 |
| 23 | 37.2 | 37.2 | 37.2 | 31.4 | 31.4 | 31.5 | 37.2 | 37.3 |
| 24 | 28.4 | 28.3 | 28.4 | 23.7 | 23.6 | 23.7 | 28.4 | 28.4 |
| 25 | 34.5 | 34.5 | 34.5 | 33.8 | 33.7 | 33.4 | 34.5 | 34.3 |
| 26 | 75.4 | 75.4 | 75.4 | 75.3 | 75.2 | 75.0 | 75.4 | 75.3 |
| 27 | 17.5 | 17.5 | 17.5 | 17.2 | 17.2 | 17.4 | 17.5 | 17.5 |
| 3-O-Glc | 3-O-Glc | 3-O-Glc | 3-O-Glc | 3-O-Glc | 3-O-Glc | 3-O-Glc | 3-O-Glc | |
| 1′ | 102.2 | 100.9 | 100.6 | 100.6 | 103.0 | 100.9 | 104.0 | 104.0 |
| 2′ | 74.8 | 77.9 | 77.3 | 77.3 | 75.3 | 77.9 | 75.0 | 75.0 |
| 3′ | 79.0 | 77.9 | 76.5 | 76.5 | 76.6 | 77.9 | 76.6 | 76.6 |
| 4′ | 71.6 | 78.5 | 81.4 | 81.4 | 78.3 | 78.7 | 78.1 | 78.1 |
| 5′ | 78.7 | 77.2 | 77.8 | 77.8 | 77.4 | 77.2 | 77.4 | 77.5 |
| 6′ | 62.4 | 61.1 | 61.4 | 61.4 | 61.3 | 61.1 | 61.3 | 61.3 |
| 26-O-Glc | 2′-O-Rha | 2′-O-Rha | 2′-O-Rha | 4′-O-Rha | 2′-O-Rha | 4′-O-Rha | 4′-O-Rha | |
| 1″ | 105.2 | 102.1 | 102.2 | 102.1 | 102.7 | 102.1 | 102.7 | 102.7 |
| 2″ | 75.3 | 72.5 | 72.4 | 72.4 | 72.6 | 72.5 | 72.6 | 72.7 |
| 3″ | 78.6 | 72.8 | 72.8 | 72.8 | 72.8 | 72.8 | 72.8 | 72.8 |
| 4″ | 71.7 | 74.1 | 74.1 | 74.1 | 74.0 | 74.1 | 74.0 | 74.0 |
| 5″ | 78.5 | 69.5 | 69.6 | 69.6 | 70.5 | 69.5 | 70.4 | 70.4 |
| 6″ | 62.8 | 18.6 | 18.6 | 18.6 | 18.6 | 18.6 | 18.6 | 18.6 |
| 4′-O-Rha | 4′-O-Xyl | 4′-O-Xyl | 26-O-Glc | 4′-O-Rha | 26-O-Glc | 26-O-Glc | ||
| 1‴ | 102.9 | 105.8 | 105.8 | 105.2 | 103.0 | 105.2 | 105.0 | |
| 2‴ | 72.6 | 75.0 | 75.0 | 75.2 | 72.6 | 75.2 | 75.2 | |
| 3‴ | 72.8 | 78.4 | 78.4 | 78.6 | 72.8 | 78.6 | 78.6 | |
| 4‴ | 73.9 | 70.8 | 70.8 | 71.7 | 73.9 | 71.7 | 71.7 | |
| 5‴ | 70.5 | 67.4 | 67.4 | 78.6 | 70.5 | 78.5 | 78.5 | |
| 6‴ | 18.5 | 62.8 | 18.6 | 62.8 | 62.8 | |||
| 26-O-Glc | 26-O-Glc | 26-O-Glc | 26-O-Glc | |||||
| 1‴′ | 105.2 | 105.2 | 105.2 | 104.9 | ||||
| 2‴′ | 75.3 | 75.3 | 75.3 | 75.2 | ||||
| 3‴′ | 78.6 | 78.7 | 78.7 | 78.7 | ||||
| 4‴′ | 71.7 | 71.7 | 71.7 | 71.7 | ||||
| 5‴′ | 78.6 | 78.5 | 78.6 | 78.6 | ||||
| 6‴′ | 62.8 | 62.8 | 62.9 | 62.9 |
| No. | 9 | 10 | 22 | 23 | 24 | 25 | 26 |
|---|---|---|---|---|---|---|---|
| 1 | 40.3 | 40.0 | 30.9 | 40.4 | 45.9 | 45.9 | 45.9 |
| 2 | 67.2 | 67.0 | 27.0 | 67.1 | 70.7 | 70.6 | 70.6 |
