Abstract
Crotoxin, from the venom of the South American rattlesnake Crotalus durissus terrificus, is a potent heterodimeric presynaptic β-neurotoxin that exists in individual snake venom as a mixture of isoforms of a basic phospholipase A2 (PLA2) subunit (CBa2, CBb, CBc, and CBd) and acidic subunit (CA1–4). Specific natural mutations in CB isoforms are implicated in functional differences between crotoxin isoforms. The three-dimensional structure of two individual CB isoforms (CBa2, CBc), and one isoform in a crotoxin (CA2CBb) complex, have been previously reported. This study concerns CBd, which by interaction with various protein targets exhibits many physiological or pharmacological functions. It binds with high affinity to presynaptic receptors showing neurotoxicity, but also interacts with human coagulation factor Xa (hFXa), exhibiting anticoagulant effect, and acts as a positive allosteric modulator and corrector of mutated chloride channel, cystic fibrosis transmembrane conductance regulator (CFTR), implicated in cystic fibrosis. Thus, CBd represents a novel family of agents that have potential in identifying new drug leads related to anticoagulant and anti-cystic fibrosis function. We determined here the X-ray structure of CBd and compare it with the three other natural isoforms of CB. The structural role of specific amino acid variations between CB isoforms are analyzed and the structural framework of CB for interaction with protein targets is described.
1. Introduction
Crotoxin from the venom of the South American rattlesnake Crotalus durissus terrificus, is a potent β-neurotoxin with phospholipase A2 (PLA2) activity. It is a heterodimeric protein complex formed by the non-covalent association of a basic PLA2 subunit of low toxicity (CB), and a nontoxic acidic subunit (CA) devoid of catalytic activity that potentiates the toxic effect of CB, by enabling the toxin to reach the specific crotoxin receptor at the neuromuscular junction [1]. The CA and CB subunits are present in individual snake venom as a mixture of isoforms (CA1–4 and CBa2, CBb, CBc, CBd) that can form up to 16 natural crotoxin complexes with different biochemical and pharmacological properties [2,3,4]. Based on these differences, two classes of crotoxin (class I and II) were determined [5]. Class I, composed of CBb, CBc, CBd with any of the CA isoforms, contains very stable complexes (half-life 10–20 min, Kd = 4.5 nM) of high toxicity (these isoforms block neuromuscular transmission of chick biventer cervicis preparations more efficiently than weakly toxic isoforms) and low PLA2 activity. Class II, composed of CBa2 with any of the CA isoforms, contains less stable complexes (half-life about 1 min, Kd = 25 nM) of low toxicity and high enzymatic activity [5].
Crotoxin acts primarily at the presynaptic level and induces complete failure of neuromuscular transmission by interacting with specific neuronal targets [6,7,8]. In addition to neurotoxicity, crotoxin exhibits other pharmacological and biological properties, including antiviral [9], cytotoxic (against tumor cells) [10,11,12], anti-inflammatory [13,14], immunomodulatory (via dendritic cells) [15], or analgesic [16] effects. It can also induce calcium-dependent glutamate release via N and P/Q calcium channels [17]. The CB PLA2 subunit purified from a crotoxin complex, may also act independently on different biological systems: (i) it blocks neuromuscular transmission by binding to presynaptic receptors (the neuronal CB target is different from that of the crotoxin receptor, and 7-fold higher doses of CB, with respect to class I crotoxin complexes, are needed to achieve neurotoxicity) [5,7,18]; (ii) it exhibits anticoagulant properties through direct interaction with human coagulation factor Xa (hFXa), leading to inhibition of the prothrombinase complex required for efficient production of thrombin [19]; (iii) it forms a complex with the ΔF508-NBD1 and WT-NBD1 (nucleotide-binding domain 1) of the cystic fibrosis transmembrane conductance regulator (CFTR), and acts as a positive allosteric modulator and corrector of the mutated chloride channel [20]; (iv) it interacts with Gloeobacter ligand-gated ion channel (GLIC), a member of the pentameric ligand-gated ion channel (pLGIC) family, as a negative allosteric modulator [21]; and (v) it forms a complex with the natural crotoxin inhibitor from Crotalus serum (CICS), which neutralizes its neurotoxicity and enzymatic activity [22,23,24]. Crotoxin and its CB PLA2 subunit are of interest because of potential therapeutic perspectives, leading to active investigation of their mechanism of interaction with various biological targets at the molecular and cellular level [25].
We previously solved the crystal structure of the class I crotoxin isoform CA2CBb and identified the binding interface between the two subunits, showing the crucial role of residue His1 in the formation of the more stable interface between Trp31 and Trp70 of CBb, with Asp99 and Asp89 of CA2 (amino acids are numbered according to Renetseder [26]) [27]. Moreover, the crystal structure of CBa2 (CB2, with Ser in position 1) and CBc (CB1, with His in position 1) in a tetrameric complex without the CA subunit, revealed conformational differences between these two isoforms [28]. A comparison of the individual CB isoform crystal structures and the correlation of their differences with the CA2CBb complex, shows how the structural changes within CB contribute to the functional differences between the two classes of crotoxin complexes [27].
In the present work, we report the crystal structure of the most abundant CB isoform of crotoxin, CBd. It is now possible to compare the crystal structure and function of the four natural CB isoforms. The structural role of specific amino acid variations between CB isoforms are discussed herein, and the structural framework of CB for interaction with protein targets is described.
2. Results
2.1. Three-Dimensional Structure of CBd Isoform-General Description
The crystal structure of CBd (Figure 1A) was determined at 1.8 Å resolution. The asymmetric unit of the crystal contains six individual CBd molecules. A summary of the crystallographic parameters and data, as well as refinement statistics, are shown in Table 1.
