From Differential Stains to Next Generation Physiology: Chemical Probes to Visualize Bacterial Cell Structure and Physiology
Abstract
1. Introduction—From 19th Century Microbiology to Modern Day Chemical Biology
2. A Guide to Structure and Mode-Of-Action of Different Chemical Probes
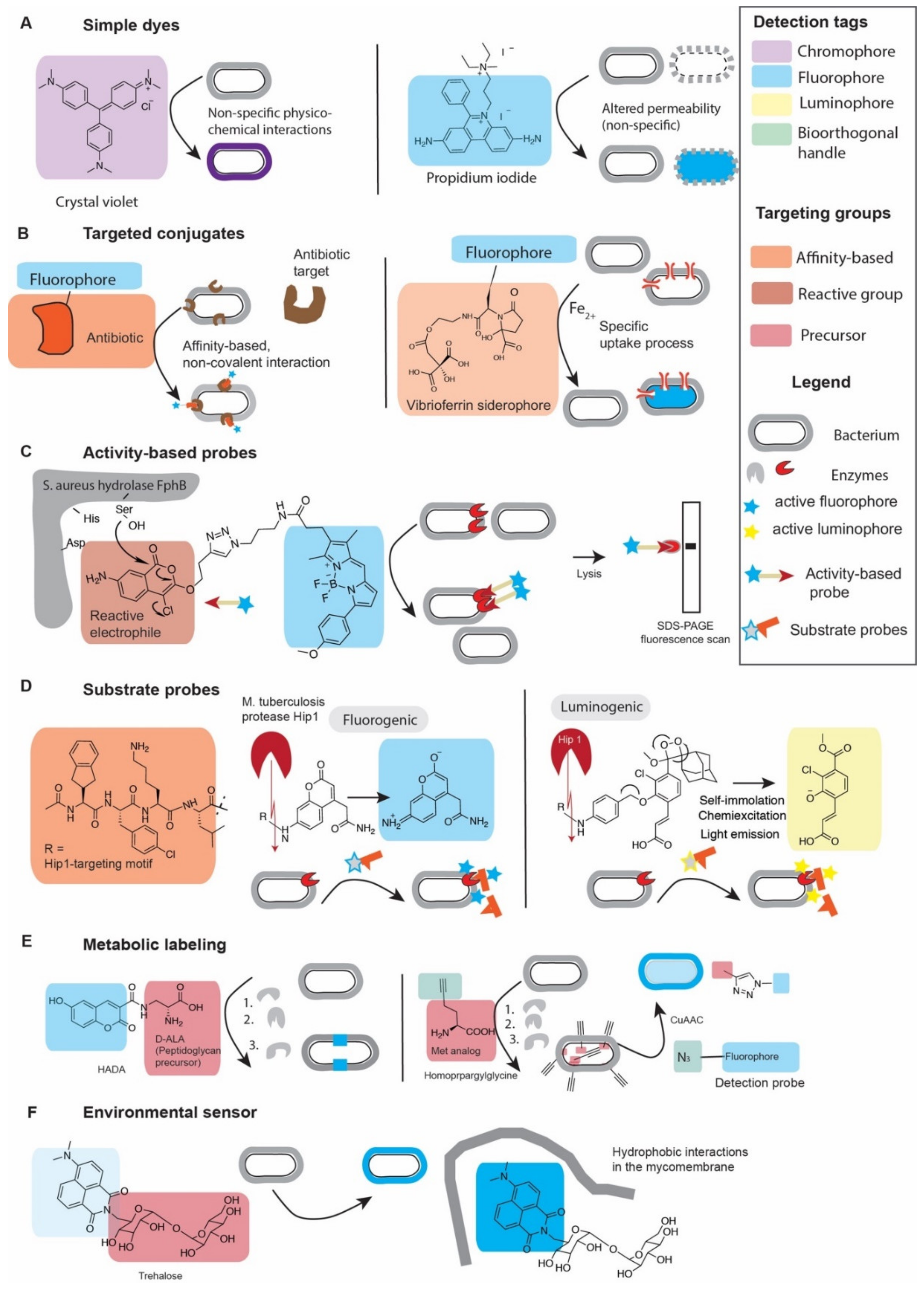
2.1. How to Detect and How to Target Probes
2.1.1. Chemical Dyes
2.1.2. Targeted Fluorescent Probes
2.1.3. Fluorogenic and Quenched Fluorescent Probes
2.1.4. Two-Step Detection Strategies Relying on Bio-Orthogonal Tags
2.1.5. Chemiluminescent Probes
2.1.6. Alkyne-Based Probes for Raman Spectroscopy
2.1.7. Radioactive Labels
2.2. Chemical Probes to Illuminate (Micro)Biological Activities
2.2.1. Cellular Permeability and Its Effect on Specific and Non-Specific Probes
2.2.2. Non-Covalent Targeted Conjugates
2.2.3. Activity-Based Probes
2.2.4. Substrate Probes
2.2.5. Metabolic Labeling
2.2.6. Environmental Sensors
2.3. The Frontier of Chemical Probe Synthesis
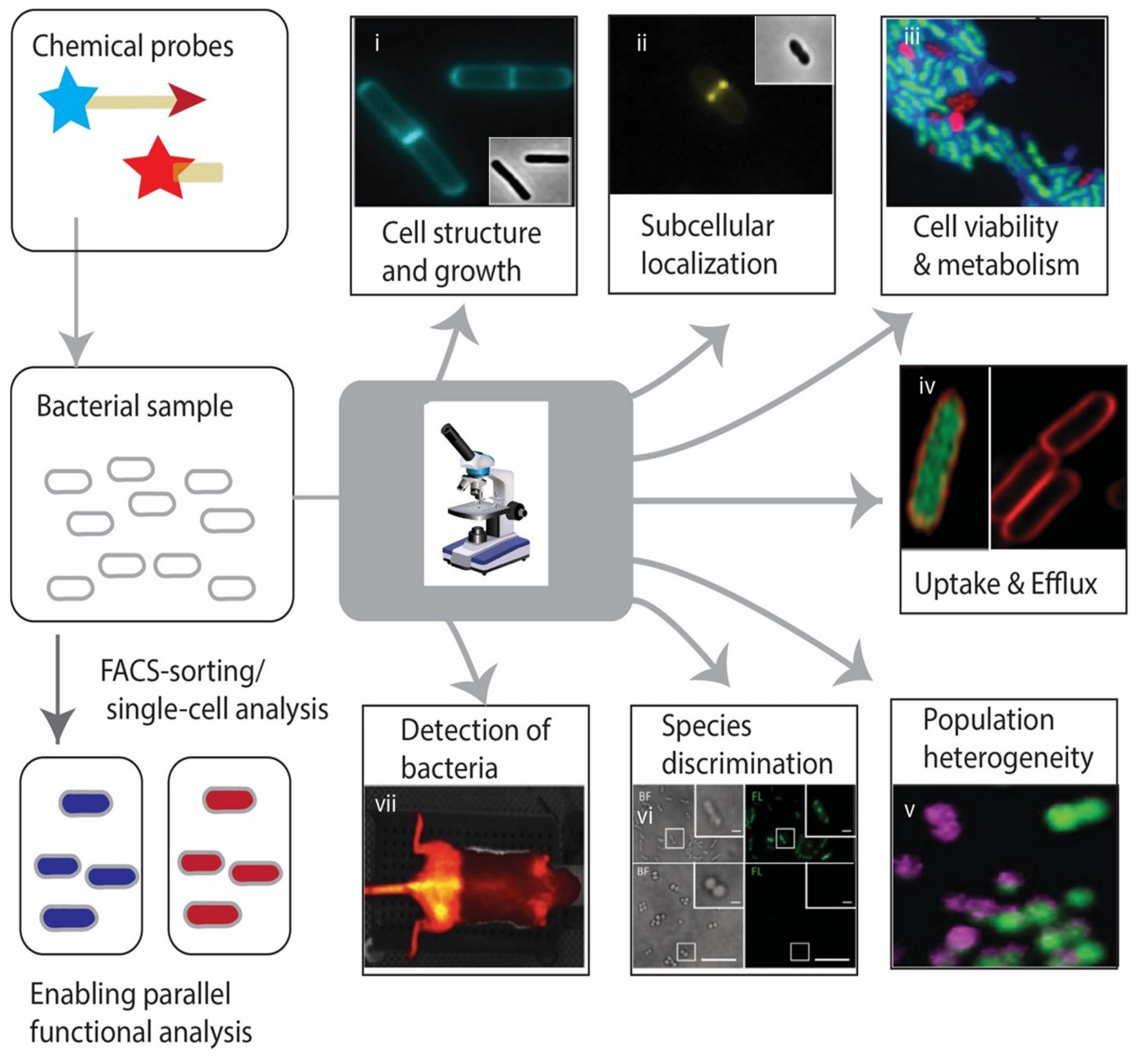
3. Chemical Probes in Action: Applications in Imaging of Bacteria
3.1. The Cell Wall
3.1.1. Metabolic Labeling of Peptidoglycan
3.1.2. Dissecting the Activity of Penicillin-Binding Proteins
3.1.3. Targeting Cell Wall Precursors
3.1.4. Targeting Other Components of the Cell Envelope
3.1.5. Trehalose and the Unique Cell Envelope of Mycobacteria
3.2. Dissecting Antibiotic Susceptibility and Resistance
3.3. Visualizing Specific Metabolic Uptake Pathways
3.3.1. Siderophores
3.3.2. Sugar Uptake
3.4. Visualizing Virulence-Associated Enzymes
3.5. Biofilms and Other Microbial Communities
4. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACC | 7-amino-4 carbamoylmethylcoumarin |
| AM | Acetoxymethyl |
| BONCAT | Bio-orthogonal non-canonical amino acid tagging |
| BODIPY | 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene |
| CuAAC | Copper-catalyzed azide-alkyne cycloaddition |
| Cy5 | Cyanine 5 |
| DAA | D-amino acid |
| DAP | mesodiaminopimelic acid |
| DDAO | 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) |
| Ddl | D-alanine-D-alanine ligase |
| DNA | Deoxyribonucleic acid |
| DMN | 4-N,N-Dimethylamino-1,8-naphthalimide |
| FACS | Fluorescence-activated cell sorting |
| FDA | Federal drug administration |
| FDAA | Fluorescent D-Amino Acid |
| FDL | Fluorescein-D-Lysine |
| FRET | Fluorescence resonance energy transfer |
| GFP | Green fluorescent protein |
| HADA | HCC-amino-D-alanine |
| Hip1 | Hydrolase important for pathogenesis 1 |
| KDO | 3-deoxy-D-manno-octulosonic acid |
| LPS | Lipopolysaccharide |
| MOA | Mode of action |
| Mtb | Mycobacterium tuberculosis |
| NADA | NBD-amino-D-alanine |
| NBD | 7-Amino-4-nitro-2,1,3-benzoxadiazole |
| NTR | nitroreductase |
| OM | Outer membrane |
| Fph | Fluorophosphonate-binding hydrolase |
| PBP | Penicillin-binding proteins |
| pNA | p-nitroaniline |
| PET | Positron-emission tomography |
| PG | Peptidoglycan |
| SDS-PAGE | Sodium dodecyl sulphate-polyacrylamide gel electrophoresis |
| STORM | Stochastic optical reconstruction microscopy |
| SuFEx | Sulfur fluoride exchange |
| TAMRA | Tetramethylrhodamine |
| TDL | TAMRA-d-lysine |
| VF-FL | Vibrioferrin-Fluorescein |
References
- Voices of chemical biology. Nat. Chem. Biol. 2015, 11, 378–379. [CrossRef]
- Ehrlich, P. Beiträge zur Kenntnis der Anilinfärbungen und ihrer Verwendung in der mikroskopischen Technik. Arch. Mikrosk. Anat. 1877, 13, 263–278. (In German) [Google Scholar] [CrossRef]
- Koch, R. Classics in infectious diseases. The etiology of tuberculosis: Robert Koch. Berlin, Germany 1882. Rev. Infect. Dis. 1982, 4, 1270–1274. [Google Scholar] [CrossRef] [PubMed]
- Gram, H.C. Über die isolierte Färbung der Schizomyceten in Schnitt- und Trockenpräparaten. Fortschritte Med. 1884, 2, 185–189. (In German) [Google Scholar]
- Yang, Y.; Xiang, Y.; Xu, M. From red to green: The propidium iodide-permeable membrane of Shewanella decolorationis S12 is repairable. Sci. Rep. 2015, 5, 18583. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Günther, S.; Hübschmann, T.; Wick, L.Y.; Harms, H.; Müller, S. Limits of propidium iodide as a cell viability indicator for environmental bacteria. Cytom. Part A 2007, 71, 592–598. [Google Scholar] [CrossRef]
- Lentz, C.S.; Sheldon, J.R.; Crawford, L.A.; Cooper, R.; Garland, M.; Amieva, M.R.; Weerapana, E.; Skaar, E.P.; Bogyo, M. Identification of a S. aureus virulence factor by activity-based protein profiling (ABPP). Nat. Chem. Biol. 2018, 14, 609–617. [Google Scholar] [CrossRef]
- Hsu, Y.-P.; Hall, E.; Booher, G.; Murphy, B.; Radkov, A.D.; Yablonowski, J.; Mulcahey, C.; Alvarez, L.; Cava, F.; Brun, Y.V.; et al. Fluorogenic d-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays. Nat. Chem. 2019, 11, 335–341. [Google Scholar] [CrossRef]
- Ellison, C.K.; Dalia, T.N.; Ceballos, A.V.; Wang, J.C.-Y.; Biais, N.; Brun, Y.V.; Dalia, A.B. Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat. Microbiol. 2018, 3, 773–780. [Google Scholar] [CrossRef]
- Ofori, L.O.; Withana, N.P.; Prestwood, T.R.; Verdoes, M.; Brady, J.J.; Winslow, M.M.; Sorger, J.; Bogyo, M. Design of protease activated optical contrast agents that exploit a latent lysosomotropic effect for use in fluorescence-guided surgery. ACS Chem. Biol. 2015, 10, 1977–1988. [Google Scholar] [CrossRef]
- Kasperkiewicz, P.; Altman, Y.; D’Angelo, M.; Salvesen, G.S.; Drag, M. Toolbox of fluorescent probes for parallel imaging reveals uneven location of serine proteases in neutrophils. J. Am. Chem. Soc. 2017, 139, 10115–10125. [Google Scholar] [CrossRef]
- Lentz, C.S.; Ordonez, A.A.; Kasperkiewicz, P.; La Greca, F.; O’Donoghue, A.J.; Schulze, C.J.; Powers, J.C.; Craik, C.S.; Drag, M.; Jain, S.K.; et al. Design of selective substrates and activity-based probes for hydrolase important for pathogenesis 1 (HIP1) from Mycobacterium tuberculosis. ACS Infect. Dis. 2016, 2, 807–815. [Google Scholar] [CrossRef]
- White, A.; Koelper, A.; Russell, A.; Larsen, E.M.; Kim, C.; Lavis, L.D.; Hoops, G.; Johnson, R.J. Fluorogenic structure activity library pinpoints molecular variations in substrate specificity of structurally homologous esterases. J. Biol. Chem. 2018, 293, 13851–13862. [Google Scholar] [CrossRef]
- Verdoes, M.; Bender, K.O.; Segal, E.; Van Der Linden, W.A.; Syed, S.; Withana, N.P.; Sanman, L.E.; Bogyo, M. Improved quenched fluorescent probe for imaging of cysteine cathepsin activity. J. Am. Chem. Soc. 2013, 135, 14726–14730. [Google Scholar] [CrossRef]
