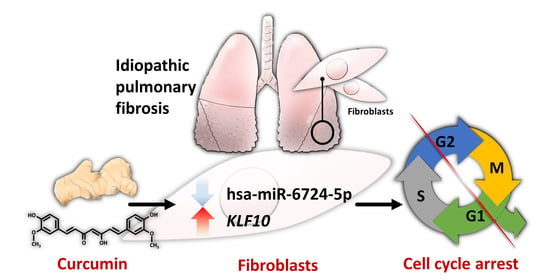
The Potential Effects of Curcumin on Pulmonary Fibroblasts of Idiopathic Pulmonary Fibrosis (IPF)—Approaching with Next-Generation Sequencing and Bioinformatics
Abstract
1. Introduction
2. Results
2.1. Curcumin Inhibited the Proliferation/Viability of IPF Fibroblasts
2.2. Gene Expression Changes in Curcumin-Treated IPF Fibroblasts
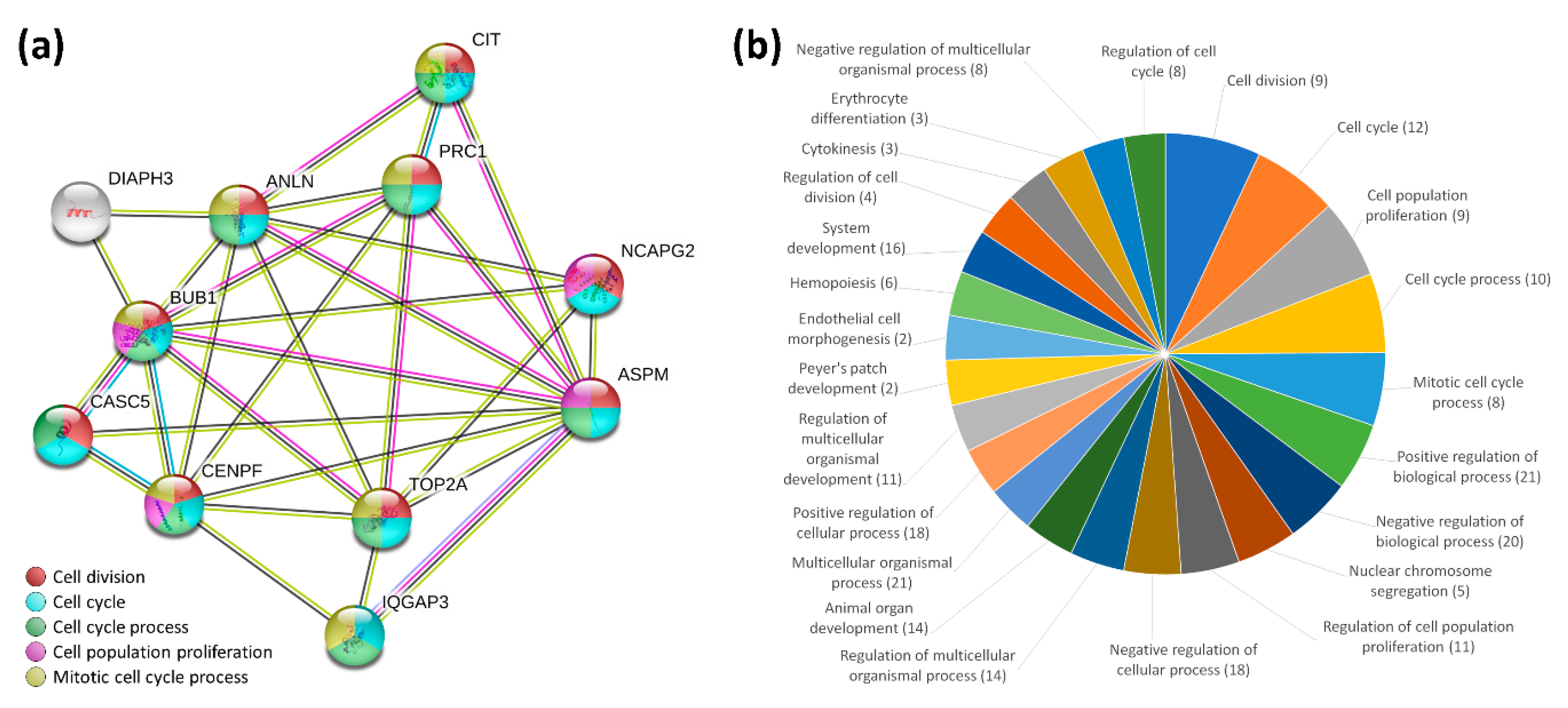
2.3. STRING Analysis for Protein-Protein Interaction and Biological Process of the Differentially Expressed Genes in Curcumin-Treated IPF Fibroblasts
2.4. IPA Analysis for Cellular Functions of the Differentially Expressed Genes in Curcumin-Treated IPF Fibroblasts
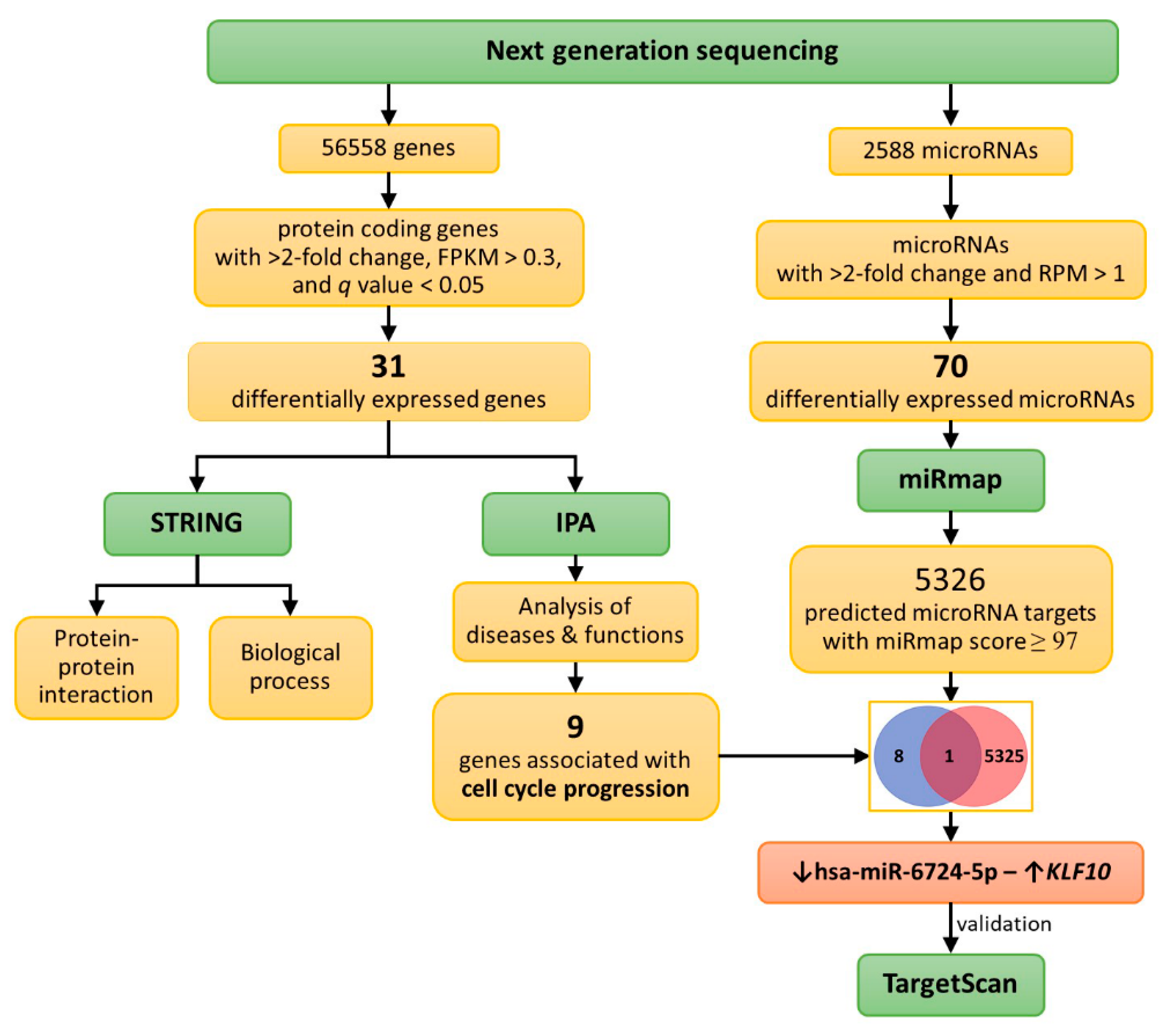
2.5. Differentially Expressed Genes with Potential microRNA-mRNA Interactions in Curcumin-Treated IPF Fibroblasts
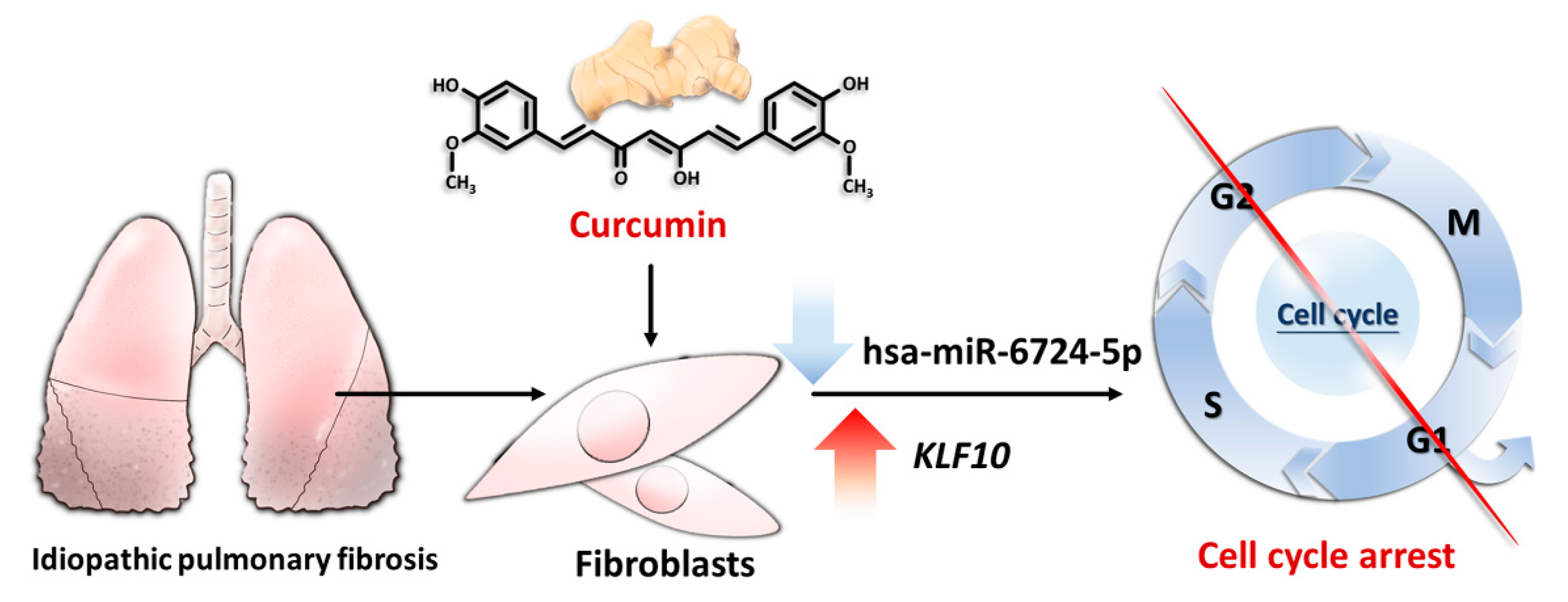
2.6. Potential Effects of Curcumin on Dysregulated Genes and microRNAs in IPF Fibroblasts
3. Discussion
4. Materials and Methods
4.1. Study Design and Analysis Strategy
4.2. Cultures of IPF Lung Fibroblasts and Curcumin Treatment
4.3. Cell Proliferation Assay
4.4. Next-Generation Sequencing (NGS)
4.5. STRING Database Analysis
4.6. The Ingenuity® Pathway Analysis (IPA)
4.7. miRmap Database Analysis
4.8. TargetScan Database Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barratt, S.L.; Creamer, A.; Hayton, C.; Chaudhuri, N. Idiopathic Pulmonary Fibrosis (IPF): An Overview. J. Clin. Med. 2018, 7, 201. [Google Scholar] [CrossRef] [PubMed]
- Lynch, D.A.; Sverzellati, N.; Travis, W.D.; Brown, K.K.; Colby, T.V.; Galvin, J.R.; Goldin, J.G.; Hansell, D.M.; Inoue, Y.; Johkoh, T.; et al. Diagnostic criteria for idiopathic pulmonary fibrosis: A Fleischner Society White Paper. Lancet Respir. Med. 2018, 6, 138–153. [Google Scholar] [CrossRef]
- Martinez, F.J.; Collard, H.R.; Pardo, A.; Raghu, G.; Richeldi, L.; Selman, M.; Swigris, J.J.; Taniguchi, H.; Wells, A.U. Idiopathic pulmonary fibrosis. Nat. Rev. Dis Primers 2017, 3, 17074. [Google Scholar] [CrossRef]
- Raghu, G.; Remy-Jardin, M.; Myers, J.L.; Richeldi, L.; Ryerson, C.J.; Lederer, D.J.; Behr, J.; Cottin, V.; Danoff, S.K.; Morell, F.; et al. Diagnosis of Idiopathic Pulmonary Fibrosis. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2018, 198, e44–e68. [Google Scholar] [CrossRef]
- Richeldi, L.; Collard, H.R.; Jones, M.G. Idiopathic pulmonary fibrosis. Lancet 2017, 389, 1941–1952. [Google Scholar] [CrossRef]
- Sheu, C.C.; Chang, W.A.; Tsai, M.J.; Liao, S.H.; Chong, I.W.; Kuo, P.L. Bioinformatic analysis of nextgeneration sequencing data to identify dysregulated genes in fibroblasts of idiopathic pulmonary fibrosis. Int. J. Mol. Med. 2019, 43, 1643–1656. [Google Scholar] [CrossRef]
- Sheu, C.C.; Chang, W.A.; Tsai, M.J.; Liao, S.H.; Chong, I.W.; Kuo, P.L. Gene Expression Changes Associated with Nintedanib Treatment in Idiopathic Pulmonary Fibrosis Fibroblasts: A Next-Generation Sequencing and Bioinformatics Study. J. Clin. Med. 2019, 8, 308. [Google Scholar] [CrossRef]