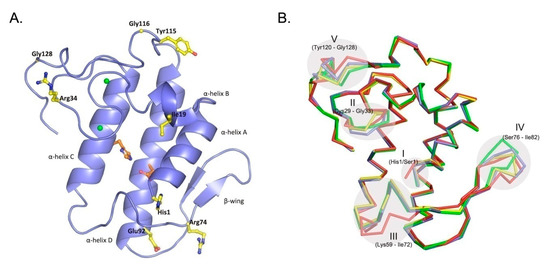
Figure 1.
Overall three-dimensional structure of the CBd isoform. (A) Crystal structure of CBd, with distinguishing residues at positions 1, 19, 34, 74, 92, 115, 116, and 128 shown in yellow stick representation. The α-helices A–D and the β-wing are labeled and the His48/Asp99 pair of the catalytic site is shown in red. (B) Superposition of the α-carbon backbones of the four natural CB isoforms which have been crystallographically resolved: CBa2 (pdb 2QOG, polypeptide chain A) in red, CBb (pdb 3R0L, polypeptide chain D) in green, CBc (pdb 2QOG, polypeptide chain B) in yellow and CBd (this study, pdb 6TMY) in blue. Five conformationally variable or flexible regions are indicated (see text).

Table 1.
Crystallographic data and refinement statistics.
As expected, the structure of CBd is very similar to the other natural CB isoforms (CBa2, CBb, and CBc) and to other group IIA sPLA2 (EC 3.1.1.4). This canonical structure contains an N-terminal α-helix (A) followed by a short helix (B), a calcium-binding loop, a long α-helix (C), an anti-parallel two-stranded beta sheet (the β-wing), a long α-helix (D) anti-parallel to helix C, and a C-terminal extension (Figure 1A). The overall structure is stabilized by seven disulfide bonds. However, comparison of the crystal structures of the four natural isoforms of CB revealed five conformationally variable or more flexible regions (Figure 1B). Region I applies to a single residue in position 1 (His/Ser, depending on the class of crotoxin) at the N-terminal of α-helix A. Region II (residues 29–33) is the canonical calcium-binding loop. Region III (residues 59–72) is located in a loop between α-helix C and the β-wing. Region IV (residues 76–82) is a part of the β-wing. Region V (residues 120–128) occurs at the C-terminal extension and contains a Gly/Glu variation at position 128 (Gly in CBb, CBc, CBd; Glu in CBa2). The crystal structure also revealed the amino acid sequence of CBd, including specific distinguishing single-residue variations (Figure 2). Amino acid residues are numbered according to Renetseder [26].
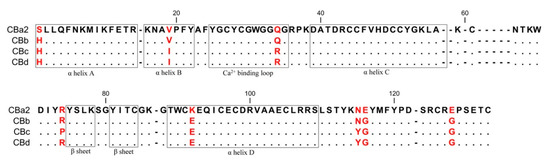
Figure 2.
Alignment of amino acid sequences of the four natural CB isoforms. Distinguishing residues of the isoforms are indicated in red. Sequences are from the GenBank/NCBI database: CBa2 (P24027.1), CBb (P0CG56.1), CBc (P62022.1) and the sequence of CBd from this study. Residues are numbered according to Renetseder et al. [26].
Two sodium ions were observed per CBd molecule in the crystal structure presented here. One Na+ binds in the canonical sPLA2 calcium-binding loop, making hydrogen bonding contacts with the carbonyl atoms of the polypeptide chain at positions Tyr28, Gly30, and Gly32, and a bidentate hydrogen bond with the side chain of Asp49. This pattern of interactions in the calcium-binding loop is also observed in the crystal structure of isoform CBb [27] and is a common feature in sPLA2 structures [29]. A second Na+ makes interactions with the backbone carbonyl oxygen of Phe24, Gly26, and Tyr120 as in the crystal structure of the heterodimeric CA2CBb crotoxin complex [27]. This second Na+ binding is weaker and lacks full coordination in two of the six individual CBd molecules in the asymmetric unit of the crystal. All six CBd molecules in the asymmetric unit contain PEG 400 (present in the crystallization solution) in the interfacial binding region near the canonical calcium-binding site.
2.2. Oligomeric Assembly of CBd Isoform
Analysis of the protein interfaces in the crystal structure of CBd using the PDBePISA web-based interactive tool [30] revealed two probable tetrameric quaternary assemblies of isoform CBd. These assemblies are composed of polypeptide chains A,E,B,C in a non-crystallographic tetramer (Figure 3A), and chains D, F in a crystallographic tetramer (2D, 2F, not shown). The two tetramers are very similar, with an rms deviation of 1.12 Å over all alpha-carbons of the superimposed structures. The total buried surface area in the AEBC and 2D2F tetramers is 9730 Å2 and 9070 Å2, respectively. The tetrameric assembly of CBd can be considered as a central dimer (A,E or D,D) associated with two peripheral monomers (B,C or F,F), respectively (Figure 3B). The most extensive contacts in the central dimeric interface are made by residues His1, Leu2, Leu3, Ile19, Phe24, Trp31, Gly32, Ala56, Lys59, Asn67, Lys69, Trp70, Phe119, Tyr120, Pro121, Asp122, and Arg127. Interestingly, the central dimer interface overlaps with the sPLA2 interfacial-binding surface [31] and the CA-binding interface of CBb in the CA2CBb crotoxin complex [27].
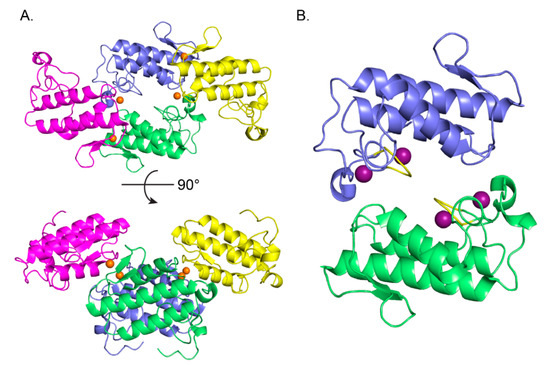
Figure 3.