- Swarts, B.M.; Holsclaw, C.M.; Jewett, J.C.; Alber, M.; Fox, D.M.; Siegrist, M.S.; Leary, J.A.; Kalscheuer, R.; Bertozzi, C.R. Probing the mycobacterial trehalome with bioorthogonal chemistry. J. Am. Chem. Soc. 2012, 134, 16123–16126. [Google Scholar] [CrossRef]
- Siegrist, M.S.; Whiteside, S.; Jewett, J.C.; Aditham, A.; Cava, F.; Bertozzi, C.R. d-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem. Biol. 2013, 8, 500–505. [Google Scholar] [CrossRef]
- Möckl, L.; Pedram, K.; Roy, A.R.; Krishnan, V.; Gustavsson, A.-K.; Dorigo, O.; Bertozzi, C.R.; Moerner, W.E. Quantitative super-resolution microscopy of the mammalian glycocalyx. Dev. Cell 2019, 50, 57–72.e6. [Google Scholar] [CrossRef]
- Barrow, A.S.; Smedley, C.J.; Zheng, Q.; Li, S.; Dong, J.; Moses, J.E. The growing applications of SuFEx click chemistry. Chem. Soc. Rev. 2019, 48, 4731–4758. [Google Scholar] [CrossRef]
- Hatzenpichler, R.; Scheller, S.; Tavormina, P.L.; Babin, B.M.; Tirrell, D.A.; Orphan, V.J. In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ. Microbiol. 2014, 16, 2568–2590. [Google Scholar] [CrossRef]
- Green, O.; Eilon, T.; Hananya, N.; Gutkin, S.; Bauer, C.R.; Shabat, D. Opening a gateway for chemiluminescence cell imaging: Distinctive methodology for design of bright chemiluminescent dioxetane probes. ACS Central Sci. 2017, 3, 349–358. [Google Scholar] [CrossRef]
- Hananya, N.; Green, O.; Blau, R.; Satchi-Fainaro, R.; Shabat, D. A highly efficient chemiluminescence probe for the detection of singlet oxygen in living cells. Angew. Chem. Int. Ed. 2017, 56, 11793–11796. [Google Scholar] [CrossRef]
- Hananya, N.; Shabat, D. Recent advances and challenges in luminescent imaging: Bright outlook for chemiluminescence of dioxetanes in water. ACS Central Sci. 2019, 5, 949–959. [Google Scholar] [CrossRef]
- Wei, L.; Hu, F.; Shen, Y.; Chen, Z.; Yu, Y.; Lin, C.-C.; Wang, M.C.; Min, W. Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat. Methods 2014, 11, 410–412. [Google Scholar] [CrossRef]
- Matanfack, G.A.; Rüger, J.; Stiebing, C.; Schmitt, M.; Popp, J. Imaging the invisible—Bioorthogonal Raman probes for imaging of cells and tissues. J. Biophotonics 2020, 13, e202000129. [Google Scholar] [CrossRef]
- Ferreira, M.S.K.; Hu, H.-Y.; Fetz, V.; Prochnow, H.; Rais, B.; Müller, P.P.; Brönstrup, M. Multivalent siderophore-DOTAM conjugates as theranostics for imaging and treatment of bacterial infections. Angew. Chem. Int. Ed. 2017, 56, 8272–8276. [Google Scholar] [CrossRef]
- Ning, X.; Seo, W.; Lee, S.; Takemiya, K.; Rafi, M.; Feng, X.; Weiss, D.; Wang, X.; Williams, L.; Camp, V.M.; et al. PET imaging of bacterial infections with fluorine-18-labeled maltohexaose. Angew. Chem. Int. Ed. 2014, 53, 14096–14101. [Google Scholar] [CrossRef] [PubMed]
- Takemiya, K.; Ning, X.; Seo, W.; Wang, X.; Mohammad, R.; Joseph, G.; Titterington, J.S.; Kraft, C.S.; Nye, J.A.; Murthy, N.; et al. Novel PET and near infrared imaging probes for the specific detection of bacterial infections associated with cardiac devices. JACC: Cardiovasc. Imaging 2019, 12, 875–886. [Google Scholar] [CrossRef]
- Spahn, C.K.; Glaesmann, M.; Grimm, J.B.; Ayala, A.X.; Lavis, L.D.; Heilemann, M. A toolbox for multiplexed super-resolution imaging of the E. coli nucleoid and membrane using novel PAINT labels. Sci. Rep. 2018, 8, 14768. [Google Scholar] [CrossRef]
- Yim, J.J.; Tholen, M.; Klaassen, A.; Sorger, J.; Bogyo, M. Optimization of a protease activated probe for optical surgical navigation. Mol. Pharm. 2017, 15, 750–758. [Google Scholar] [CrossRef]
- Levine, S.R.; Beatty, K.E. Synthesis of a far-red fluorophore and its use as an esterase probe in living cells. Chem. Commun. 2016, 52, 1835–1838. [Google Scholar] [CrossRef]
- Tallman, K.R.; Levine, S.R.; Beatty, K.E. Profiling esterases in Mycobacterium tuberculosis using far-red fluorogenic substrates. ACS Chem. Biol. 2016, 11, 1810–1815. [Google Scholar] [CrossRef]
- Zheng, Q.; Ayala, A.X.; Chung, I.; Weigel, A.V.; Ranjan, A.; Falco, N.; Grimm, J.B.; Tkachuk, A.N.; Wu, C.; Lippincott-Schwartz, J.; et al. Rational design of fluorogenic and spontaneously blinking labels for super-resolution imaging. ACS Cent. Sci. 2019, 5, 1602–1613. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, X.; Sun, Y.; Yang, X.; Zhang, L.; Zhang, Q.; Hu, Z.-Q.; Hu, H.-Y. Ratiometric fluorescent probes for selective and sensitive visualization of bacterial microenvironment protease activity. Chem. Commun. 2019, 55, 5064–5067. [Google Scholar] [CrossRef]
- Gordon, C.G.; Mackey, J.L.; Jewett, J.C.; Sletten, E.M.; Houk, K.N.; Bertozzi, C.R. Reactivity of biarylazacyclooctynones in copper-free click chemistry. J. Am. Chem. Soc. 2012, 134, 9199–9208. [Google Scholar] [CrossRef]
- Saxon, E. Cell surface engineering by a modified Staudinger reaction. Science 2000, 287, 2007–2010. [Google Scholar] [CrossRef]
- Wu, H.; Devaraj, N.K. Advances in tetrazine bioorthogonal chemistry driven by the synthesis of novel tetrazines and dienophiles. Acc. Chem. Res. 2018, 51, 1249–1259. [Google Scholar] [CrossRef]
- Karver, M.R.; Weissleder, R.; Hilderbrand, S.A. Bioorthogonal reaction pairs enable simultaneous, selective, multi-target imaging. Angew. Chem. Int. Ed. 2011, 51, 920–922. [Google Scholar] [CrossRef]
- Dong, J.; Krasnova, L.; Finn, M.G.; Sharpless, K.B. Sulfur(VI) fluoride exchange (SuFEx): Another good reaction for click chemistry. Angew. Chem. Int. Ed. 2014, 53, 9430–9448. [Google Scholar] [CrossRef]
- Baranczak, A.; Liu, Y.; Connelly, S.; Du, W.-G.H.; Greiner, E.R.; Genereux, J.C.; Wiseman, R.L.; Eisele, Y.S.; Bradbury, N.C.; Dong, J.; et al. A fluorogenic aryl fluorosulfate for intraorganellar transthyretin imaging in living cells and in Caenorhabditis elegans. J. Am. Chem. Soc. 2015, 137, 7404–7414. [Google Scholar] [CrossRef]
- Chen, W.; Dong, J.; Plate, L.; Mortenson, D.E.; Brighty, G.J.; Li, S.; Liu, Y.; Galmozzi, A.; Lee, P.S.; Hulce, J.J.; et al. Arylfluorosulfates inactivate intracellular lipid binding protein(s) through chemoselective SuFEx reaction with a binding site Tyr residue. J. Am. Chem. Soc. 2016, 138, 7353–7364. [Google Scholar] [CrossRef]