- Tsai, M.J.; Chang, W.A.; Liao, S.H.; Chang, K.F.; Sheu, C.C.; Kuo, P.L. The Effects of Epigallocatechin Gallate (EGCG) on Pulmonary Fibroblasts of Idiopathic Pulmonary Fibrosis (IPF)-A Next-Generation Sequencing and Bioinformatic Approach. Int. J. Mol. Sci. 2019, 20, 1958. [Google Scholar] [CrossRef]
- Hutchinson, J.; Fogarty, A.; Hubbard, R.; McKeever, T. Global incidence and mortality of idiopathic pulmonary fibrosis: A systematic review. Eur. Respir. J. 2015, 46, 795–806. [Google Scholar] [CrossRef]
- Vancheri, C.; Failla, M.; Crimi, N.; Raghu, G. Idiopathic pulmonary fibrosis: A disease with similarities and links to cancer biology. Eur. Respir. J. 2010, 35, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Pardo, A.; Selman, M. Lung Fibroblasts, Aging, and Idiopathic Pulmonary Fibrosis. Ann. Am. Thorac. Soc. 2016, 13, S417–S421. [Google Scholar] [CrossRef] [PubMed]
- Pierce, E.M.; Carpenter, K.; Jakubzick, C.; Kunkel, S.L.; Evanoff, H.; Flaherty, K.R.; Martinez, F.J.; Toews, G.B.; Hogaboam, C.M. Idiopathic pulmonary fibrosis fibroblasts migrate and proliferate to CC chemokine ligand 21. Eur. Respir. J. 2007, 29, 1082–1093. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.; Montano, M.; Garcia-Alvarez, J.; Ruiz, V.; Uhal, B.D.; Selman, M.; Pardo, A. Fibroblasts from idiopathic pulmonary fibrosis and normal lungs differ in growth rate, apoptosis, and tissue inhibitor of metalloproteinases expression. Am. J. Respir. Cell Mol. Biol. 2001, 24, 591–598. [Google Scholar] [CrossRef]
- Vuga, L.J.; Ben-Yehudah, A.; Kovkarova-Naumovski, E.; Oriss, T.; Gibson, K.F.; Feghali-Bostwick, C.; Kaminski, N. WNT5A is a regulator of fibroblast proliferation and resistance to apoptosis. Am. J. Respir. Cell Mol. Biol. 2009, 41, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Oldham, J.M.; Ma, S.F.; Martinez, F.J.; Anstrom, K.J.; Raghu, G.; Schwartz, D.A.; Valenzi, E.; Witt, L.; Lee, C.; Vij, R.; et al. TOLLIP, MUC5B, and the Response to N-Acetylcysteine among Individuals with Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2015, 192, 1475–1482. [Google Scholar] [CrossRef]
- Shimizu, Y.; Dobashi, K.; Sano, T.; Yamada, M. ROCK activation in lung of idiopathic pulmonary fibrosis with oxidative stress. Int. J. Immunopathol. Pharmacol. 2014, 27, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Veith, C.; Drent, M.; Bast, A.; van Schooten, F.J.; Boots, A.W. The disturbed redox-balance in pulmonary fibrosis is modulated by the plant flavonoid quercetin. Toxicol. Appl. Pharmacol. 2017, 336, 40–48. [Google Scholar] [CrossRef]
- Azuma, A.; Nukiwa, T.; Tsuboi, E.; Suga, M.; Abe, S.; Nakata, K.; Taguchi, Y.; Nagai, S.; Itoh, H.; Ohi, M.; et al. Double-blind, placebo-controlled trial of pirfenidone in patients with idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2005, 171, 1040–1047. [Google Scholar] [CrossRef]
- King, T.E., Jr.; Bradford, W.Z.; Castro-Bernardini, S.; Fagan, E.A.; Glaspole, I.; Glassberg, M.K.; Gorina, E.; Hopkins, P.M.; Kardatzke, D.; Lancaster, L.; et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2014, 370, 2083–2092. [Google Scholar] [CrossRef]
