Hetiamacin E and F, New Amicoumacin Antibiotics from Bacillus subtilis PJS Using MS/MS-Based Molecular Networking
Abstract
1. Introduction
2. Results
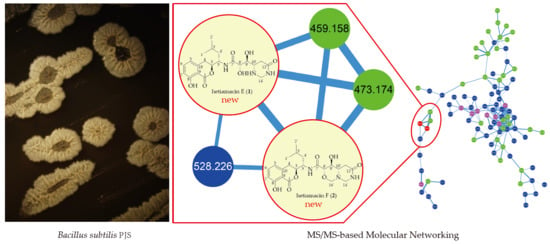
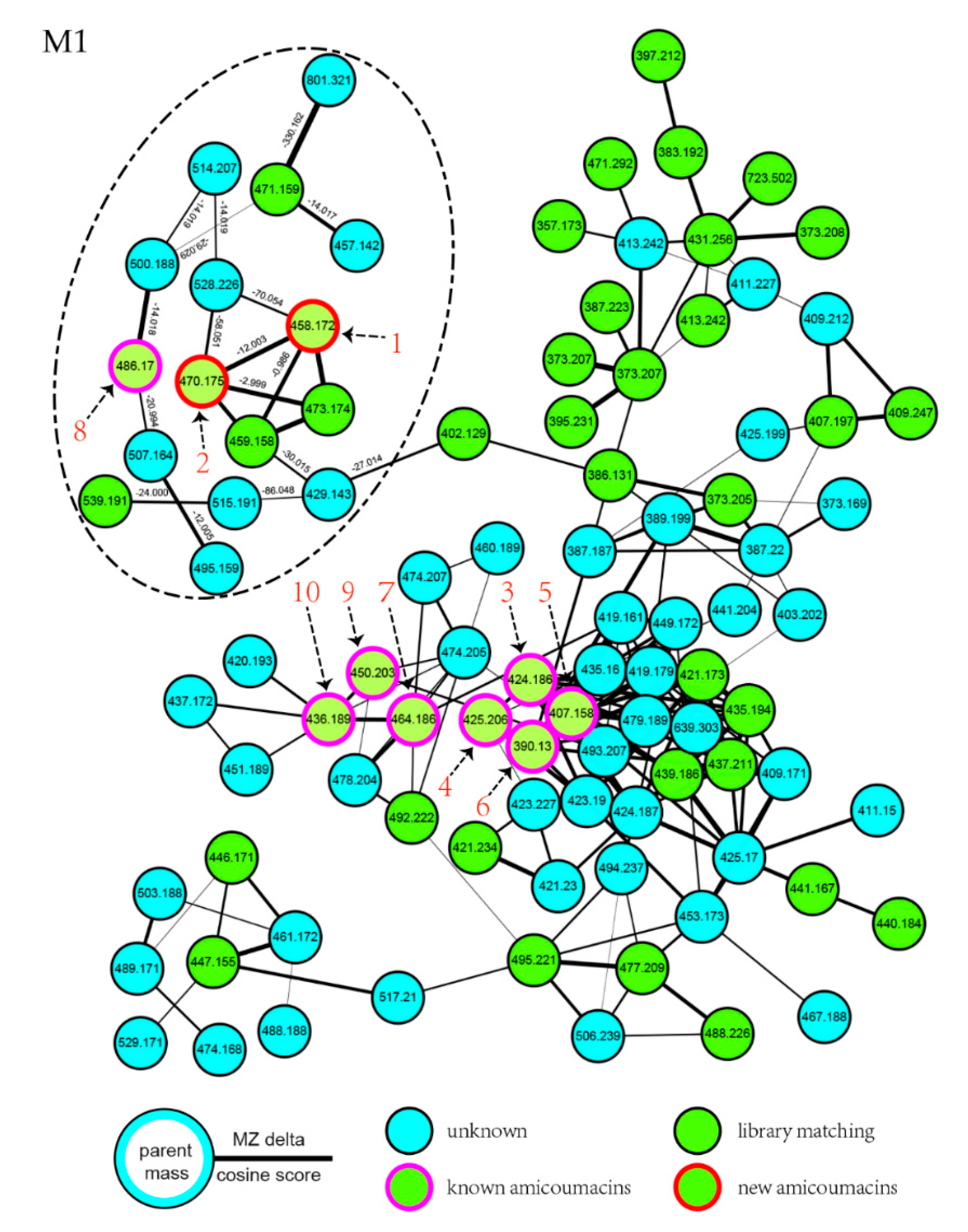
2.1. The Profile of Amicoumacins in Strain PJS Using MS/MS-Based Molecular Networking
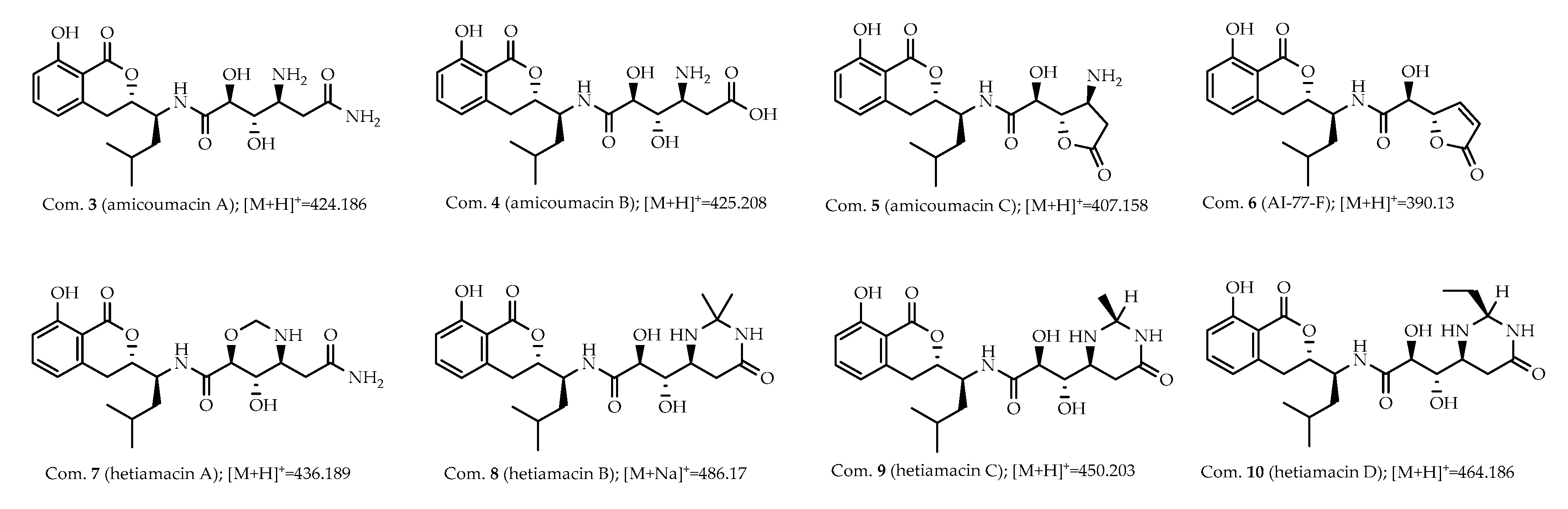
2.2. Structure-Based Purification of Molecules Detected by Molecular Networking
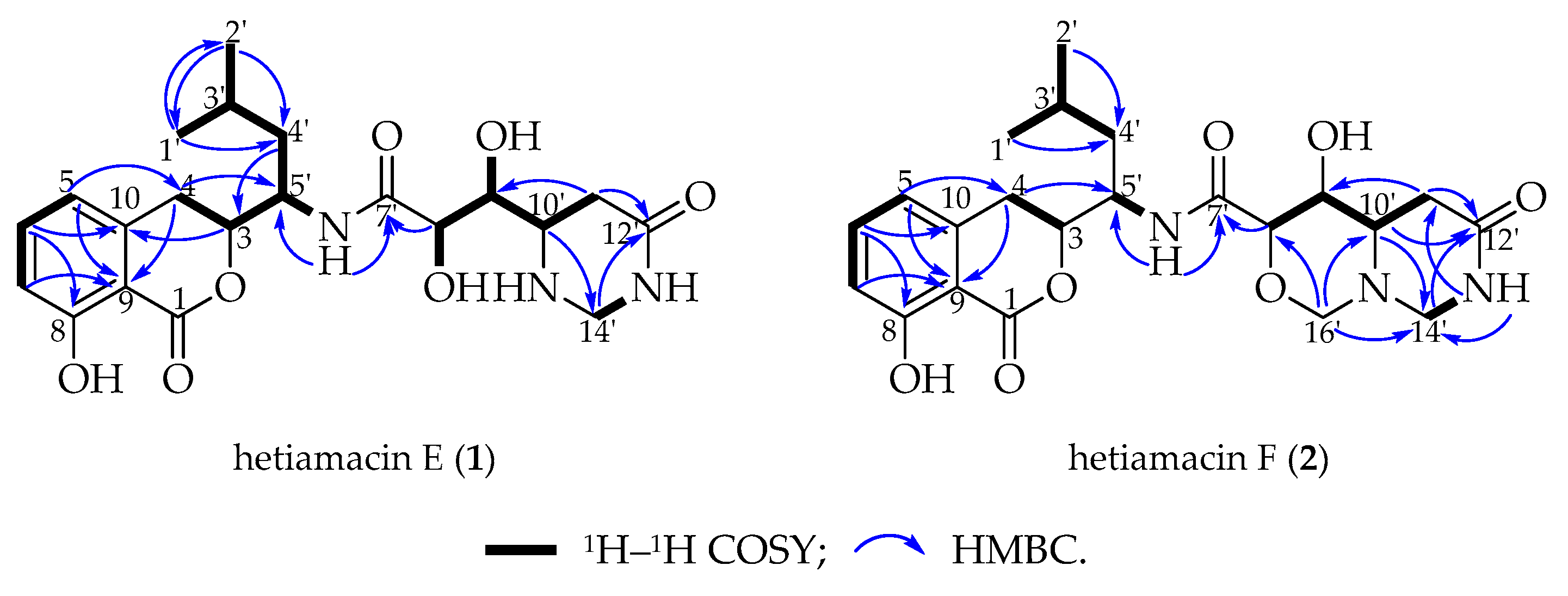
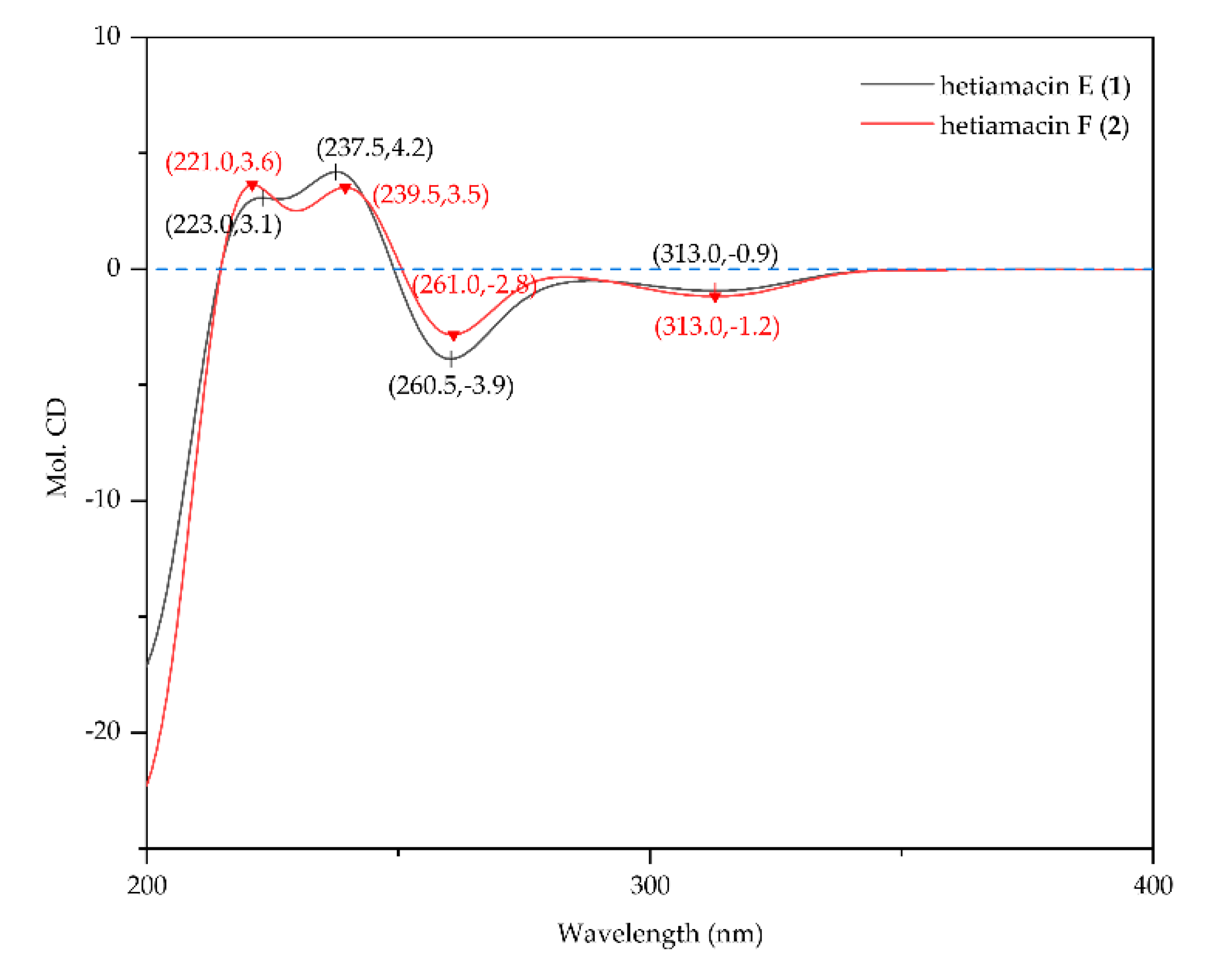
2.3. Structural Elucidation of Compounds
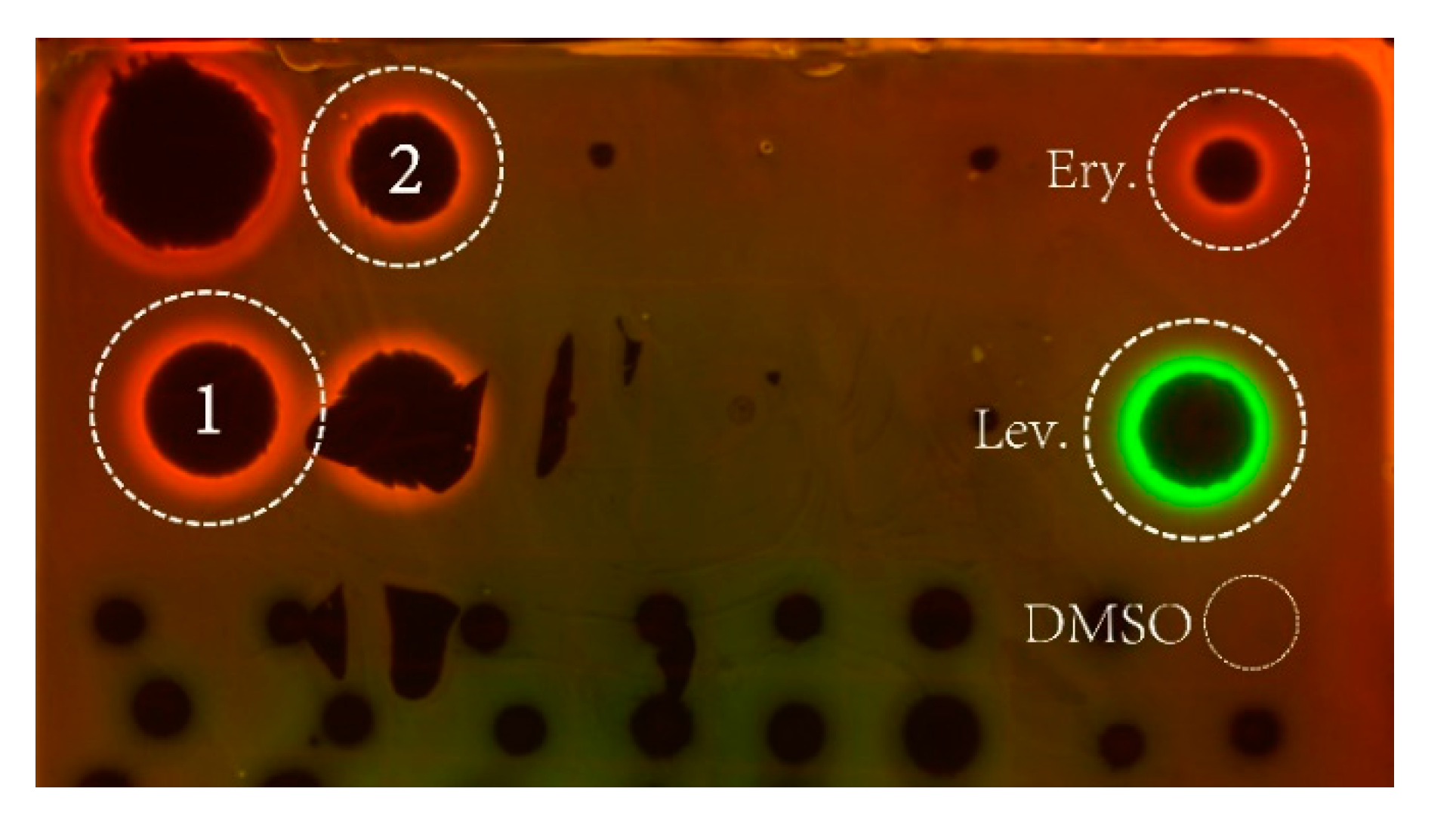
2.4. Antibacterial Activity
2.5. Mechanism of Action Determination
3. Discussion
4. Materials and Methods
4.1. General Procedures
4.2. Small-Scale Fermentation and Preliminary UPLC-UV Analysis
4.3. Mass Spectrometry Analysis
4.4. Molecular Networking
4.5. Large-Scale Fermentation, Extraction, and Targeted Isolation