- Wang, N.; Yang, B.; Fu, C.; Zhu, H.; Zheng, F.; Kobayashi, T.; Liu, J.; Li, S.; Ma, C.; Wang, P.G.; et al. Genetically Encoding Fluorosulfate-l-tyrosine To React with Lysine, Histidine, and Tyrosine via SuFEx in proteins in vivo. J. Am. Chem. Soc. 2018, 140, 4995–4999. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Yang, X.; Jia, S.; Weeks, A.M.; Hornsby, M.; Lee, P.S.; Nichiporuk, R.V.; Iavarone, A.T.; Wells, J.A.; Toste, F.D.; et al. Redox-based reagents for chemoselective methionine bioconjugation. Science 2017, 355, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Ohata, J.; Krishnamoorthy, L.; Gonzalez, M.A.; Xiao, T.; Iovan, D.A.; Toste, F.D.; Miller, E.W.; Chang, C.J. An activity-based methionine bioconjugation approach to developing proximity-activated imaging reporters. ACS Cent. Sci. 2020, 6, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Roth-Konforti, M.; Green, O.; Hupfeld, M.; Fieseler, L.; Heinrich, N.; Ihssen, J.; Vorberg, R.; Wick, L.; Spitz, U.; Shabat, D. Ultrasensitive detection of Salmonella and Listeria monocytogenes by small-molecule chemiluminescence probes. Angew. Chem. Int. Ed. 2019, 58, 10361–10367. [Google Scholar] [CrossRef] [PubMed]
- Babin, B.M.; Fernandez-Cuervo, G.; Sheng, J.; Green, O.; Ordonez, A.A.; Turner, M.L.; Keller, L.J.; Jain, S.K.; Shabat, D.; Bogyo, M. A chemiluminescent protease probe for rapid, sensitive, and inexpensive detection of live Mycobacterium tuberculosis. bioRxiv 2020. [Google Scholar] [CrossRef]
- Son, S.; Won, M.; Green, O.; Hananya, N.; Sharma, A.; Jeon, Y.; Kwak, J.H.; Sessler, J.L.; Shabat, D.; Kim, J.S. Chemiluminescent probe for the in vitro and in vivo imaging of cancers over-expressing NQO1. Angew. Chem. Int. Ed. 2019, 58, 1739–1743. [Google Scholar] [CrossRef]
- Ye, S.; Hananya, N.; Green, O.; Chen, H.; Zhao, A.Q.; Shen, J.; Shabat, D.; Yang, D. A highly selective and sensitive chemiluminescent probe for real-time monitoring of hydrogen peroxide in cells and animals. Angew. Chem. Int. Ed. Engl. 2020. [Google Scholar] [CrossRef]
- Bruemmer, K.J.; Green, O.; Su, T.A.; Shabat, D.; Chang, C.J. Chemiluminescent probes for activity-based sensing of formaldehyde released from folate degradation in living mice. Angew. Chem. Int. Ed. 2018, 57, 7508–7512. [Google Scholar] [CrossRef]
- Jamieson, L.E.; Greaves, J.; McLellan, J.A.; Munro, K.R.; Tomkinson, N.C.; Chamberlain, L.H.; Faulds, K.; Graham, D. Tracking intracellular uptake and localisation of alkyne tagged fatty acids using Raman spectroscopy. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2018, 197, 30–36. [Google Scholar] [CrossRef]
- El-Mashtoly, S.F.; Petersen, D.; Yosef, H.K.; Mosig, A.; Reinacher-Schick, A.; Kötting, C.; Gerwert, K. Label-free imaging of drug distribution and metabolism in colon cancer cells by Raman microscopy. Analyst 2014, 139, 1155–1161. [Google Scholar] [CrossRef]
- Hu, F.; Chen, Z.; Zhang, L.; Shen, Y.; Wei, L.; Min, W. Vibrational imaging of glucose uptake activity in live cells and tissues by stimulated Raman scattering. Angew. Chem. Int. Ed. 2015, 54, 9821–9825. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.T.; Tipping, W.J.; Jamieson, L.E.; Wetherill, C.; Henley, Z.; Faulds, K.; Graham, D.; Mackay, S.P.; Tomkinson, N.C. A new class of ratiometric small molecule intracellular pH sensors for Raman microscopy. Analyst 2020, 145, 5289–5298. [Google Scholar] [CrossRef]
- Ordonez, A.A.; Weinstein, E.A.; Bambarger, L.E.; Saini, V.; Chang, Y.S.; Demarco, V.P.; Klunk, M.H.; Urbanowski, M.E.; Moulton, K.L.; Murawski, A.M.; et al. A systematic approach for developing bacteria-specific imaging tracers. J. Nucl. Med. 2016, 58, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Auletta, S.; Galli, F.; Lauri, C.; Martinelli, D.; Santino, I.; Signore, A. Imaging bacteria with radiolabelled quinolones, cephalosporins and siderophores for imaging infection: A systematic review. Clin. Transl. Imaging 2016, 4, 229–252. [Google Scholar] [CrossRef]
- Ordonez, A.A.; Jain, S.K. Pathogen-specific bacterial imaging in nuclear medicine. Semin. Nucl. Med. 2018, 48, 182–194. [Google Scholar] [CrossRef]
- Signore, A.; Artiko, V.; Conserva, M.; Ferro-Flores, G.; Welling, M.M.; Jain, S.K.; Hess, S.; Sathekge, M. Imaging bacteria with radiolabelled probes: Is it feasible? J. Clin. Med. 2020, 9, 2372. [Google Scholar] [CrossRef]
- Brock, T.D. Milestones in Microbiology 1546–1940, 2nd ed.; ASM Press: Washington, DC, USA, 1999; pp. 215–218. [Google Scholar]
- Moyes, R.B.; Reynolds, J.; Breakwell, D.P. Differential staining of bacteria: Gram stain. Curr. Protoc. Microbiol. 2009. [Google Scholar] [CrossRef]
- O’Toole, G.A. Classic spotlight: How the gram stain works. J. Bacteriol. 2016, 198, 3128. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-H.C.; Ho, S.-Y.; Chen, P.-L.; Hung, T.-C.; Liang, A.-J.; Kuo, T.-F.; Huang, H.-C.; Wang, T.-S.A. Selective targeting of vibrios by fluorescent siderophore-based probes. ACS Chem. Biol. 2017, 12, 2720–2724. [Google Scholar] [CrossRef]
- Hsu, Y.P.; Rittichier, J.; Kuru, E.; Yablonowski, J.; Pasciak, E.; Tekkam, S.; Hall, E.; Murphy, B.; Lee, T.K.; Garner, E.C.; et al. Full color palette of fluorescent d-amino acids for in situ labeling of bacterial cell walls. Chem. Sci. 2017, 8, 6313–6321. [Google Scholar] [CrossRef] [PubMed]
- Reichart, N.J.; Jay, Z.J.; Krukenberg, V.; Parker, A.E.; Spietz, R.L.; Hatzenpichler, R. Activity-based cell sorting reveals responses of uncultured archaea and bacteria to substrate amendment. ISME J. 2020, 14, 2851–2861. [Google Scholar] [CrossRef] [PubMed]
- Kamariza, M.; Shieh, P.; Ealand, C.S.; Peters, J.S.; Chu, B.; Rodriguez-Rivera, F.P.; Sait, M.R.B.; Treuren, W.V.; Martinson, N.; Kalscheuer, R.; et al. Rapid detection of Mycobacterium tuberculosis in sputum with a solvatochromic trehalose probe. Sci. Transl. Med. 2018, 10, eaam6310. [Google Scholar] [CrossRef] [PubMed]
- Tiyanont, K.; Doan, T.; Lazarus, M.B.; Fang, X.; Rudner, D.Z.; Walker, S. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. USA 2006, 103, 11033–11038. [Google Scholar] [CrossRef]
- Newton, B.A. A fluorescent derivative of polymyxin: Its preparation and use in studying the site of action of the antibiotic. J. Gen. Microbiol. 1955, 12, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Langlois, R.; Cantor, C.R.; Vince, R.; Pestka, S. Interaction between the erythromycin and chloramphenicol binding sites on the Escherichia coli ribosome. Biochemistry 1977, 16, 2349–2356. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kim, I.H.; Roche, E.D.; Beeman, D.; Lynch, A.S.; Ding, C.Z.; Ma, Z. Design, synthesis, and biological evaluation of BODIPY®–erythromycin probes for bacterial ribosomes. Bioorganic Med. Chem. Lett. 2006, 16, 794–797. [Google Scholar] [CrossRef] [PubMed]
- Vince, R.; Weiss, D.; Pestka, S. Binding of N-substituted erythromycylamines to ribosomes. Antimicrob. Agents Chemother. 1976, 9, 131–136. [Google Scholar] [CrossRef]
- Kao, J.C.; Geroski, D.H.; Edelhauser, H.F. Transscleral permeability of fluorescent-labeled antibiotics. J. Ocul. Pharmacol. Ther. 2005, 21, 1–10. [Google Scholar] [CrossRef]