- Kolb, M.; Raghu, G.; Wells, A.U.; Behr, J.; Richeldi, L.; Schinzel, B.; Quaresma, M.; Stowasser, S.; Martinez, F.J.; Investigators, I. Nintedanib plus Sildenafil in Patients with Idiopathic Pulmonary Fibrosis. N. Engl. J. Med. 2018, 379, 1722–1731. [Google Scholar] [CrossRef]
- Kropski, J.A.; Blackwell, T.S. Progress in Understanding and Treating Idiopathic Pulmonary Fibrosis. Annu. Rev. Med. 2019, 70, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Nathan, S.D.; Costabel, U.; Glaspole, I.; Glassberg, M.K.; Lancaster, L.H.; Lederer, D.J.; Pereira, C.A.; Trzaskoma, B.; Morgenthien, E.A.; Limb, S.L.; et al. Efficacy of Pirfenidone in the Context of Multiple Disease Progression Events in Patients With Idiopathic Pulmonary Fibrosis. Chest 2018. [Google Scholar] [CrossRef] [PubMed]
- Richeldi, L.; Costabel, U.; Selman, M.; Kim, D.S.; Hansell, D.M.; Nicholson, A.G.; Brown, K.K.; Flaherty, K.R.; Noble, P.W.; Raghu, G.; et al. Efficacy of a tyrosine kinase inhibitor in idiopathic pulmonary fibrosis. N. Engl. J. Med. 2011, 365, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Richeldi, L.; du Bois, R.M.; Raghu, G.; Azuma, A.; Brown, K.K.; Costabel, U.; Cottin, V.; Flaherty, K.R.; Hansell, D.M.; Inoue, Y.; et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N. Engl. J. Med. 2014, 370, 2071–2082. [Google Scholar] [CrossRef]
- Saidi, A.; Kasabova, M.; Vanderlynden, L.; Wartenberg, M.; Kara-Ali, G.H.; Marc, D.; Lecaille, F.; Lalmanach, G. Curcumin inhibits the TGF-beta1-dependent differentiation of lung fibroblasts via PPARgamma-driven upregulation of cathepsins B and L. Sci. Rep. 2019, 9, 491. [Google Scholar] [CrossRef]
- Tsai, C.F.; Hsieh, T.H.; Lee, J.N.; Hsu, C.Y.; Wang, Y.C.; Kuo, K.K.; Wu, H.L.; Chiu, C.C.; Tsai, E.M.; Kuo, P.L. Curcumin Suppresses Phthalate-Induced Metastasis and the Proportion of Cancer Stem Cell (CSC)-like Cells via the Inhibition of AhR/ERK/SK1 Signaling in Hepatocellular Carcinoma. J. Agric. Food Chem. 2015, 63, 10388–10398. [Google Scholar] [CrossRef]
- Lelli, D.; Sahebkar, A.; Johnston, T.P.; Pedone, C. Curcumin use in pulmonary diseases: State of the art and future perspectives. Pharmacol. Res. 2017, 115, 133–148. [Google Scholar] [CrossRef]
- Abidi, A.; Gupta, S.; Agarwal, M.; Bhalla, H.L.; Saluja, M. Evaluation of Efficacy of Curcumin as an Add-on therapy in Patients of Bronchial Asthma. J. Clin. Diagn. Res. 2014, 8, HC19-24. [Google Scholar] [CrossRef]
- Hu, Y.; Li, M.; Zhang, M.; Jin, Y. Inhalation treatment of idiopathic pulmonary fibrosis with curcumin large porous microparticles. Int. J. Pharm. 2018, 551, 212–222. [Google Scholar] [CrossRef]
- Jiang, A.; Wang, X.; Shan, X.; Li, Y.; Wang, P.; Jiang, P.; Feng, Q. Curcumin Reactivates Silenced Tumor Suppressor Gene RARbeta by Reducing DNA Methylation. Phytother. Res. 2015, 29, 1237–1245. [Google Scholar] [CrossRef]