Oligomeric assembly of the CBd isoform. (A) Tetrameric quaternary organization of CBd (polypeptide chains A in green, E in blue, B in magenta, C in yellow). Chloride ions are shown as orange spheres. (B) Structure of the central CBd-CBd dimer (polypeptide chains A,E). Two Na+ binding sites in each monomer are shown as purple spheres and the canonical calcium-binding loop is shown in yellow.
Extensive crystal contacts are made between a central and peripheral monomer in the tetrameric assembly, between the β-wing and α-helix A. Chloride ions in the structures reinforce these contacts. One Cl− binding site is located between the β-wings of two adjacent monomers, and makes binding interactions with the polypeptide chains of Tyr75 from the two monomers. (Figure 4A). A second chloride site is also coordinated by binding interactions with the side chains of His1 from a central and a peripheral monomer of the tetrameric assembly (chains A,B) (Figure 4B). A complementary hydrophobic interface from a third monomer (chain E) stabilizes interactions in region of the second Cl− binding site. Residues Ile19, Pro20, Phe24, Phe119 are within 4 Å of the Cl− binding site (Figure 4B). Other nearby hydrophobic residues include Met118 from chain E and Trp70 from chain B.
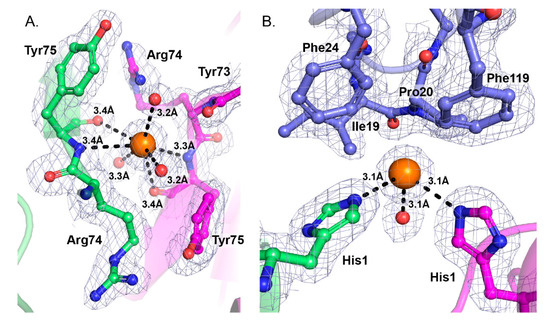
Figure 4.
Interactions of chloride ions contribute to the stability of the tetrameric quaternary assembly. (A) Cl− binds to the polypeptide chains at Tyr75 from two adjacent monomers (chains A and B). Chains are colored as in Figure 3. (B) Cl− binds to the side chains of His1 from the two adjacent CB monomers (chains A and B). Additional bonds are made with nearby water molecules. Residues within 4 Å are labeled and shown as sticks. The bond distances are indicated. Chloride ions are represented as orange spheres and coordinating water molecules are shown as red spheres. The electron density map is contoured at the 1.0 sigma level.
Additional crystal contacts are made between the central and peripheral monomers principally through interactions of Arg14 with either Glu12 or Phe11, and interactions of the tip of the β-wing.
3. Discussion
3.1. Residue Variations in CB Isoforms Distinguish between Class I and II Crotoxin Complexes
Classification of crotoxin complexes was made according to their stability, toxicity, and enzymatic activity, which depend solely on the isoform of the CB subunit in the complex [5]. Isoform CBd was previously classified to class I since it possesses biological properties similar to CBb and CBc in complex with CA [5]. Class I forms very stable and toxic crotoxin complexes with low enzymatic activity. The sequence determination of CBd from the crystal structure, and its comparison to the other CB isoforms clearly reveal four residues that distinguish between two classes (I and II) of crotoxin complexes. These are His1Ser, Glu92Lys, Gly116Glu, Gly128Glu in CBd, CBc, CBb (class I) and CBa2 (class II), respectively (Table 2).

Table 2.
Distinguishing residues in CB isoforms form two classes of crotoxin complexes.
His1Ser: The presence of His at position 1 stabilizes the N-terminal region through the formation of a hydrogen bond network. This network originates from a hydrogen bond between the πN of His1 and the backbone amine of Leu3. This interaction stabilizes the N-terminal amine and allows for formation of two hydrogen bonds with the side chain oxygen of Gln4 and the carbonyl backbone of Asp71. Gln4 is then locked in place and thereby forms two respective hydrogen bonds between its side chain amide atoms and the main chain amide of Tyr73. Additional hydrogen bonds are observed between the backbone carbonyl of Lys69 and the backbone amine of Leu2. These arrangements create the observed conformational structure in region III for CBb, CBc, and CBd, and facilitate the direct interaction of Trp70 with Asp89 of the acidic CA subunit, leading to increased stability of the crotoxin complexes [27] (from Kd = 28 nM for CBa2 to Kd = 5 nM for CBb, CBc, and CBd [5]).
Glu92Lys: In CBa2, which lacks N-terminal stability through the presence of Ser1, the Lys92 could make a hydrogen bond to the side chain of Asp71 leading to further destabilization of the N-terminal and a conformational change in region III with respect to class I CB isoforms. In class I complexes the presence of Glu92 does not interfere with the above-mentioned hydrogen bond network.
Gly116Glu, Gly128Glu: The C-terminal region of CB, does not participate in the formation of the crotoxin complex [27], but might be involved in the neurotoxic activity of crotoxin [32]. This region in CBd, CBc, and CBb isoforms (of class I) is strongly positively charged. The presence of two negatively charged residues: Glu116 and Glu128 in CBa2 (instead of Gly116 and 128) results in a significant change in surface charge of the molecule, which may affect the binding to specific receptors (such as CAPT-48 from Torpedo marmorata [8]) and the overall toxicity of the crotoxin.