- Matijašić, M.; Kostrun, S.; Nujić, K.; Čužić, S.; Padovan, J.; Kragol, G.; Alihodzic, S.; Mildner, B.; Verbanac, D.; Haber, V.E. Fluorescently labeled macrolides as a tool for monitoring cellular and tissue distribution of azithromycin. Pharmacol. Res. 2012, 66, 332–342. [Google Scholar] [CrossRef]
- Pogliano, J.; Silverman, J.A. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J. Bacteriol. 2012, 194, 4494–4504. [Google Scholar] [CrossRef]
- Phetsang, W.; Blaskovich, M.A.T.; Butler, M.S.; Huang, J.X.; Zuegg, J.; Mamidyala, S.K.; Ramu, S.; Kavanagh, A.M.; Cooper, M.A. An azido-oxazolidinone antibiotic for live bacterial cell imaging and generation of antibiotic variants. Bioorganic Med. Chem. 2014, 22, 4490–4498. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Lee, S.; Wang, Z.; Kim, D.; Stubblefield, B.; Gilbert, E.; Murthy, N. Maltodextrin-based imaging probes detect bacteria in vivo with high sensitivity and specificity. Nat. Mater. 2011, 10, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Faucher, F.; Bennett, J.M.; Bogyo, M.; Lovell, S. Strategies for tuning the selectivity of chemical probes that target serine hydrolases. Cell Chem. Biol. 2020, 27, 937–952. [Google Scholar] [CrossRef]
- Sharifzadeh, S.; Shirley, J.D.; Carlson, E.E. Activity-based protein profiling methods to study bacteria: The power of small-molecule electrophiles. Future HIV-1 Ther. 2018, 420, 23–48. [Google Scholar] [CrossRef]
- Deng, H.; Lei, Q.; Wu, Y.; He, Y.; Li, W. Activity-based protein profiling: Recent advances in medicinal chemistry. Eur. J. Med. Chem. 2020, 191, 112151. [Google Scholar] [CrossRef] [PubMed]
- Niphakis, M.J.; Cravatt, B.F. Enzyme inhibitor discovery by activity-based protein profiling. Annu. Rev. Biochem. 2014, 83, 341–377. [Google Scholar] [CrossRef]
- Van Kasteren, S.I.; Florea, B.I.; Overkleeft, H.S. Activity-based protein profiling: From chemical novelty to biomedical stalwart. Adv. Struct. Saf. Stud. 2016, 1491, 1–8. [Google Scholar]
- Lentz, C.S. What you see is what you get: Activity-based probes in single-cell analysis of enzymatic activities. Biol. Chem. 2020, 401, 233–248. [Google Scholar] [CrossRef]
- Liu, Y.; Patricelli, M.P.; Cravatt, B.F. Activity-based protein profiling: The serine hydrolases. Proc. Natl. Acad. Sci. USA 1999, 96, 14694–14699. [Google Scholar] [CrossRef]
- Cognetta, A.B.; Niphakis, M.J.; Lee, H.-C.; Martini, M.L.; Hulce, J.J.; Cravatt, B.F. Selective N-hydroxyhydantoin carbamate inhibitors of mammalian serine hydrolases. Chem. Biol. 2015, 22, 928–937. [Google Scholar] [CrossRef]
- Böttcher, T.; Sieber, S.A. β-Lactams and β-lactones as activity-based probes in chemical biology. MedChemComm 2012, 3, 408–417. [Google Scholar] [CrossRef]
- Adibekian, A.; Martin, B.R.; Wang, C.; Hsu, K.-L.; Bachovchin, D.A.; Niessen, S.; Hoover, H.; Cravatt, B.F. Click-generated triazole ureas as ultrapotent in vivo—Active serine hydrolase inhibitors. Nat. Chem. Biol. 2011, 7, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Keller, L.J.; Cordasco, E.A.; Bogyo, M.; Lentz, C.S. Fluorescent triazole urea activity-based probes for the single-cell phenotypic characterization of Staphylococcus aureus. Angew. Chem. Int. Ed. 2019, 58, 5643–5647. [Google Scholar] [CrossRef]
- Fellner, M.; Lentz, C.S.; Jamieson, S.A.; Brewster, J.L.; Chen, L.; Bogyo, M.; Mace, P.D. Structural basis for the inhibitor and substrate specificity of the unique Fph serine hydrolases of Staphylococcus aureus. ACS Infect. Dis. 2020, 6, 2771–2782. [Google Scholar] [CrossRef]
- Weerapana, E.; Simon, G.M.; Cravatt, B.F. Disparate proteome reactivity profiles of carbon electrophiles. Nat. Chem. Biol. 2008, 4, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Armstrong, Z.; Schröder, S.P.; De Boer, C.; Artola, M.; Aerts, J.M.; Overkleeft, H.S.; Davies, G.J. An overview of activity-based probes for glycosidases. Curr. Opin. Chem. Biol. 2019, 53, 25–36. [Google Scholar] [CrossRef]
- Palmer, J.T.; Rasnick, D.; Klaus, J.L.; Bromme, D. Vinyl sulfones as mechanism-based cysteine protease inhibitors. J. Med. Chem. 1995, 38, 3193–3196. [Google Scholar] [CrossRef]
- Patricelli, M.P.; Giang, D.K.; Stamp, L.M.; Burbaum, J.J. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics 2001, 1, 1067–1071. [Google Scholar] [CrossRef]
- Keller, L.J.; Lentz, C.S.; Chen, Y.E.; Metivier, R.J.; Weerapana, E.; Fischbach, M.A.; Bogyo, M. Characterization of serine hydrolases across clinical isolates of commensal skin bacteria Staphylococcus epidermidis using activity-based protein profiling. ACS Infect. Dis. 2020, 6, 930–938. [Google Scholar] [CrossRef]
- Kato, D.; Boatright, K.M.; Berger, A.B.; Nazif, T.; Blum, G.; Ryan, C.A.; Chehade, K.A.H.; Salvesen, G.S.; Bogyo, M. Activity-based probes that target diverse cysteine protease families. Nat. Chem. Biol. 2005, 1, 33–38. [Google Scholar] [CrossRef]
- Serim, S.; Haedke, U.; Verhelst, S.H.L. Activity-based probes for the study of proteases: Recent advances and developments. ChemMedChem 2012, 7, 1146–1159. [Google Scholar] [CrossRef]
- Saghatelian, A.; Jessani, N.; Joseph, A.; Humphrey, M.; Cravatt, B.F. Activity-based probes for the proteomic profiling of metalloproteases. Proc. Natl. Acad. Sci. USA 2004, 101, 10000–10005. [Google Scholar] [CrossRef]
- Pace, N.J.; Pimental, D.R.; Weerapana, E. An inhibitor of glutathione s-transferase omega 1 that selectively targets apoptotic cells. Angew. Chem. Int. Ed. 2012, 51, 8365–8368. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.T.; Cravatt, B.F. Chemical proteomic probes for profiling cytochrome p450 activities and drug interactions in vivo. Chem. Biol. 2007, 14, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Sadler, N.C.; Angel, T.E.; Lewis, M.P.; Pederson, L.M.; Chauvigné-Hines, L.M.; Wiedner, S.D.; Zink, E.M.; Smith, R.D.; Wright, A.T. Activity-based protein profiling reveals mitochondrial oxidative enzyme impairment and restoration in diet-induced obese mice. PLoS ONE 2012, 7, e47996. [Google Scholar] [CrossRef]
- Kocaoglu, O.; Calvo, R.A.; Sham, L.-T.; Cozy, L.M.; Lanning, B.R.; Francis, S.; Winkler, M.E.; Kearns, D.B.; Carlson, E.E. Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem. Biol. 2012, 7, 1746–1753. [Google Scholar] [CrossRef] [PubMed]
- Sharifzadeh, S.; Brown, N.W.; Shirley, J.D.; Bruce, K.E.; Winkler, M.E.; Carlson, E.E. Chemical tools for selective activity profiling of bacterial penicillin-binding proteins. Methods Enzymol. 2020, 638, 27–55. [Google Scholar] [CrossRef]