- Kim, D.H.; Phillips, J.F.; Lockey, R.F. Oral curcumin supplementation in patients with atopic asthma. Allergy Rhinol (Providence) 2011, 2, e51–e53. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.P.; Niti, M.; Yap, K.B.; Tan, W.C. Curcumins-rich curry diet and pulmonary function in Asian older adults. PLoS ONE 2012, 7, e51753. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R.; Gangireddy, S.R.; Narala, V.R.; Hogaboam, C.M.; Standiford, T.J.; Christensen, P.J.; Kondapi, A.K.; Reddy, R.C. Curcumin inhibits fibrosis-related effects in IPF fibroblasts and in mice following bleomycin-induced lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2010, 298, L616–L625. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Yang, M.; Sun, D.; Sun, S. Curcumin protects against sepsis-induced acute lung injury in rats. J. Surg. Res. 2012, 176, e31–e39. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.S.; Dash, D.; Singh, R. Intranasal Curcumin Inhibits Pulmonary Fibrosis by Modulating Matrix Metalloproteinase-9 (MMP-9) in Ovalbumin-Induced Chronic Asthma. Inflammation 2017, 40, 248–258. [Google Scholar] [CrossRef]
- Lee, J.C.; Kinniry, P.A.; Arguiri, E.; Serota, M.; Kanterakis, S.; Chatterjee, S.; Solomides, C.C.; Javvadi, P.; Koumenis, C.; Cengel, K.A.; et al. Dietary curcumin increases antioxidant defenses in lung, ameliorates radiation-induced pulmonary fibrosis, and improves survival in mice. Radiat. Res. 2010, 173, 590–601. [Google Scholar] [CrossRef]
- Liu, D.; Gong, L.; Zhu, H.; Pu, S.; Wu, Y.; Zhang, W.; Huang, G. Curcumin Inhibits Transforming Growth Factor beta Induced Differentiation of Mouse Lung Fibroblasts to Myofibroblasts. Front. Pharmacol. 2016, 7, 419. [Google Scholar] [CrossRef]
- Tourkina, E.; Gooz, P.; Oates, J.C.; Ludwicka-Bradley, A.; Silver, R.M.; Hoffman, S. Curcumin-induced apoptosis in scleroderma lung fibroblasts: Role of protein kinase cepsilon. Am. J. Respir. Cell Mol. Biol. 2004, 31, 28–35. [Google Scholar] [CrossRef]
- Tyagi, N.; Dash, D.; Singh, R. Curcumin inhibits paraquat induced lung inflammation and fibrosis by extracellular matrix modifications in mouse model. Inflammopharmacology 2016, 24, 335–345. [Google Scholar] [CrossRef]
- Ho, C.; Hsu, Y.C.; Lei, C.C.; Mau, S.C.; Shih, Y.H.; Lin, C.L. Curcumin Rescues Diabetic Renal Fibrosis by Targeting Superoxide-Mediated Wnt Signaling Pathways. Am. J. Med. Sci. 2016, 351, 286–295. [Google Scholar] [CrossRef]
- Pivari, F.; Mingione, A.; Brasacchio, C.; Soldati, L. Curcumin and Type 2 Diabetes Mellitus: Prevention and Treatment. Nutrients 2019, 11, 1837. [Google Scholar] [CrossRef] [PubMed]
- Bachmeier, B.E.; Melchart, D. Therapeutic Effects of Curcumin-From Traditional Past to Present and Future Clinical Applications. Int. J. Mol. Sci. 2019, 20, 3757. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Patchva, S.; Aggarwal, B.B. Therapeutic roles of curcumin: Lessons learned from clinical trials. AAPS J. 2013, 15, 195–218. [Google Scholar] [CrossRef] [PubMed]
- Burge, K.; Gunasekaran, A.; Eckert, J.; Chaaban, H. Curcumin and Intestinal Inflammatory Diseases: Molecular Mechanisms of Protection. Int. J. Mol. Sci. 2019, 20, 1912. [Google Scholar] [CrossRef] [PubMed]
- Bielak-Zmijewska, A.; Grabowska, W.; Ciolko, A.; Bojko, A.; Mosieniak, G.; Bijoch, L.; Sikora, E. The Role of Curcumin in the Modulation of Ageing. Int. J. Mol. Sci. 2019, 20, 1239. [Google Scholar] [CrossRef]