3.2. Role of the Differences in Amino Acid Sequences between CBd, CBc, and CBb on the Properties of Class I Crotoxin Complexes
Our previous studies showed that the biological properties of CBb, CBc, and CBd, and the respective crotoxin complexes, are very similar [5]. The amino acid sequences of the CB isoforms of class I are also highly similar. CBd differs from CBc by only one amino acid (Arg74Pro, respectively) and by three amino acids from CBb (Ile19Val, Arg34Gln, and Tyr115Asn, respectively). Comparative analysis of the crystal structures of all four CB isoforms suggests that these differences do not affect the catalytic activity of CB or the toxicity of crotoxin; however, there may be slight effects upon the interaction of CB with the CA subunit. Specific evaluation of each position is as follows:
Ile19Val: In the region around position 19 (Ile/Val) the structures of all isoforms are similar. The side chain is solvent exposed and centrally located in the CA binding interface. Steric bulk differences between Ile and Val could have a small effect on binding, but there is no observed correlation with toxicity [27].
Arg34Gln: The Arg side chain at position 34 appears to be highly flexible in the resolved crystal structures, whereas the main chain conformation is similar in all CB isoforms. Since this solvent-exposed residue does not participate in any binding, the mutation to Gln (in CBb) does not appear to play any significant role in the functional properties of CB and crotoxin complexes. However, changes in the intrinsic properties of CB through the presence of Arg and the addition of a positive charge on the protein surface may have other significant consequences.
Arg74Pro: Comparison of the available CB structures, in which only CBc contains Pro at position 74, shows the two adjacent Tyr residues (at position 73, 75), along with the main chain at position 74, in the same conformation in all isoforms. However the presence of Pro74 in CBc, instead of Arg, could lead to reduced flexibility of the polypeptide chain in this region and could lead to a distinct change in properties between the two protein variants.
Tyr115Asn: In the crystal structure of the CA2CBb complex [27], Asn115 (in CBb) is solvent exposed and situated near Gly35 of the CA subunit α-chain. However, the distance between these two residues (greater than 6 Å) is too large to generate any significant interaction. In contrast, Tyr115 (in CBc and CBd) could form a hydrogen bond with the polypeptide chain or the side chain of Gln34 of the CA α-chain. Indeed, Fortes-Dias et al. confirmed the importance of Tyr115 in protein-protein interactions [33]. They showed that the region of CBc containing Tyr115 plays an important role in CICS/CNF binding (a natural crotoxin inhibitor from snake blood). According to these studies, even 10 molar excess of CA did not lead to dissociation of the inhibitor bound to this region. In the CBa2 isoform, in which Asn115 is present, this region did not participate in CICS binding [33].
3.3. The Natural CA-Binding Domain on CB Serves as the Structural Framework for Interaction with Other Biological Targets: Human Coagulation FXa and Human CFTR
It was previously shown that the CB subunit of crotoxin, acting independently of the crotoxin complex can exhibit various pathophysiological and pharmacological functions [7,19,20]. We showed that at presynaptic membranes, the isolated CB can recognize similar protein receptors, such as monomeric ammodytoxin (Atx). since CB was able to completely inhibit the binding activity of radio-iodinated ammodytoxin [7]. Several other protein targets, including human coagulation FXa and human CFTR, were identified and characterized as a target for this PLA2, showing its anticoagulant or anti-cystic fibrosis activity [19,20]. All of these previously studied mechanisms are independent of catalytic activity of PLA2 and are based on direct protein-protein interaction. CB can affect hemostasis by binding to human FXa, and can inhibit formation of the prothrombinase complex required for efficient production of thrombin by the non-enzymatic, phospholipid-independent, anticoagulant mode of action [19]. More recently, we showed that CB forms a high nanomolar affinity complex with NBD1, as well as its mutated form ΔF508-NBD1 of CFTR [20]. CB could act as a positive allosteric modulator increasing Cl− channel current of CFTR and as corrector of the ΔF508-CFTR mutant implicated in cystic fibrosis disorder [20].
The three-dimensional structure of the crotoxin complex revealed the nature of the binding interface between the CA and CB subunits and allowed us to identify key amino acid residues responsible for significant differences in the stability, toxicity, and enzymatic activity of the two classes of crotoxin complexes [27]. Molecular docking analysis supported by hydrogen deuterium exchange mass spectrometry (HDX-MS) and surface plasmon resonance (SPR) competition studies [20]. showed that regions of CB participating in the interaction with ΔF508-NBD1 overlap with those involved in forming the natural CA-CB complex (residues His1, Leu3, Asn6, Asn17, Ala18, Val19, Ala23, Phe24, Trp31, Gly32, Lys69, Trp70, Met118, Phe119, Tyr120, Pro121, Ser124) [20]. Similar studies using SPR showed that the hFXa-binding site partially overlaps with the CA-binding site of CB (residues His1, Leu3, Phe24, Trp31, Gly32, Lys69, Trp70, Phe119, Pro121, Ser124) [19,34,35]. Interestingly, it was shown by analytical ultracentrifugation that CA and hFXa prevent oligomerization of CB [36]. Interfacial binding surface (IBS) residues of CB (Leu2, Leu3, Ile19, Phe24 and Lys69, identified by homology with Atx C [37], overlap with the interface of complexes CA-CB and CB-hFXa, but also CB-NBD1 and CB-CB. Protein targets (CA, hFXa, or NBD1) therefore prevent CB contact with phospholipids. The CA-, hFXa-, and ΔF508-NBD1-binding sites of CB are shown in Figure 5 and Figure 6. Three regions containing six conserved amino acid residues appear to play an important role in protein-protein interactions. These are α-helix A (His1 and Leu3), the calcium-binding loop (Trp31 and Gly32), and a loop preceding the β-wing (Lys69 and Trp70). Additionally, the C-terminal region of CB, despite the lack of conservation, also actively participates in the aforementioned interactions. The side-chains of these residues are exposed on the surface of the CB molecule, presenting favorable orientations for interaction with CFTR or hFXa. Interestingly, our previous study showed that hFXa-binding site of PLA2 (Atx A and C) partly overlaps with the CaM-binding site [38]. Characterization of this basal CB pharmacological platform offers promising perspectives for structure-based design of new anti-cystic fibrosis and anticoagulant therapeutics.