- Hatzios, S.K.; Abel, S.; Martell, J.; Hubbard, T.; Sasabe, J.; Munera, D.; Clark, L.; Bachovchin, D.A.; Qadri, F.; Ryan, E.T.; et al. Chemoproteomic profiling of host and pathogen enzymes active in cholera. Nat. Chem. Biol. 2016, 12, 268–274. [Google Scholar] [CrossRef]
- Tallman, K.R.; Levine, S.R.; Beatty, K.E. Small-molecule probes reveal esterases with persistent activity in dormant and reactivating Mycobacterium tuberculosis. ACS Infect. Dis. 2016, 2, 936–944. [Google Scholar] [CrossRef]
- Lukowski, J.K.; Savas, C.P.; Gehring, A.M.; McKary, M.G.; Adkins, C.T.; Lavis, L.D.; Hoops, G.C.; Johnson, R.J. Distinct substrate selectivity of a metabolic hydrolase from Mycobacterium tuberculosis. Biochemistry 2014, 53, 7386–7395. [Google Scholar] [CrossRef]
- Bassett, B.; Waibel, B.; White, A.; Hansen, H.; Stephens, D.; Koelper, A.; Larsen, E.M.; Kim, C.; Glanzer, A.; Lavis, L.D.; et al. Measuring the global substrate specificity of mycobacterial serine hydrolases using a library of fluorogenic ester substrates. ACS Infect. Dis. 2018, 4, 904–911. [Google Scholar] [CrossRef] [PubMed]
- Rego, E.H.; Audette, R.E.; Rubin, E.J. Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nat. Cell Biol. 2017, 546, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Orman, M.A.; Brynildsen, M.P. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob. Agents Chemother. 2013, 57, 3230–3239. [Google Scholar] [CrossRef]
- Orman, M.A.; Brynildsen, M.P. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat. Commun. 2015, 6, 7983. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Williams, J.; Twieg, R.J.; Rao, J.; Moerner, W.E. Enzymatic activation of nitro-aryl fluorogens in live bacterial cells for enzymatic turnover-activated localization microscopy. Chem. Sci. 2013, 4, 220–225. [Google Scholar] [CrossRef]
- Chyan, W.; Raines, R.T. Enzyme-activated fluorogenic probes for live-cell and in vivo imaging. ACS Chem. Biol. 2018, 13, 1810–1823. [Google Scholar] [CrossRef]
- Siegrist, M.S.; Swarts, B.M.; Fox, D.M.; Lim, S.A.; Bertozzi, C.R. Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol. Rev. 2015, 39, 184–202. [Google Scholar] [CrossRef]
- Dube, D.H.; Champasa, K.; Wang, B. Chemical tools to discover and target bacterial glycoproteins. Chem. Commun. 2011, 47, 87–101. [Google Scholar] [CrossRef]
- Bunschoten, A.; Welling, M.M.; TerMaat, M.F.; Sathekge, M.M.; Van Leeuwen, F.W.B. Development and prospects of dedicated tracers for the molecular imaging of bacterial infections. Bioconjugate Chem. 2013, 24, 1971–1989. [Google Scholar] [CrossRef]
- Grammel, M.; Hang, H.C. Chemical reporters for biological discovery. Nat. Chem. Biol. 2013, 9, 475–484. [Google Scholar] [CrossRef]
- Kuru, E.; Hughes, H.V.; Brown, P.J.; Hall, E.; Tekkam, S.; Cava, F.; De Pedro, M.A.; Brun, Y.V.; VanNieuwenhze, M.S. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescentd-amino acids. Angew. Chem. Int. Ed. 2012, 51, 12519–12523. [Google Scholar] [CrossRef]
- Backus, K.M.; Boshoff, H.I.; Barry, C.S.; Boutureira, O.; Patel, M.K.; D’Hooge, F.; Lee, S.S.; Via, L.E.; Tahlan, K.; Barry, C.E.; et al. Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nat. Chem. Biol. 2011, 7, 228–235. [Google Scholar] [CrossRef]
- Prindle, A.; Liu, J.; Asally, M.; Ly, S.; Garcia-Ojalvo, J.; Süel, G.M. Ion channels enable electrical communication in bacterial communities. Nat. Cell Biol. 2015, 527, 59–63. [Google Scholar] [CrossRef]
- Cernak, T.; Dykstra, K.D.; Tyagarajan, S.; Vachal, P.; Krska, S.W. The medicinal chemist’s toolbox for late stage functionalization of drug-like molecules. Chem. Soc. Rev. 2016, 45, 546–576. [Google Scholar] [CrossRef]
- DiRocco, D.A.; Dykstra, K.; Krska, S.; Vachal, P.; Conway, D.V.; Tudge, M. Late-stage functionalization of biologically active heterocycles through photoredox catalysis. Angew. Chem. Int. Ed. 2014, 53, 4802–4806. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Engle, K.M.; Wang, N.-H.; Yu, J.-Q. ChemInform abstract: Palladium(ii)-catalyzed c-h activation/c-c cross-coupling reactions: Versatility and practicality. Angew. Chem. Int. Ed. 2009, 40, 5094–5115. [Google Scholar] [CrossRef] [PubMed]
- De Moliner, F.; Kielland, N.; Lavilla, R.; Vendrell, M. Modern synthetic avenues for the preparation of functional fluorophores. Angew. Chem. Int. Ed. 2017, 56, 3758–3769. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.; Luo, T.; Lei, X. Late-stage diversification of natural products. ACS Cent. Sci. 2020, 6, 622–635. [Google Scholar] [CrossRef]
- Hesp, K.D.; Xiao, J.; West, G.M. Late-stage synthesis and application of photoreactive probes derived from direct benzoylation of heteroaromatic C–H bonds. Org. Biomol. Chem. 2020, 18, 3669–3673. [Google Scholar] [CrossRef] [PubMed]
- Fadeyi, O.O.; Parikh, M.D.; Chen, M.Z.; Kyne, R.E.; Taylor, A.P.; O’Doherty, I.; Kaiser, S.E.; Barbas, S.; Niessen, S.; Shi, M.; et al. Chemoselective preparation of clickable aryl sulfonyl fluoride monomers: A toolbox of highly functionalized intermediates for chemical biology probe synthesis. ChemBioChem 2016, 17, 1925–1930. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, H.; Li, S.; Bare, G.A.L.; Chen, X.; Wang, C.; Moses, J.E.; Wu, P.; Sharpless, K.B. Biocompatible SuFEx click chemistry: Thionyl tetrafluoride (sof 4)-derived connective hubs for bioconjugation to dna and proteins. Angew. Chem. Int. Ed. 2019, 58, 8029–8033. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, J.; Li, S.; Li, G.; Sharpless, K.B.; Wu, P. SuFEx click chemistry enabled late-stage drug functionalization. J. Am. Chem. Soc. 2018, 140, 2919–2925. [Google Scholar] [CrossRef] [PubMed]
- Dumont, E.; Vergalli, J.; Pajovic, J.; Bhamidimarri, S.P.; Morante, K.; Wang, J.; Lubriks, D.; Suna, E.; Stavenger, R.A.; Winterhalter, M.; et al. Mechanistic aspects of maltotriose-conjugate translocation to the Gram-negative bacteria cytoplasm. Life Sci. Alliance 2018, 2, e201800242. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.S.; Okuda, M.; Cyrenne, M.; Bourge, M.; Heck, M.-P.; Yoshizawa, S.; Fourmy, D. Fluorescent aminoglycoside antibiotics and methods for accurately monitoring uptake by bacteria. ACS Infect. Dis. 2020, 6, 1008–1017. [Google Scholar] [CrossRef]
- Stone, M.R.L.; Masi, M.; Phetsang, W.; Pagès, J.-M.; Cooper, M.A.; Blaskovich, M.A.T. Fluoroquinolone-derived fluorescent probes for studies of bacterial penetration and efflux. MedChemComm 2019, 10, 901–906. [Google Scholar] [CrossRef]
- Daniel, R.A.; Errington, J. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell 2003, 113, 767–776. [Google Scholar] [CrossRef]