4.6. Antibacterial Assay
4.7. Mechanism of Action Determination
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baker, D.D.; Chu, M.; Oza, U.; Rajgarhia, V. The value of natural products to future pharmaceutical discovery. Nat. Prod. Rep. 2007, 24, 1225–1244. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Fischbach, M.A. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015, 11, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H. Computational genomics of specialized metabolism: From natural product discovery to microbiome ecology. mSystems 2018, 3. [Google Scholar] [CrossRef]
- Quinn, R.A.; Nothias, L.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular networking as a drug discovery, drug metabolism, and precision medicine strategy. Trends Pharmacol. Sci. 2017, 38, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef]
- Zhu, G.; Hou, C.; Yuan, W.; Wang, Z.; Zhang, J.; Jiang, L.; Karthik, L.; Li, B.; Ren, B.; Lv, K.; et al. Molecular networking assisted discovery and biosynthesis elucidation of the antimicrobial spiroketals epicospirocins. Chem. Commun. (Camb.) 2020, 56, 10171–10174. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Dewapriya, P.; Li, F.; Blumel, M.; Tasdemir, D. Pyrenosetins A–C, new decalinoylspirotetramic acid derivatives isolated by bioactivity-based molecular networking from the seaweed-derived fungus Pyrenochaetopsis sp. FVE-001. Mar. Drugs 2020, 18, 47. [Google Scholar] [CrossRef]
- Lee, J.; Yang, H.S.; Jeong, H.; Kim, J.H.; Yang, H. Targeted isolation of lignans from Trachelospermum asiaticum using molecular networking and hierarchical clustering analysis. Biomolecules 2020, 10, 378. [Google Scholar] [CrossRef]