Figure 5.
Sequences of CB isoforms showing the key residues in the interface regions of the specified complexes. Green-shadowed characters denote residues at the interface of CBa2/hFXa [19], CBb/CA [27], CBb/ΔF508-NBD1 [20] of CFTR, CBc/hFXa [19], and CBd/CBd [this study].
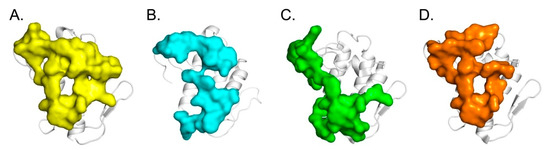
Figure 6.
Functional sites of CB, a basal platform for protein-protein interaction. (A) CA-binding site identified by crystallography (in yellow) [27], (B) CBd-CBd dimer site identified by crystallography (present study, in blue), (C) hFXa-binding site generated by molecular docking (in green) [19] and (D) ΔF508-NBD1-binding site identified by molecular docking and HDX-MS experiments (in orange) [20].
3.4. Pathophysiological Relevance of Dimer of CBd
The CBd crystal structure was resolved as a homogenous oligomer of four monomers, which form a central dimer connected with two peripheral molecules. A tetrameric structure was also obtained by Salvador-Marchi and colleagues [28]; however, in this case it contained two heterodimers each composed of CB1 (CBc) and CB2 (CBa2) isoforms [28]. The interface between the heterodimers, the CB1 (CBc) homodimer, resembles the central dimer described in the present structure. The physiological or pathophysiological relevance of the oligomeric formation of CB subunits is still unclear. In rattlesnake venom, all CB isoforms are present in complex with the CA subunit, which prevents oligomerization of CB. The region of CB participating in the formation of the central dimer overlaps with the region involved in forming the natural CA-CB complex (Figure 5 and Figure 6). Therefore, the formation of oligomer structures does not seem to be a natural conformation and may be due to crystallization conditions, as variations in pH have been shown to affect the CB oligomerization state [39].
However, we still do not know the mechanism of crotoxin-receptor interactions at the atomic level. Formation of the ternary complex, CA-CB-Receptor, and dissociation of CA at equilibrium (with only CB attached to the receptor) have been previously proposed by biochemical and SPR studies [8].
Similarly to the CA-CB interaction, the human coagulation factor Xa also prevents oligomerization of CB, showing the importance of the monomeric state of this PLA2 for anticoagulant action [36]. According to this data, the formation of CB oligomers does not seem to be a physiological process. However, the CB dimer could create more accessible surface areas for interaction with specific PLA2-receptors by increasing the number of binding sites [28,38,40]. According to this hypothesis, tetrameric or dimeric configurations may play an important role in neurotoxicity or other functions of human inflammatory sPLA2GIIA. Moreover, in pathological conditions, where high concentrations of sPLA2 have been detected [41,42,43], formation of sPLA2 oligomers could be expected. Additional in vivo studies are needed to explain the role of oligomerization of sPLA2 in pathophysiological processes.
4. Materials and Methods
4.1. Purification of Isoform CBd
Four CB isoforms were isolated from the crude venom of Crotalus durissus terrificus, as described previously [3]. In brief, whole venom was fractionated on a Sephadex G-75 column and the major peak of crotoxin (a mixture of crotoxin isoforms) was isolated. The CA and CB subunits were separated by anion exchange chromatography in 6 M urea and dialyzed. The isoforms of CA subunit were purified on a Mono Q column and the isoforms of CB were purified on a Mono S column [3] followed by reversed phase chromatography for each isoform on a Vydac C4 column [44]. The purity of the CB isoforms was confirmed by electrospray mass spectrometry [45], or non-denaturing electrophoresis after reconstitution of the crotoxin complexes (one CB isoform with one CA isoform) using 20% homogenous PhastGel [3]. Protein concentration was determined spectrophotometrically as described [3]. A single, homogenous CBd isoform was used for crystallization assays.
4.2. Crystallization
The lyophilized CBd was dissolved in 20 mM Tris (pH 7.4), 300 mM NaCl and 0.02% DDM, with a final protein concentration of 4.2 mg/mL. Initial crystallization conditions were screened at the Crystallography Platform of the Institut Pasteur [46] using MemGold screen kits (Molecular Dimensions), and gave small crystals after 8–10 days. Optimization was pursued on 24-well Linbro plates at 4 °C using the hanging-drop vapor-diffusion method. In the drop, 1 µL of crystallization buffer was mixed with 1 µL of CBd, giving a final protein concentration of 2.1 mg/mL. The best crystals were obtained with a solution containing 21% (v/v) PEG 400, 0.1 M NaCl, 0.18 M sodium acetate, and 0.1 M Tris (pH 8.0). The crystals grew within seven weeks at 4 °C.
4.3. Data Collection and Refinement
Single crystals of CBd were rinsed briefly in solutions containing the crystallization buffer with 30% (v/v) PEG 400 as cryoprotectant and flash-cooled in liquid nitrogen for data collection at cryogenic temperature. X-ray diffraction data were measured on beamline Proxima-1 at synchrotron SOLEIL (Saint-Aubin, France). The diffraction images were integrated with XDS [47] and crystallographic calculations carried out with the ccp4 program suite [48]. The structure was solved by molecular replacement with the Phaser [49] using crotoxin isoform CBb (pdb 3R0L, polypeptide chain D) as a template. The structure was refined with Buster [50] and manual adjustments were made to the model with Coot [51]. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (pdb) with accession code 6TMY.