- Gee, K.R.; Kang, H.C.; Meier, T.I.; Zhao, G.; Blaszcak, L.C. Fluorescent bocillins: Synthesis and application in the detection of penicillin-binding proteins. Electrophoresis 2001, 22, 960–965. [Google Scholar] [CrossRef]
- Zhao, G.; Meier, T.I.; Kahl, S.D.; Gee, K.R.; Blaszczak, L.C. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 1999, 43, 1124–1128. [Google Scholar] [CrossRef]
- David, B.; Duchêne, M.-C.; Haustenne, G.L.; Pérez-Núñez, D.; Chapot-Chartier, M.-P.; De Bolle, X.; Guédon, E.; Hols, P.; Hallet, B. PBP2b plays a key role in both peripheral growth and septum positioning in Lactococcus lactis. PLoS ONE 2018, 13, e0198014. [Google Scholar] [CrossRef]
- Sharifzadeh, S.; Boersma, M.J.; Kocaoglu, O.; Shokri, A.; Brown, C.L.; Shirley, J.D.; Winkler, M.E.; Carlson, E.E. Novel electrophilic scaffold for imaging of essential penicillin-binding proteins in Streptococcus pneumoniae. ACS Chem. Biol. 2017, 12, 2849–2857. [Google Scholar] [CrossRef]
- Sharifzadeh, S.; Dempwolff, F.; Kearns, D.B.; Carlson, E.E. Harnessing beta-Lactam antibiotics for illumination of the activity of penicillin-binding proteins in Bacillus subtilis. ACS Chem. Biol. 2020, 15, 1242–1251. [Google Scholar] [CrossRef] [PubMed]
- Whidbey, C.; Sadler, N.C.; Nair, R.N.; Volk, R.; DeLeon, A.J.; Bramer, L.M.; Fansler, S.J.; Hansen, J.R.; Shukla, A.K.; Jansson, J.K.; et al. A probe-enabled approach for the selective isolation and characterization of functionally active subpopulations in the gut microbiome. J. Am. Chem. Soc. 2018, 141, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.W.; Chamessian, A.G.; McEnaney, P.J.; Murelli, R.P.; Kazmiercak, B.I.; Spiegel, D.A.; Kazmierczak, B.I. A biosynthetic strategy for re-engineering the Staphylococcus aureus cell wall with non-native small molecules. ACS Chem. Biol. 2010, 5, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Maňásková, S.H.; Nazmi, K.; Van Belkum, A.; Bikker, F.J.; Van Wamel, W.J.B.; Veerman, E.C.I. Synthetic LPETG-containing peptide incorporation in the Staphylococcus aureus cell-wall in a sortase a- and growth phase-dependent manner. PLoS ONE 2014, 9, e89260. [Google Scholar] [CrossRef]
- Xie, H.; Mire, J.; Kong, Y.; Chang, M.; Hassounah, H.A.; Thornton, C.N.; Sacchettini, J.C.; Cirillo, J.D.; Rao, J. Rapid point-of-care detection of the tuberculosis pathogen using a BlaC-specific fluorogenic probe. Nat. Chem. 2012, 4, 802–809. [Google Scholar] [CrossRef]
- Cheng, Y.; Xie, H.; Sule, P.; Hassounah, H.; Graviss, E.A.; Kong, Y.; Cirillo, J.D.; Rao, J. Fluorogenic probes with substitutions at the 2 and 7 positions of cephalosporin are highly blac-specific for rapid Mycobacterium tuberculosis detection. Angew. Chem. Int. Ed. 2014, 53, 9360–9364. [Google Scholar] [CrossRef]
- Hernandez, F.J.; Huang, L.; Olson, M.E.; Powers, K.M.; Hernandez, L.I.; Meyerholz, D.K.; Thedens, D.R.; Behlke, M.A.; Horswill, A.R.; McNamara, J.O. Noninvasive imaging of Staphylococcus aureus infections with a nuclease-activated probe. Nat. Med. 2014, 20, 301–306. [Google Scholar] [CrossRef]
- Di Guilmi, A.; Bonnet, J.; Peiβert, S.; Durmort, C.; Gallet, B.; Vernet, T.; Gisch, N.; Wong, Y.-S. Specific and spatial labeling of choline-containing teichoic acids in Streptococcus pneumoniae by click chemistry. Chem. Commun. 2017, 53, 10572–10575. [Google Scholar] [CrossRef]
- Kuru, E.; Tekkam, S.; Hall, E.K.; Brun, Y.V.; Van Nieuwenhze, M.S. Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat. Protoc. 2014, 10, 33–52. [Google Scholar] [CrossRef]
- Liechti, G.W.; Kuru, E.; Hall, E.; Kalinda, A.; Brun, Y.V.; VanNieuwenhze, M.S.; Maurelli, A.T. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nat. Cell Biol. 2013, 506, 507–510. [Google Scholar] [CrossRef]
- Kuru, E.; Radkov, A.; Meng, X.; Egan, A.; Alvarez, L.; Dowson, A.; Booher, G.; Breukink, E.; Roper, D.I.; Cava, F.; et al. Mechanisms of incorporation for d-amino acid probes that target peptidoglycan biosynthesis. ACS Chem. Biol. 2019, 14, 2745–2756. [Google Scholar] [CrossRef] [PubMed]
- Dumont, A.; Malleron, A.; Awwad, M.; Dukan, S.; Vauzeilles, B. Click-mediated labeling of bacterial membranes through metabolic modification of the lipopolysaccharide inner core. Angew. Chem. Int. Ed. 2012, 51, 3143–3146. [Google Scholar] [CrossRef] [PubMed]
- Hatzenpichler, R.; Connon, S.A.; Goudeau, D.; Malmstrom, R.R.; Woyke, T.; Orphan, V.J. Visualizing in situ translational activity for identifying and sorting slow-growing archaeal–bacterial consortia. Proc. Natl. Acad. Sci. USA 2016, 113, E4069–E4078. [Google Scholar] [CrossRef] [PubMed]
- Hatzenpichler, R.; Krukenberg, V.; Spietz, R.L.; Jay, Z.J. Next-generation physiology approaches to study microbiome function at single cell level. Nat. Rev. Genet. 2020, 18, 241–256. [Google Scholar] [CrossRef]
- Van Oosten, M.; Schäfer, T.; Gazendam, J.A.C.; Ohlsen, K.; Tsompanidou, E.; De Goffau, M.C.; Harmsen, H.J.M.; Crane, L.M.A.; Lim, E.; Francis, K.P.; et al. Real-time in vivo imaging of invasive- and biomaterial-associated bacterial infections using fluorescently labelled vancomycin. Nat. Commun. 2013, 4, 2584. [Google Scholar] [CrossRef]
- Turner, R.D.; Vollmer, W.; Foster, S.J. Different walls for rods and balls: The diversity of peptidoglycan. Mol. Microbiol. 2014, 91, 862–874. [Google Scholar] [CrossRef]
- Cava, F.; De Pedro, M.A. Peptidoglycan plasticity in bacteria: Emerging variability of the murein sacculus and their associated biological functions. Curr. Opin. Microbiol. 2014, 18, 46–53. [Google Scholar] [CrossRef]
- Teo, A.C.K.; Roper, D.I. Core Steps of membrane-bound peptidoglycan biosynthesis: Recent advances, insight and opportunities. Antibiotics 2015, 4, 495–520. [Google Scholar] [CrossRef]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef]
- Egan, A.J.F.; Errington, J.; Vollmer, W. Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Genet. 2020, 18, 446–460. [Google Scholar] [CrossRef]
- Morè, N.; Martorana, A.M.; Biboy, J.; Otten, C.; Winkle, M.; Serrano, C.K.G.; Silva, A.M.; Atkinson, L.; Yau, H.; Breukink, E.; et al. Peptidoglycan remodeling enables Escherichia coli to survive severe outer membrane assembly defect. mBio 2019, 10, e02729-18. [Google Scholar] [CrossRef] [PubMed]
- Baquero, M.R.; Bouzon, M.; Quintela, J.C.; Ayala, J.A.; Moreno, F. dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with DD-carboxypeptidase activity. J. Bacteriol. 1996, 178, 7106–7111. [Google Scholar] [CrossRef]
- Nelson, D.E.; Young, K.D. Contributions of PBP 5 and dd-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 2001, 183, 3055–3064. [Google Scholar] [CrossRef]