- Tomeh, M.A.; Hadianamrei, R.; Zhao, X. A Review of Curcumin and Its Derivatives as Anticancer Agents. Int. J. Mol. Sci. 2019, 20, 1033. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Bueso-Ramos, C.; Chatterjee, D.; Pantazis, P.; Aggarwal, B.B. Curcumin downregulates cell survival mechanisms in human prostate cancer cell lines. Oncogene 2001, 20, 7597–7609. [Google Scholar] [CrossRef]
- Su, C.C.; Lin, J.G.; Chen, G.W.; Lin, W.C.; Chung, J.G. Down-regulation of Cdc25c, CDK1 and Cyclin B1 and Up-regulation of Wee1 by Curcumin Promotes Human Colon Cancer Colo 205 Cell Entry into G2/M-phase of Cell Cycle. Cancer Genom. Proteom. 2006, 3, 55–61. [Google Scholar]
- Maiti, P.; Scott, J.; Sengupta, D.; Al-Gharaibeh, A.; Dunbar, G.L. Curcumin and Solid Lipid Curcumin Particles Induce Autophagy, but Inhibit Mitophagy and the PI3K-Akt/mTOR Pathway in Cultured Glioblastoma Cells. Int. J. Mol. Sci. 2019, 20, 399. [Google Scholar] [CrossRef]
- Rodriguez, L.R.; Bui, S.N.; Beuschel, R.T.; Ellis, E.; Liberti, E.M.; Chhina, M.K.; Cannon, B.; Lemma, M.; Nathan, S.D.; Grant, G.M. Curcumin induced oxidative stress attenuation by N-acetylcysteine co-treatment: A fibroblast and epithelial cell in-vitro study in idiopathic pulmonary fibrosis. Mol. Med. 2019, 25, 27. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, C.; Yang, C.; Liu, R.J.; Wang, J.; Niu, J.; Bromme, D. Antifibrotic effects of curcumin are associated with overexpression of cathepsins K and L in bleomycin treated mice and human fibroblasts. Respir. Res. 2011, 12, 154. [Google Scholar] [CrossRef] [PubMed]
- Punithavathi, D.; Venkatesan, N.; Babu, M. Protective effects of curcumin against amiodarone-induced pulmonary fibrosis in rats. Br. J. Pharmacol. 2003, 139, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, R.; Tang, Y.; Fu, X. Effects of curcumin on artery blood gas index of rats with pulmonary fibrosis caused by paraquat poisoning and the expression of Smad 4, Smurf 2, interleukin-4 and interferon-gamma. Exp. Ther. Med. 2019, 17, 3664–3670. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.J.; Yi, C.O.; Jeon, B.T.; Jeong, Y.Y.; Kang, G.M.; Lee, J.E.; Roh, G.S.; Lee, J.D. Curcumin attenuates radiation-induced inflammation and fibrosis in rat lungs. Korean J. Physiol. Pharmacol. 2013, 17, 267–274. [Google Scholar] [CrossRef]
- Epstein Shochet, G.; Wollin, L.; Shitrit, D. Fibroblast-matrix interplay: Nintedanib and pirfenidone modulate the effect of IPF fibroblast-conditioned matrix on normal fibroblast phenotype. Respirology 2018, 23, 756–763. [Google Scholar] [CrossRef]
- Wollin, L.; Maillet, I.; Quesniaux, V.; Holweg, A.; Ryffel, B. Antifibrotic and anti-inflammatory activity of the tyrosine kinase inhibitor nintedanib in experimental models of lung fibrosis. J. Pharmacol. Exp. Ther. 2014, 349, 209–220. [Google Scholar] [CrossRef]
- Wollin, L.; Wex, E.; Pautsch, A.; Schnapp, G.; Hostettler, K.E.; Stowasser, S.; Kolb, M. Mode of action of nintedanib in the treatment of idiopathic pulmonary fibrosis. Eur. Respir. J. 2015, 45, 1434–1445. [Google Scholar] [CrossRef]
- Schaefer, C.J.; Ruhrmund, D.W.; Pan, L.; Seiwert, S.D.; Kossen, K. Antifibrotic activities of pirfenidone in animal models. Eur. Respir. Rev. 2011, 20, 85–97. [Google Scholar] [CrossRef]