| 3 | 81.7 | 80.1 | 75.3 | 81.1 | 85.4 | 85.0 | 85.1 |
| 4 | 31.6 | 31.7 | 30.7 | 31.2 | 33.6 | 33.5 | 33.5 |
| 5 | 36.5 | 36.4 | 36.7 | 32.1 | 44.6 | 44.6 | 44.6 |
| 6 | 26.3 | 26.2 | 26.8 | 26.3 | 28.2 | 28.2 | 28.2 |
| 7 | 26.8 | 26.8 | 26.8 | 26.8 | 32.1 | 32.1 | 32.1 |
| 8 | 35.6 | 35.2 | 35.5 | 35.5 | 34.6 | 34.6 | 34.6 |
| 9 | 41.5 | 36.9 | 40.3 | 41.4 | 54.4 | 54.4 | 54.4 |
| 10 | 37.1 | 36.9 | 35.3 | 37.0 | 36.9 | 36.9 | 36.9 |
| 11 | 21.4 | 40.5 | 21.2 | 21.3 | 21.4 | 21.5 | 21.5 |
| 12 | 40.6 | 40.5 | 40.2 | 40.2 | 40.0 | 40.0 | 40.0 |
| 13 | 41.2 | 43.8 | 40.9 | 40.8 | 40.7 | 40.8 | 40.8 |
| 14 | 56.3 | 54.6 | 56.5 | 56.3 | 56.3 | 56.3 | 56.3 |
| 15 | 32.4 | 34.4 | 32.2 | 32.1 | 32.3 | 32.3 | 32.3 |
| 16 | 81.2 | 84.5 | 81.3 | 81.4 | 81.2 | 81.3 | 81.3 |
| 17 | 64.0 | 64.6 | 62.9 | 63.0 | 62.9 | 62.9 | 62.9 |
| 18 | 16.7 | 14.4 | 16.6 | 16.5 | 16.5 | 16.6 | 16.6 |
| 19 | 23.8 | 23.8 | 24.0 | 23.9 | 13.5 | 13.6 | 13.6 |
| 20 | 40.7 | 103.6 | 42.1 | 42.4 | 42.4 | 42.4 | 42.4 |
| 21 | 16.5 | 11.8 | 15.1 | 14.9 | 14.8 | 14.8 | 14.8 |
| 22 | 110.6 | 152.4 | 109.7 | 109.7 | 109.7 | 109.7 | 109.7 |
| 23 | 37.3 | 31.4 | 31.6 | 27.1 | 27.1 | 27.1 | 27.1 |
| 24 | 28.4 | 23.6 | 24.1 | 21.4 | 21.4 | 21.5 | 21.5 |
| 25 | 34.5 | 33.7 | 39.2 | 33.5 | 33.5 | 33.5 | 33.5 |
| 26 | 75.4 | 75.2 | 64.4 | 61.0 | 60.9 | 60.9 | 60.9 |
| 27 | 17.5 | 17.2 | 64.1 | 69.5 | 69.5 | 69.5 | 69.5 |
| 3-O-Glc | 3-O-Glc | 3-O-Glc | 3-O-Glc | 3-O-Glc | 3-O-Glc | 3-O-Glc | |
| 1′ | 102.7 | 104.0 | 101.9 | 102.3 | 101.2 | 100.8 | 100.5 |
| 2′ | 83.1 | 75.4 | 82.8 | 82.3 | 78.1 | 77.9 | 77.8 |
| 3′ | 78.1 | 76.6 | 77.1 | 77.1 | 79.6 | 78.0 | 76.5 |
| 4′ | 71.4 | 78.1 | 77.3 | 77.3 | 71.9 | 78.5 | 81.4 |
| 5′ | 78.4 | 77.5 | 76.4 | 76.3 | 78.4 | 77.2 | 77.3 |
| 6′ | 62.4 | 61.3 | 61.3 | 61.0 | 62.5 | 61.1 | 61.5 |
| 2′-O-Glc | 4′-O-Rha | 2′-O-Glc | 2′-O-Glc | 2′-O-Rha | 2′-O-Rha | 2′-O-Rha | |
| 1″ | 106.2 | 102.7 | 105.7 | 105.6 | 102.2 | 102.2 | 102.2 |
| 2″ | 77.1 | 72.7 | 77.1 | 77.0 | 72.5 | 72.5 | 72.4 |
| 3″ | 77.9 | 72.8 | 77.9 | 78.0 | 72.8 | 72.8 | 72.8 |
| 4″ | 71.8 | 74.0 | 71.8 | 71.9 | 74.2 | 74.1 | 74.1 |