- Hugonnet, J.-E.; Mengin-Lecreulx, D.; Monton, A.; Blaauwen, T.D.; Carbonnelle, E.; Veckerlé, C.; Yves, V.B.; Van Nieuwenhze, M.; Bouchier, C.; Tu, K.; et al. Factors essential for L,D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. eLife 2016, 5, e19469. [Google Scholar] [CrossRef] [PubMed]
- Mainardi, J.L.; Fourgeaud, M.; Hugonnet, J.E.; Dubost, L.; Brouard, J.P.; Ouazzani, J.; Rice, L.B.; Gutmann, L.; Arthur, M. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J. Biol. Chem. 2005, 280, 38146–38152. [Google Scholar] [CrossRef] [PubMed]
- Radkov, A.D.; Hsu, Y.-P.; Booher, G.; VanNieuwenhze, M.S. Imaging bacterial cell wall biosynthesis. Annu. Rev. Biochem. 2018, 87, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Soon, R.L.; Velkov, T.; Chiu, F.; Thompson, P.E.; Kancharla, R.; Roberts, K.; Larson, I.; Nation, R.L.; Li, J. Design, synthesis, and evaluation of a new fluorescent probe for measuring polymyxin–lipopolysaccharide binding interactions. Anal. Biochem. 2011, 409, 273–283. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, Z.; Yang, J.; Peng, K.; Ab Baker, M.; Bai, F.; Lo, C.-J. Length-dependent flagellar growth of Vibrio alginolyticus revealed by real time fluorescent imaging. eLife 2017, 6, e22140. [Google Scholar] [CrossRef]
- Ellison, C.K.; Kan, J.; Dillard, R.S.; Kysela, D.T.; Ducret, A.; Berne, C.; Hampton, C.M.; Ke, Z.; Wright, E.R.; Biais, N.; et al. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 2017, 358, 535–538. [Google Scholar] [CrossRef]
- Skerker, J.M.; Berg, H.C. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 2001, 98, 6901–6904. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhao, Y.; Zhuang, X.-Y.; Lo, W.-C.; Baker, M.A.B.; Lo, C.-J.; Bai, F. Frequent pauses in Escherichia coli flagella elongation revealed by single cell real-time fluorescence imaging. Nat. Commun. 2018, 9, 1885. [Google Scholar] [CrossRef]
- Maitra, A.; Munshi, T.; Healy, J.; Martin, L.T.; Vollmer, W.; Keep, N.H.; Bhakta, S. Cell wall peptidoglycan in Mycobacterium tuberculosis: An Achilles’ heel for the TB-causing pathogen. FEMS Microbiol. Rev. 2019, 43, 548–575. [Google Scholar] [CrossRef] [PubMed]
- Nobre, A.; Alarico, S.; Maranha, A.; Mendes, V.; Empadinhas, N. The molecular biology of mycobacterial trehalose in the quest for advanced tuberculosis therapies. Microbiology 2014, 160, 1547–1570. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef]
- Swaney, S.M.; Aoki, H.; Ganoza, M.C.; Shinabarger, D.L. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 1998, 42, 3251–3255. [Google Scholar] [CrossRef] [PubMed]
- Phetsang, W.; Pelingon, R.; Butler, M.S.; Kc, S.; Pitt, M.E.; Kaeslin, G.; Cooper, M.A.; Blaskovich, M.A.T. Fluorescent trimethoprim conjugate probes to assess drug accumulation in wild type and mutant Escherichia coli. ACS Infect. Dis. 2016, 2, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Lin, Y.-M.; Miller, P.A.; Möllmann, U.; Boggess, W.C.; Miller, M.J. Siderophore conjugates of daptomycin are potent inhibitors of carbapenem resistant strains of Acinetobacter baumannii. ACS Infect. Dis. 2018, 4, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Klahn, P.; Brönstrup, M. Bifunctional antimicrobial conjugates and hybrid antimicrobials. Nat. Prod. Rep. 2017, 34, 832–885. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Yamawaki, K. Cefiderocol: Discovery, chemistry, and in vivo profiles of a novel siderophore cephalosporin. Clin. Infect. Dis. 2019, 69, S538–S543. [Google Scholar] [CrossRef]
- Chakraborty, R.; Storey, E.; Van Der Helm, D. Molecular mechanism of ferricsiderophore passage through the outer membrane receptor proteins of Escherichia coli. BioMetals 2006, 20, 263–274. [Google Scholar] [CrossRef]
- Kiedrowski, M.R.; Crosby, H.A.; Hernandez, F.J.; Malone, C.L.; Ii, J.O.M.; Horswill, A.R. Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PLoS ONE 2014, 9, e95574. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Na Goh, B.; Teh, W.K.; Jiang, Z.; Goh, J.P.Z.; Goh, A.; Wu, G.; Hoon, S.S.; Raida, M.; Camattari, A.; et al. Skin commensal malassezia globosa secreted protease attenuates Staphylococcus aureus biofilm formation. J. Investig. Dermatol. 2018, 138, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Müsken, M.; Klimmek, K.; Sauer-Heilborn, A.; Donnert, M.; Sedlacek, L.; Suerbaum, S.; Häussler, S. Towards individualized diagnostics of biofilm-associated infections: A case study. NPJ Biofilms Microbiomes 2017, 3, 22. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Genet. 2019, 17, 441–448. [Google Scholar] [CrossRef]
- Imdahl, F.; Vafadarnejad, E.; Homberger, C.; Saliba, A.-E.; Vogel, J. Single-cell RNA-sequencing reports growth-condition-specific global transcriptomes of individual bacteria. Nat. Microbiol. 2020, 1–5. [Google Scholar] [CrossRef]
- Park, L.M.; Lannigan, J.; Jaimes, M.C. OMIP-069: Forty-color full spectrum flow cytometry panel for deep immunophenotyping of major cell subsets in human peripheral blood. Cytometry A 2020, 97, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Poreba, M.; Groborz, K.; Rut, W.; Pore, M.; Snipas, S.J.; Vizovisek, M.; Turk, B.; Kuhn, P.; Drag, M.; Salvesen, G.S. Multiplexed probing of proteolytic enzymes using mass cytometry-compatible activity-based probes. J. Am. Chem. Soc. 2020. [Google Scholar] [CrossRef] [PubMed]
| Probe Name | Probe Type | Targeted Species | Molecular Target | Detection Tag(s) | Application | References |
|---|---|---|---|---|---|---|
| Vibrioferrin-FL | Non-covalent targeted conjugate | V. parahaemolyticus, V. cholerae, and V. vulnificus | Siderophore uptake pathway | Fluorophore/Bio-orthogonal handle | Visualization of vibrioferrin uptake and selective detection of Vibrios under iron-limited conditions | [60] |
| DOTAM–FL | Non-covalent targeted conjugate | P. aeruginosa and E. coli | Siderophore uptake pathway | Fluorophore | Visualization of iron transport and detection of bacterial infections | [25] |
| MDPs | Non-covalent targeted conjugate | E. coli, P. aeruginosa, B. subtilis, and S. aureus | Maltodextrin uptake pathway | Fluorophore/Radiolabel | Visualization of maltodextrin uptake and high-sensitivity detection of bacteria in vivo | [27,73,124] |
| Neo–Cy5 | Non-covalent targeted conjugate | P. aeruginosa, A. baumannii, K. pneumoniae, S. typhimurium, and S. aureus | Aminoglycoside antibiotics uptake pathway | Fluorophore/Bio-orthogonal handle | Visualization of aminoglycoside uptake and mode of action | [125] |