4.4. Bioinformatics Analysis of Sequences
For bioinformatics analysis amino acid sequences of Crotalus durissus terrificus CB isoforms deposited in GenBank/National Center for Biotechnology Information (NCBI) data base (P24027.1 (CBa2), P0CG56.1 (CBb), P62022.1 (CBc)) were used. Multiple alignment analysis of the sequences was performed using CLC Sequence Viewer 6.8 software.
5. Conclusions
Snake venoms are an important source of biologically active molecules that, in addition to toxicity, can reveal a therapeutic potential. Detailed characterization of these proteins is crucial for the possibility of their use in medical and pharmacological applications. Here we presented the three-dimensional structure of CBd, the most abundant isoform of the basic PLA2 subunit of crotoxin. Comparison of its three-dimensional (3D) structure and quaternary organization, as well as amino acid sequence, with the three other CB isoforms indicated specific single residue variations that are important for the functional diversity of crotoxin complexes. We also described the functional sites of CB that allow for its interaction with other proteins. These functional sites were previously determined experimentally by biophysical and biochemical methods (protein-protein interaction using SPR, affinity binding studies and competition experiments, physiological and electrophysiological functional analysis, hydrogen-deuterium exchange mass spectrometry, molecular docking simulations and crystallographic studies). The binding interface of four complexes (CA2-CBb, CBc-FXa, CBb-∆F508NBD1) and CBd-CBd (this study), identified at the level of specific amino acid residues, significantly overlap. Determination of such interaction platforms may be useful for developing new anti-cystic fibrosis, as well as anticoagulant, therapeutics.
Author Contributions
Project conceptualization (G.F., M.O.); Project supervision (G.F.); Protein purification and characterization (G.F., M.R., M.O.); Crystalogenesis (M.O., G.F.); Crystal data collection and refinement (F.S.); bioinformatics (D.N.); Original draft (D.N.); Review and editing (G.F., D.N., F.S.). All authors have read and agreed to the published version of the manuscript.
Funding
M.O. and D.N. received research fellowships within project “Enhancing Educational Potential of Nicolaus Copernicus University in the Disciplines of Mathematical and Natural Sciences” (project no. POKL.04.01.01-00-081/10).
Acknowledgments
We thank Ahmed Haouz and Patrick Weber of the crystallography platform at the Institut Pasteur for assistance and access to automated crystallization screening facilities. We acknowledge SOLEIL for provision of synchrotron radiation facilities and we would like to thank the personnel of beamlines Proxima-1 and Proxima-2 for assistance. We are grateful to Ákos Nemecz for critical reading of the manuscript and for helpful suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hendon, R.A.; Fraenkel-Conrat, H. Biological roles of the two components of crotoxin. Proc. Natl. Acad. Sci. USA 1971, 68, 1560–1563. [Google Scholar] [CrossRef] [PubMed]
- Faure, G.; Bon, C. Several isoforms of crotoxin are present in individual venoms from the South American rattlesnake Crotalus durissus terrificus. Toxicon 1987, 25, 229–234. [Google Scholar] [CrossRef]
- Faure, G.; Bon, C. Crotoxin, a phospholipase A2 neurotoxin from the South American rattlesnake crotalus durissus terrificus: Purification of several isoforms and comparison of their molecular structure and of their biological activities. Biochemistry 1988, 27, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Faure, G.; Guillaume, J.L.; Camoin, L.; Saliou, B.; Bon, C. Multiplicity of acidic subunit isoforms of crotoxin, the phospholipase A2 neurotoxin from Crotalus durissus terrificus venom, results from posttranslational modifications. Biochemistry 1991, 30, 8074–8083. [Google Scholar] [CrossRef] [PubMed]
- Faure, G.; Harvey, A.L.; Thomson, E.; Saliou, B.; Radvanyi, F.; Bon, C. Comparison of crotoxin isoforms reveals that stability of the complex plays a major role in its pharmacological action. Eur. J. Biochem. 1993, 214, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Hawgood, B.J.; Smith, J.W. The mode of action at the mouse neuromuscular junction of the phospholipase A—Crotapotin complex isolated from venom of the South American rattlesnake. Br. J. Pharmacol. 1977, 61, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Krizaj, I.; Faure, G.; Gubensek, F.; Bon, C. Neurotoxic phospholipases A2 ammodytoxin and crotoxin bind to distinct high-affinity protein acceptors in Torpedo marmorata electric organ. Biochemistry 1997, 36, 2779–2787. [Google Scholar] [CrossRef]
- Faure, G.; Copic, A.; Le Porrier, S.; Gubensek, F.; Bon, C.; Krizaj, I. Crotoxin acceptor protein isolated from Torpedo electric organ: Binding properties to crotoxin by surface plasmon resonance. Toxicon 2003, 41, 509–517. [Google Scholar] [CrossRef]