- Azuma, A. Pirfenidone treatment of idiopathic pulmonary fibrosis. Ther. Adv. Respir. Dis. 2012, 6, 107–114. [Google Scholar] [CrossRef]
- Chiambaretta, F.; De Graeve, F.; Turet, G.; Marceau, G.; Gain, P.; Dastugue, B.; Rigal, D.; Sapin, V. Cell and tissue specific expression of human Kruppel-like transcription factors in human ocular surface. Mol. Vis. 2004, 10, 901–909. [Google Scholar]
- Leduc, C.; Sobilo, L.; Toumi, H.; Mondon, P.; Lespessailles, E.; Ossant, F.; Kurfurst, R.; Pichon, C. TGF-beta-induced early gene-1 overexpression promotes oxidative stress protection and actin cytoskeleton rearrangement in human skin fibroblasts. Biochim. Biophys. Acta 2016, 1860, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Song, K.D.; Kim, D.J.; Lee, J.E.; Yun, C.H.; Lee, W.K. KLF10, transforming growth factor-beta-inducible early gene 1, acts as a tumor suppressor. Biochem. Biophys. Res. Commun. 2012, 419, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Lin, S.Y.; Chang, H.W.; Ko, L.J.; Tseng, Y.S.; Chang, V.H.; Yu, W.C. CDK2 phosphorylation regulates the protein stability of KLF10 by interfering with binding of the E3 ligase SIAH1. Biochim. Biophys. Acta 2015, 1853, 1174–1181. [Google Scholar] [CrossRef] [PubMed]
- Ellenrieder, V. TGFbeta regulated gene expression by Smads and Sp1/KLF-like transcription factors in cancer. Anticancer Res. 2008, 28, 1531–1539. [Google Scholar] [PubMed]
- Spittau, G.; Happel, N.; Behrendt, M.; Chao, T.I.; Krieglstein, K.; Spittau, B. Tieg1/Klf10 is upregulated by NGF and attenuates cell cycle progression in the pheochromocytoma cell line PC12. J. Neurosci. Res. 2010, 88, 2017–2025. [Google Scholar] [CrossRef]
- Parakati, R.; DiMario, J.X. Repression of myoblast proliferation and fibroblast growth factor receptor 1 promoter activity by KLF10 protein. J. Biol. Chem. 2013, 288, 13876–13884. [Google Scholar] [CrossRef]
- Xu, Y.L.; Zhang, M.H.; Guo, W.; Xue, Y.; Du, X.; Zhang, T.; Wu, N.; Wu, Y. MicroRNA-19 restores vascular endothelial cell function in lower limb ischemia-reperfusion injury through the KLF10-dependent TGF-beta1/Smad signaling pathway in rats. J. Cell Biochem. 2018, 119, 9303–9315. [Google Scholar] [CrossRef]
- Huang, L.T.; Chang, H.W.; Wu, M.J.; Lai, Y.T.; Wu, W.C.; Yu, W.C.Y.; Chang, V.H.S. Klf10 deficiency in mice exacerbates pulmonary inflammation by increasing expression of the proinflammatory molecule NPRA. Int. J. Biochem. Cell Biol. 2016, 79, 231–238. [Google Scholar] [CrossRef]
- Iwata, Y.; Yoshizaki, A.; Ogawa, F.; Komura, K.; Hara, T.; Muroi, E.; Takenaka, M.; Shimizu, K.; Hasegawa, M.; Fujimoto, M.; et al. Increased serum pentraxin 3 in patients with systemic sclerosis. J. Rheumatol 2009, 36, 976–983. [Google Scholar] [CrossRef]
- Gao, P.; Tang, K.; Lu, Y.; Huang, Z.; Wang, S.; Wang, M.; Wang, J.; Zhao, J.; Xie, J. Pentraxin 3 promotes airway inflammation in experimental asthma. Respir. Res. 2020, 21, 237. [Google Scholar] [CrossRef]