| Cipro-azide | Non-covalent targeted conjugate | E. coli and S. aureus | Antibiotics uptake pathway | Fluorophore/Bio-orthogonal handle | Understanding the bacterial penetration and efflux pump mechanisms | [126] |
| Van-FL | Non-covalent targeted conjugate | B. subtilis, S. pneumoniae, S. coelicolor and C. glutamicum | PG stem peptide (D-Ala-D-ALA) | Fluorophore | Visualize nascent PG biosynthesis in live cells | [64,127] |
| BOCILLIN-FL | Activity-based probe | E. coli, P. aeruginosa, and S. pneumoniae | Active PBPs (broad spectrum) | Fluorophore | Broad-spectrum detection of PBP activities in live cells. | [128,129,130] |
| Ceph C-T | Activity-based probe | B. subtilis and S. pneumoniae | PBPs 1a/1b, 2b, 2c, and 4 (B. subtilis) and PBP1b and 3b (S. pneumoniae) | Fluorophore | Visualize involvement of different PBP subsets in live cells | [97] |
| β-lactone probes | Activity-based probe | S. pneumoniae | PBP1a, PBP1b, PBP2x, and PBP2a | Fluorophore | Visualize the catalytic activity of PBP subsets in live cells | [131] |
| Meropenem-derived probe MEM-FL | Activity-based probe | B. subtilis | PBP3 and 5 | Fluorophore/Bio-orthogonal handle | Visualize PBP3 activity in single cells during cell division | [132] |
| Fluoro-phosphonates (FP-TMR) | Activity-based probe | S. aureus | Serine hydrolases | Fluorophore/Biotin | Identification of serine hydrolase activities | [7,89] |
| JCP251-bT | Activity-based probe | S. aureus | Fluorophos-phonate-binding serine hydrolase B (FphB) | Fluorophore | Visualize subcellular FphB localization and distribution across cell population | [7] |
| Triazole urea probes | Activity-based probe | S. aureus | Fluorophos-phonate-binding serine hydrolases and lipases | Fluorophore/Bio-orthogonal handle | Assessment of specific cellular serine hydrolase activity levels | [84] |
| GlcA-ABP | Activity-based probe | Mouse gastrointestinal microbes | β-glucuronidase | Fluorophore/Bio-orthogonal handle | Detection, isolation and identification of microbial subpopulations in the gut microbiome | [133] |
| CSL174 | Substrate probe | M. tuberculosis | Hydrolase-important for pathogenesis 1 (Hip1) | Fluorophore | Specific detection of Hip1 protease activity | [12] |
| FLASH | Substrate probe | M. tuberculosis | Hydrolase-important for pathogenesis 1 (Hip1) | Chemiluminescent | Detection of live M. tuberculosis | [45] |
| Calcein- AM | Substrate probe | M. tuberculosis and Mycobacterium smegmatis | Esterases | Fluorophore | Single-cell assessment of esterase activity and probe uptake | [103] |
| Redox Sensor Green (RSG) | Substrate probe | E. coli | Bacterial reductase | Fluorophore | Assessment of cellular redox activity | [104,105] |
| LPETG-derived peptides | Metabolic labeling | S. aureus | Sortase A–dependent cell wall anchoring | Fluorophore/Bio-orthogonal handle | Imaging of cellular Sortase A levels, cell wall re-engineering | [134,135] |
| CLSP and CLLP | Substrate probe | Salmonella spp. and L. monocytogenes | Esterase and phosphatidylinositol-specific phospholipase C (PI-PLC) | Luminophore | Selective detection of Salmonella spp. and L.monocytogenes from food samples | [44] |
| Nitro-aryl fluorogen | Substrate probe | B. subtilis | Nitroreductase activity | Fluorophore | Visualization subcellular localization of nitroreductases | [106] |
| CDG-OMe | Substrate probe | M. tuberculosis | β-lactamase (Bla) C | Fluorophore | Detection of live M. tuberculosis | [136,137] |
| Cy5. 5-TT | Substrate probe | S. aureus | Micrococcal nuclease (MN) | Quenched fluorophore | Noninvasive detection of S. aureus infections in mouse pyomyositis model | [138] |
| D-alanine analogues | Metabolic labeling | L. monocytogenes | Peptidoglycan | Bio-orthogonal handle | Visualization of PG dynamics | [16] |
| Propargyl-choline | Metabolic labeling | S. pneumoniae | Teichoic acid | Bio-orthogonal handle | Visualization of pneumococcal teichoic acid biosynthesis. | [139] |
| FDAA | Metabolic labeling | B. subtilis, E. coli, S. aureus, S. pneumoniae, Agrobacterium tumefaciens and C. crescentus | PG stem peptide | Fluorophore/Bio-orthogonal handle | Visualization of PG biosynthesis and illustration of bacterial growth and division | [61,112,140,141,142] |
| KDO | Metabolic labeling | E. coli and Salmonella typhimurium | LPS | Fluorophore/Bio-orthogonal handle | Visualization of LPS structure and location | [143] |
| Homopropargylglycine (HPG) | Metabolic labeling | Sulfate-reducing bacteria, uncultured microbes | Protein synthesis | Bio-orthogonal handle | Single-cell assessment of translational activity | [144,145] |
| L-azidohomo-alanine (AHA) | Metabolic labeling | E. coli, single environmental bacterial strains and complex samples | Protein synthesis | Bio-orthogonal handle | Single-cell assessment of translational activity | [19] |
| Azido-modified trehalose | Metabolic labeling | M. tuberculosis | Cell surface glycolipids | Bio-orthogonal handle | Detection and visualization of cell-surface glycolipids | [15] |
| Trehalose analogs | Metabolic labeling | Mycobacterium spp | Myco-membrane | Fluorophore/Bio-orthogonal handle | Determination of the envelope structure of Mycobacterium | [113] |
| Thioflavin T (ThT) | Non-specific fluorescent dye | B. subtilis | Membrane | Fluorophore | Quantification of membrane potential | [114] |
| DMN-Tre | Environmental sensor/Metabolic labeling | M. tuberculosis | Myco-membrane | Fluorophore | Detection of M. tuberculosis | [63] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hira, J.; Uddin, M.J.; Haugland, M.M.; Lentz, C.S. From Differential Stains to Next Generation Physiology: Chemical Probes to Visualize Bacterial Cell Structure and Physiology. Molecules 2020, 25, 4949. https://doi.org/10.3390/molecules25214949
Hira J, Uddin MJ, Haugland MM, Lentz CS. From Differential Stains to Next Generation Physiology: Chemical Probes to Visualize Bacterial Cell Structure and Physiology. Molecules. 2020; 25(21):4949. https://doi.org/10.3390/molecules25214949
Chicago/Turabian StyleHira, Jonathan, Md. Jalal Uddin, Marius M. Haugland, and Christian S. Lentz. 2020. "From Differential Stains to Next Generation Physiology: Chemical Probes to Visualize Bacterial Cell Structure and Physiology" Molecules 25, no. 21: 4949. https://doi.org/10.3390/molecules25214949
APA StyleHira, J., Uddin, M. J., Haugland, M. M., & Lentz, C. S. (2020). From Differential Stains to Next Generation Physiology: Chemical Probes to Visualize Bacterial Cell Structure and Physiology. Molecules, 25(21), 4949. https://doi.org/10.3390/molecules25214949