- Shimizu, J.F.; Pereira, C.M.; Bittar, C.; Batista, M.N.; Campos, G.R.F.; da Silva, S.; Cintra, A.C.O.; Zothner, C.; Harris, M.; Sampaio, S.V.; et al. Multiple effects of toxins isolated from Crotalus durissus terrificus on the hepatitis C virus life cycle. PLoS ONE 2017, 12, e0187857. [Google Scholar] [CrossRef]
- Cura, J.E.; Blanzaco, D.P.; Brisson, C.; Cura, M.A.; Cabrol, R.; Larrateguy, L.; Mendez, C.; Sechi, J.C.; Silveira, J.S.; Theiller, E.; et al. Phase I and pharmacokinetics study of crotoxin (cytotoxic PLA2, NSC-624244) in patients with advanced cancer. Clin. Cancer Res. 2002, 8, 1033–1041. [Google Scholar]
- Wang, J.; Qin, X.; Zhang, Z.; Chen, M.; Wang, Y.; Gao, B. Crotoxin suppresses the tumorigenic properties and enhances the antitumor activity of Iressa® (gefinitib) in human lung adenocarcinoma SPCA-1 cells. Mol. Med. Rep. 2014, 10, 3009–3014. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Yang, Y.; Qin, Z.; Gu, Z.; Reid, P.; Liang, Z. Autophagy is involved in cytotoxic effects of crotoxin in human breast cancer cell line MCF-7 cells. Acta Pharmacol. Sin. 2007, 28, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, S.C.; Rangel-Santos, A.C.; Peres, C.M.; Curi, R.; Cury, Y. Inhibitory effect of phospholipase A2 isolated from crotalus durissus terrificus venom on macrophage function. Toxicon 2005, 45, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Zambelli, V.O.; Sampaio, S.C.; Sudo-Hayashi, L.S.; Greco, K.; Britto, L.R.G.; Alves, A.S.; Zychar, B.C.; Gonçalves, L.R.C.; Spadacci-Morena, D.D.; Otton, R.; et al. Crotoxin alters lymphocyte distribution in rats: Involvement of adhesion molecules and lipoxygenase-derived mediators. Toxicon 2008, 51, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.P.; Favoretto, B.C.; Clissa, P.B.; Sampaio, S.C.; Faquim-Mauro, E.L. Crotoxin isolated from crotalus durissus terrificus venom modulates the functional activity of dendritic cells via formyl peptide receptors. J. Immunol. Res. 2018, 2018, 7873257. [Google Scholar] [CrossRef]
- Zhang, H.-L.; Han, R.; Chen, Z.-X.; Chen, B.-W.; Gu, Z.-L.; Reid, P.F.; Raymond, L.N.; Qin, Z.-H. Opiate and acetylcholine-independent analgesic actions of crotoxin isolated from crotalus durissus terrificus venom. Toxicon 2006, 48, 175–182. [Google Scholar] [CrossRef]
- da Silva Lomeo, R.; de Faria Gonçalves, A.P.; da Silva, C.N.; de Paula, A.T.; Santos, D.O.C.; Fortes-Dias, C.L.; Gomes, D.A.; de Lima, M.E. Crotoxin from crotalus durissus terrificus snake venom induces the release of glutamate from cerebrocortical synaptosomes via N and P/Q calcium channels. Toxicon 2014, 85, 5–16. [Google Scholar] [CrossRef]
- Krizaj, I.; Faure, G.; Gubensek, F.; Bon, C. Re-examination of crotoxin-membrane interactions. Toxicon 1996, 34, 1003–1009. [Google Scholar] [CrossRef]
- Faure, G.; Gowda, V.T.; Maroun, R.C. Characterization of a human coagulation factor Xa-binding site on Viperidae snake venom phospholipases A2 by affinity binding studies and molecular bioinformatics. BMC Struct. Biol. 2007, 7, 82. [Google Scholar] [CrossRef]
- Faure, G.; Bakouh, N.; Lourdel, S.; Odolczyk, N.; Premchandar, A.; Servel, N.; Hatton, A.; Ostrowski, M.K.; Xu, H.; Saul, F.A.; et al. Rattlesnake phospholipase A2 increases CFTR-chloride channel current and corrects ∆F508CFTR dysfunction: Impact in cystic fibrosis. J. Mol. Biol. 2016, 428, 2898–2915. [Google Scholar] [CrossRef]
- Ostrowski, M.; Porowinska, D.; Prochnicki, T.; Prevost, M.; Raynal, B.; Baron, B.; Sauguet, L.; Corringer, P.-J.; Faure, G. Neurotoxic phospholipase A2 from rattlesnake as a new ligand and new regulator of prokaryotic receptor GLIC (proton-gated ion channel from G. violaceus). Toxicon 2016, 116, 63–71. [Google Scholar] [CrossRef]
- Fortes-Dias, C.L.; Lin, Y.; Ewell, J.; Diniz, C.R.; Liu, T.Y. A phospholipase A2 inhibitor from the plasma of the South American rattlesnake (Crotalus durissus terrificus). Protein structure, genomic structure, and mechanism of action. J. Biol. Chem. 1994, 269, 15646–15651. [Google Scholar]
- Perales, J.; Villela, C.; Domont, G.B.; Choumet, V.; Saliou, B.; Moussatché, H.; Bon, C.; Faure, G. Molecular structure and mechanism of action of the crotoxin inhibitor from crotalus durissus terrificus serum. Eur. J. Biochem. 1995, 227, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Faure, G. Natural inhibitors of toxic phospholipases A2. Biochimie 2000, 82, 833–840. [Google Scholar] [CrossRef]
- Faure, G.; Porowinska, D.; Saul, F. Crotoxin from crotalus durissus terrificus and crotoxin-related proteins: Structure and function relationship. In Toxins and Drug Discovery; Gopalakrishnakone, P., Cruz, L.J., Luo, S., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 3–20. ISBN 978-94-007-6452-1. [Google Scholar]
- Renetseder, R.; Brunie, S.; Dijkstra, B.W.; Drenth, J.; Sigler, P.B. A comparison of the crystal structures of phospholipase A2 from bovine pancreas and Crotalus atrox venom. J. Biol. Chem. 1985, 260, 11627–11634. [Google Scholar]
- Faure, G.; Xu, H.; Saul, F.A. Crystal structure of crotoxin reveals key residues involved in the stability and toxicity of this potent heterodimeric β-neurotoxin. J. Mol. Biol. 2011, 412, 176–191. [Google Scholar] [CrossRef]