- Djalali, M.; Djalali, M.; Abdolahi, M.; Mohammadi, H.; Heidari, H.; Hosseini, S.; Sadeghizadeh, M. The Effect of Nano-Curcumin Supplementation on Pentraxin 3 Gene Expression and Serum Level in Migraine Patients. Rep. Biochem. Mol. Biol. 2020, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Camelo, A.; Dunmore, R.; Sleeman, M.A.; Clarke, D.L. The epithelium in idiopathic pulmonary fibrosis: Breaking the barrier. Front. Pharmacol. 2014, 4, 173. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Bonchev, D. A survey of current software for network analysis in molecular biology. Hum. Genom. 2010, 4, 353–360. [Google Scholar] [CrossRef]
- Vejnar, C.E.; Zdobnov, E.M. MiRmap: Comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012, 40, 11673–11683. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.J.; Chang, W.A.; Tsai, P.H.; Wu, C.Y.; Ho, Y.W.; Yen, M.C.; Lin, Y.S.; Kuo, P.L.; Hsu, Y.L. Montelukast Induces Apoptosis-Inducing Factor-Mediated Cell Death of Lung Cancer Cells. Int. J. Mol. Sci. 2017, 18, 1353. [Google Scholar] [CrossRef]
- Chang, W.A.; Sheu, C.C.; Liu, K.T.; Shen, J.H.; Yen, M.C.; Kuo, P.L. Identification of mutations in SLC4A1, GP1BA and HFE in a family with venous thrombosis of unknown cause by next-generation sequencing. Exp. Ther. Med. 2018, 16, 4172–4180. [Google Scholar] [CrossRef]
- Chang, W.A.; Tsai, M.J.; Jian, S.F.; Sheu, C.C.; Kuo, P.L. Systematic analysis of transcriptomic profiles of COPD airway epithelium using next-generation sequencing and bioinformatics. Int. J. Chron. Obstruct Pulmon Dis. 2018, 13, 2387–2398. [Google Scholar] [CrossRef]
- Sheu, C.C.; Tsai, M.J.; Chen, F.W.; Chang, K.F.; Chang, W.A.; Chong, I.W.; Kuo, P.L.; Hsu, Y.L. Identification of novel genetic regulations associated with airway epithelial homeostasis using next-generation sequencing data and bioinformatics approaches. Oncotarget 2017, 8, 82674–82688. [Google Scholar] [CrossRef]
- Tsai, M.J.; Chang, W.A.; Jian, S.F.; Chang, K.F.; Sheu, C.C.; Kuo, P.L. Possible mechanisms mediating apoptosis of bronchial epithelial cells in chronic obstructive pulmonary disease - A next-generation sequencing approach. Pathol Res. Pract. 2018, 214, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.J.; Tsai, Y.C.; Chang, W.A.; Lin, Y.S.; Tsai, P.H.; Sheu, C.C.; Kuo, P.L.; Hsu, Y.L. Deducting MicroRNA-Mediated Changes Common in Bronchial Epithelial Cells of Asthma and Chronic Obstructive Pulmonary Disease-A Next-Generation Sequencing-Guided Bioinformatic Approach. Int. J. Mol. Sci. 2019, 20, 553. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. Royal. Stat. Soc. Ser. B (Method.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]

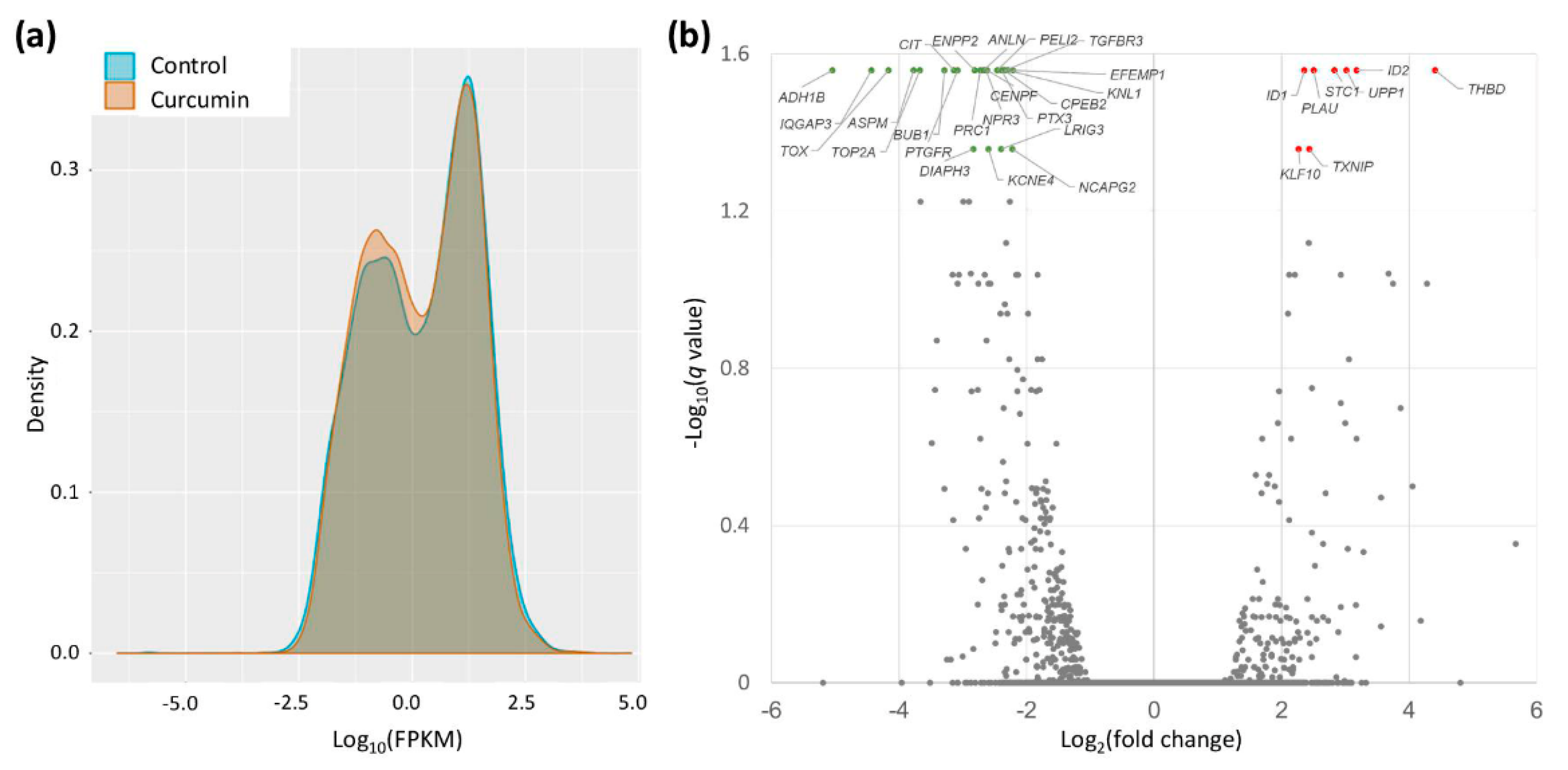

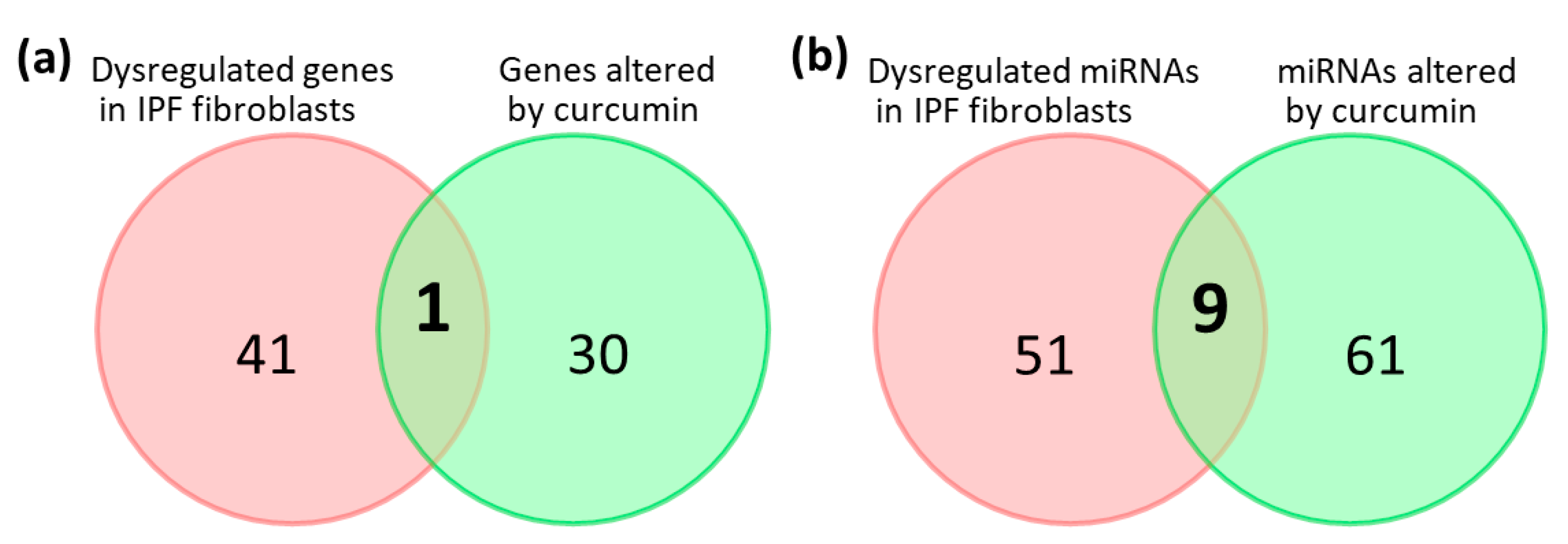
| Gene Symbol | Curcumin 10 μM (FPKM) | Control (FPKM) | Ratio (Curcumin/Control) | p-Value | q-Value * |
|---|---|---|---|---|---|
| ADH1B | 2.83 | 93.89 | 0.03 | 0.00005 | 0.0276 |
| IQGAP3 | 0.45 | 9.71 | 0.05 | 0.00005 | 0.0276 |
| TOX | 0.72 | 13.08 | 0.06 | 0.00005 | 0.0276 |
| ASPM | 1.70 | 23.23 | 0.07 | 0.00005 | 0.0276 |
| TOP2A | 5.57 | 71.17 | 0.08 | 0.00005 | 0.0276 |
| BUB1 | 2.59 | 25.37 | 0.10 | 0.00005 | 0.0276 |
| CIT | 2.23 | 19.76 | 0.11 | 0.00005 | 0.0276 |
| PTGFR | 7.94 | 67.35 | 0.12 | 0.00005 | 0.0276 |
| ENPP2 | 28.62 | 201.09 | 0.14 | 0.00005 | 0.0276 |
| DIAPH3 | 2.88 | 20.55 | 0.14 | 0.00010 | 0.0439 |