| 5″ | 78.5 | 70.4 | 78.6 | 78.5 | 69.5 | 69.6 | 69.6 |
| 6″ | 62.8 | 18.6 | 63.2 | 62.9 | 18.6 | 18.6 | 18.6 |
| 26-O-Glc | 26-O-Glc | 4′-O-Rha | 4′-O-Rha | 27-O-Glc | 4′-O-Rha | 4′-O-Xyl | |
| 1‴ | 105.2 | 105.2 | 102.4 | 102.4 | 105.0 | 103.0 | 105.8 |
| 2‴ | 75.3 | 75.0 | 72.6 | 72.5 | 75.3 | 72.6 | 75.4 |
| 3‴ | 78.7 | 78.6 | 72.8 | 72.8 | 78.6 | 72.8 | 78.4 |
| 4‴ | 71.7 | 71.7 | 74.0 | 74.0 | 71.8 | 74.0 | 70.8 |
| 5‴ | 78.6 | 78.6 | 70.3 | 70.3 | 78.6 | 70.5 | 67.4 |
| 6‴ | 62.8 | 62.8 | 18.5 | 18.6 | 62.8 | 18.5 | |
| 27-O-Glc | 27-O-Glc | 27-O-Glc | |||||
| 1‴′ | 105.0 | 105.1 | 105.1 | ||||
| 2‴′ | 75.3 | 75.4 | 75.0 | ||||
| 3‴′ | 78.6 | 78.7 | 78.6 | ||||
| 4‴′ | 71.7 | 71.8 | 71.8 | ||||
| 5‴′ | 78.6 | 78.7 | 78.5 | ||||
| 6‴′ | 62.9 | 62.8 | 62.8 |
| Compound | EC50 (μM) | Compounds | EC50 (μM) | Compound | EC50 (μM) | Compounds | EC50 (μM) |
|---|---|---|---|---|---|---|---|
| 1 | 1.0 | 8 | 1.6 | 15 | 1.1 | 22 | >50 |
| 2 | 2.6 | 9 | >50 | 16 | >50 | 23 | 2.1 |
| 3 | >50 | 10 | 1.4 | 17 | >50 | 24 | >50 |
| 4 | >50 | 11 | >50 | 18 | >50 | 25 | >50 |
| 5 | >50 | 12 | 1.8 | 19 | >50 | 26 | 4.5 |
| 6 | >50 | 13 | 1.0 | 20 | 2.0 | 27 | >50 |
| 7 | 4.4 | 14 | 1.6 | 21 | >50 | Forskolin | 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.-B.; Wei, X.-Y. Steroidal Glycosides from Allium tuberosum Seeds and Their Roles in Promoting Testosterone Production of Rat Leydig Cells. Molecules 2020, 25, 5464. https://doi.org/10.3390/molecules25225464
Zhang D-B, Wei X-Y. Steroidal Glycosides from Allium tuberosum Seeds and Their Roles in Promoting Testosterone Production of Rat Leydig Cells. Molecules. 2020; 25(22):5464. https://doi.org/10.3390/molecules25225464
Chicago/Turabian StyleZhang, Da-Bing, and Xian-Yong Wei. 2020. "Steroidal Glycosides from Allium tuberosum Seeds and Their Roles in Promoting Testosterone Production of Rat Leydig Cells" Molecules 25, no. 22: 5464. https://doi.org/10.3390/molecules25225464
APA StyleZhang, D.-B., & Wei, X.-Y. (2020). Steroidal Glycosides from Allium tuberosum Seeds and Their Roles in Promoting Testosterone Production of Rat Leydig Cells. Molecules, 25(22), 5464. https://doi.org/10.3390/molecules25225464