- Marchi-Salvador, D.P.; Corrêa, L.C.; Magro, A.J.; Oliveira, C.Z.; Soares, A.M.; Fontes, M.R.M. Insights into the role of oligomeric state on the biological activities of crotoxin: Crystal structure of a tetrameric phospholipase A2 formed by two isoforms of crotoxin B from Crotalus durissus terrificus venom. Proteins 2008, 72, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.L.; White, S.P.; Otwinowski, Z.; Yuan, W.; Gelb, M.H.; Sigler, P.B. Interfacial catalysis: The mechanism of phospholipase A2. Science 1990, 250, 1541–1546. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Snitko, Y.; Koduri, R.S.; Han, S.K.; Othman, R.; Baker, S.F.; Molini, B.J.; Wilton, D.C.; Gelb, M.H.; Cho, W. Mapping the interfacial binding surface of human secretory group IIa phospholipase A2. Biochemistry 1997, 36, 14325–14333. [Google Scholar] [CrossRef]
- Zambelli, V.O.; Picolo, G.; Fernandes, C.A.H.; Fontes, M.R.M.; Cury, Y. Secreted phospholipases A2 from animal venoms in pain and analgesia. Toxins 2017, 9, 406. [Google Scholar] [CrossRef]
- Fortes-Dias, C.L.; dos Santos, R.M.M.; Magro, A.J.; de Mattos Fontes, M.R.; Chávez-Olórtegui, C.; Granier, C. Identification of continuous interaction sites in PLA2-based protein complexes by peptide arrays. Biochimie 2009, 91, 1482–1492. [Google Scholar] [CrossRef] [PubMed]
- Faure, G.; Saul, F. Structural and functional characterization of anticoagulant, FXa-binding Viperidae snake venom phospholipases A2. Acta Chim. Slov. 2011, 58, 671–677. [Google Scholar] [PubMed]
- Faure, G.; Xu, H.; Saul, F. Anticoagulant phospholipases A2 which bind to the specific soluble receptor coagulation factor Xa. In Toxins and Hemostasis: From Bench to Bedside; Kini, R.M., Clemetson, K.J., Markland, F.S., McLane, M.A., Morita, T., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 201–217. ISBN 978-90-481-9295-3. [Google Scholar]
- Ostrowski, M.; Žnidaršič, P.P.; Raynal, B.; Saul, F.; Faure, G. Human coagulation factor Xa prevents oligomerization of anti-coagulant phospholipases A2. Toxin Rev. 2014, 33, 42–47. [Google Scholar] [CrossRef]
- Petan, T.; Križaj, I.; Gelb, M.H.; Pungerčar, J. Ammodytoxins, potent presynaptic neurotoxins, are also highly efficient phospholipase A2 enzymes. Biochemistry 2005, 44, 12535–12545. [Google Scholar] [CrossRef]
- Saul, F.A.; Prijatelj-Znidarsic, P.; Vulliez-le Normand, B.; Villette, B.; Raynal, B.; Pungercar, J.; Krizaj, I.; Faure, G. Comparative structural studies of two natural isoforms of ammodytoxin, phospholipases A2 from Vipera ammodytes ammodytes which differ in neurotoxicity and anticoagulant activity. J. Struct. Biol. 2010, 169, 360–369. [Google Scholar] [CrossRef]
- Radvanyi, F.R.; Bon, C. Catalytic activity and reactivity with p-bromophenacyl bromide of the phospholipase subunit of crotoxin. Influence of dimerization and association with the noncatalytic subunit. J. Biol. Chem. 1982, 257, 12616–12623. [Google Scholar]
- Faure, G.; Saul, F. Crystallographic characterization of functional sites of crotoxin and ammodytoxin, potent β-neurotoxins from Viperidae venom. Toxicon 2012, 60, 531–538. [Google Scholar] [CrossRef]
- Šribar, J.; Kovačič, L.; Oberčkal, J.; Ivanušec, A.; Petan, T.; Fox, J.W.; Križaj, I. The neurotoxic secreted phospholipase A2 from the Vipera a. ammodytes venom targets cytochrome c oxidase in neuronal mitochondria. Sci. Rep. 2019, 9, 283. [Google Scholar] [CrossRef]
- Mounier, C.M.; Luchetta, P.; Lecut, C.; Koduri, R.S.; Faure, G.; Lambeau, G.; Valentin, E.; Singer, A.; Ghomashchi, F.; Béguin, S.; et al. Basic residues of human group IIA phospholipase A2 are important for binding to factor Xa and prothrombinase inhibition comparison with other mammalian secreted phospholipases A2. Eur. J. Biochem. 2000, 267, 4960–4969. [Google Scholar] [CrossRef]
- Vadas, P.; Browning, J.; Edelson, J.; Pruzanski, W. Extracellular phospholipase A2 expression and inflammation: The relationship with associated disease states. J. Lipid Mediat. 1993, 8, 1–30. [Google Scholar] [PubMed]
- Faure, G.; Saul, F. Caractéristiques structurales et fonctionnelles de deuxβ-neurotoxines: L’ammodytoxine et la crotoxine. In Proceedings of the Toxines et Fonctions Cholinergiques Neuronales et non Neuronales, Paris, France, 20–21 November 2008; pp. 155–160. [Google Scholar]
- Faure, G.; Choumet, V.; Bouchier, C.; Camoin, L.; Guillaume, J.L.; Monegier, B.; Vuilhorgne, M.; Bon, C. The origin of the diversity of crotoxin isoforms in the venom of Crotalus durissus terrificus. Eur. J. Biochem. 1994, 223, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Weber, P.; Pissis, C.; Navaza, R.; Mechaly, A.E.; Saul, F.; Alzari, P.M.; Haouz, A. High-throughput crystallization pipeline at the crystallography core facility of the institut pasteur. Molecules 2019, 24, 4451. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 133–144. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Bricogne, G.; Blanc, E.; Brandl, M.; Flensburg, C.; Keller, P.; Paciorek, P.; Roversi, P.; Sharff, A.; Smart, O.; Vonrhein, C.; et al. BUSTER; Global Phasing Ltd.: Cambridge, UK, 2010. [Google Scholar]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).