| PRC1 | 6.26 | 41.46 | 0.15 | 0.00005 | 0.0276 |
| CENPF | 3.41 | 20.98 | 0.16 | 0.00005 | 0.0276 |
| NPR3 | 8.38 | 52.27 | 0.16 | 0.00005 | 0.0276 |
| ANLN | 8.44 | 54.04 | 0.16 | 0.00005 | 0.0276 |
| KCNE4 | 3.31 | 19.98 | 0.17 | 0.00010 | 0.0439 |
| PELI2 | 4.86 | 26.82 | 0.18 | 0.00005 | 0.0276 |
| PTX3 | 78.97 | 415.71 | 0.19 | 0.00005 | 0.0276 |
| LRIG3 | 4.42 | 23.31 | 0.19 | 0.00010 | 0.0439 |
| KNL1 | 2.13 | 10.42 | 0.20 | 0.00005 | 0.0276 |
| TGFBR3 | 8.78 | 44.60 | 0.20 | 0.00005 | 0.0276 |
| CPEB2 | 29.38 | 149.72 | 0.20 | 0.00005 | 0.0276 |
| NCAPG2 | 5.19 | 24.29 | 0.21 | 0.00010 | 0.0439 |
| EFEMP1 | 42.08 | 195.48 | 0.22 | 0.00005 | 0.0276 |
| KLF10 | 77.02 | 16.02 | 4.81 | 0.00010 | 0.0439 |
| ID1 | 154.13 | 30.14 | 5.11 | 0.00005 | 0.0276 |
| TXNIP | 41.05 | 7.59 | 5.41 | 0.00010 | 0.0439 |
| PLAU | 84.67 | 14.96 | 5.66 | 0.00005 | 0.0276 |
| STC1 | 79.28 | 11.17 | 7.10 | 0.00005 | 0.0276 |
| UPP1 | 80.27 | 9.93 | 8.08 | 0.00005 | 0.0276 |
| ID2 | 68.30 | 7.57 | 9.02 | 0.00005 | 0.0276 |
| THBD | 25.91 | 1.22 | 21.30 | 0.00005 | 0.0276 |
| microRNA | IPF Fibroblasts vs. NHLF | Curcumin-Treated IPF Fibroblasts vs. Control IPF Fibroblasts | ||
|---|---|---|---|---|
| Fold Change | Up/Down | Fold Change | Up/Down | |
| hsa-miR-3613-5p | 2.76 | up | −5.15 | down |
| hsa-miR-182-5p | −2.01 | down | 3.24 | up |
| hsa-miR-664a-3p | −2.46 | down | 2.14 | up |
| hsa-miR-4461 | −3.05 | down | 2.13 | up |
| hsa-miR-9-5p | −3.05 | down | 5.23 | up |
| hsa-miR-204-5p | −3.77 | down | −2.96 | down |
| hsa-miR-4521 | 3.67 | up | 4.08 | up |
| hsa-miR-619-5p | 2.94 | up | 4.27 | up |
| hsa-miR-668-3p | 2.06 | up | 3.79 | up |
Sample Availability: The compound, curcumin, is available from Sigma-Aldrich, Co. (St. Louis, MO, USA). | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, W.-A.; Chen, C.-M.; Sheu, C.-C.; Liao, S.-H.; Hsu, Y.-L.; Tsai, M.-J.; Kuo, P.-L. The Potential Effects of Curcumin on Pulmonary Fibroblasts of Idiopathic Pulmonary Fibrosis (IPF)—Approaching with Next-Generation Sequencing and Bioinformatics. Molecules 2020, 25, 5458. https://doi.org/10.3390/molecules25225458
Chang W-A, Chen C-M, Sheu C-C, Liao S-H, Hsu Y-L, Tsai M-J, Kuo P-L. The Potential Effects of Curcumin on Pulmonary Fibroblasts of Idiopathic Pulmonary Fibrosis (IPF)—Approaching with Next-Generation Sequencing and Bioinformatics. Molecules. 2020; 25(22):5458. https://doi.org/10.3390/molecules25225458
Chicago/Turabian StyleChang, Wei-An, Chia-Min Chen, Chau-Chyun Sheu, Ssu-Hui Liao, Ya-Ling Hsu, Ming-Ju Tsai, and Po-Lin Kuo. 2020. "The Potential Effects of Curcumin on Pulmonary Fibroblasts of Idiopathic Pulmonary Fibrosis (IPF)—Approaching with Next-Generation Sequencing and Bioinformatics" Molecules 25, no. 22: 5458. https://doi.org/10.3390/molecules25225458
APA StyleChang, W.-A., Chen, C.-M., Sheu, C.-C., Liao, S.-H., Hsu, Y.-L., Tsai, M.-J., & Kuo, P.-L. (2020). The Potential Effects of Curcumin on Pulmonary Fibroblasts of Idiopathic Pulmonary Fibrosis (IPF)—Approaching with Next-Generation Sequencing and Bioinformatics. Molecules, 25(22), 5458. https://doi.org/10.3390/molecules25225458