- Alcover, C.F.; Bernadat, G.; Kabran, F.A.; Le Pogam, P.; Leblanc, K.; Fox, R.A.; Gallard, J.F.; Mouray, E.; Grellier, P.; Poupon, E.; et al. Molecular networking reveals serpentinine-related bisindole alkaloids from Picralima nitida, a previously well-investigated species. J. Nat. Prod. 2020, 83, 1207–1216. [Google Scholar] [CrossRef]
- Fox, R.A.; Alcover, C.; Evanno, L.; Maciuk, A.; Litaudon, M.; Duplais, C.; Bernadat, G.; Gallard, J.F.; Jullian, J.C.; Mouray, E.; et al. Revisiting previously investigated plants: A molecular networking-based study of geissospermum laeve. J. Nat. Prod. 2017, 80, 1007–1014. [Google Scholar]
- Enomoto, M.; Kuwahara, S. Concise synthesis of PM-94128 and Y-05460M-A. J. Org. Chem. 2009, 74, 7566–7569. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Han, X.; Jiang, Z.; Wu, G.; Hu, X.; You, X.; Jiang, J.; Zhang, Y.; Sun, C. Hetiamacin B-D, new members of amicoumacin group antibiotics isolated from Bacillus subtilis PJS. J. Antibiot. (Tokyo) 2016, 69, 769–772. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Sun, C.; Hu, X.; You, X.; Wang, M.; Liu, S. Damxungmacin A and B, two new amicoumacins with rare heterocyclic cores isolated from Bacillus subtilis XZ-7. Molecules 2016, 21, 1601. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jin, J.; Chen, C.; Liu, J.; Li, J.; Wang, F.; Jiang, Z.; Hu, J.; Gao, Z.; Yao, F.; et al. PJS, a novel isocoumarin with hexahydropyrimidine ring from Bacillus subtilis PJS. J. Antibiot. (Tokyo) 2013, 66, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, Y.; Liu, L.; Han, Z.; Lai, P.Y.; Guo, X.; Zhang, X.; Lin, W.; Qian, P. Five new amicoumacins isolated from a marine-derived bacterium Bacillus subtilis. Mar. Drugs 2012, 10, 319–328. [Google Scholar] [CrossRef]
- McInerney, B.V.; Taylor, W.C.; Lacey, M.J.; Akhurst, R.J.; Gregson, R.P. Biologically active metabolites from Xenorhabdus spp., Part 2. Benzopyran-1-one derivatives with gastroprotective activity. J. Nat. Prod. 1991, 54, 785–795. [Google Scholar] [CrossRef]
- Canedo, L.M.; Fernandez, P.J.; Perez, B.J.; Acebal, C.; de la Calle, F.; Garcia, G.D.; Garcia, D.Q.T. PM-94128, a new isocoumarin antitumor agent produced by a marine bacterium. J. Antibiot. (Tokyo) 1997, 50, 175–176. [Google Scholar] [CrossRef]
- Sato, T.; Nagai, K.; Suzuki, K.; Morioka, M.; Saito, T.; Nohara, C.; Susaki, K.; Takebayashi, Y. A new isocoumarin antibiotic, Y-05460M-A. J. Antibiot. (Tokyo) 1992, 45, 1949–1952. [Google Scholar] [CrossRef]
- Huang, Y.F.; Li, L.H.; Tian, L.; Qiao, L.; Hua, H.M.; Pei, Y.H. Sg17-1-4, A novel isocoumarin from a marine fungus Alternaria tenuis Sg17-1. J. Antibiot. (Tokyo) 2006, 59, 355–357. [Google Scholar] [CrossRef]
- Itoh, J.; Omoto, S.; Shomura, T.; Nishizawa, N.; Miyado, S.; Yuda, Y.; Shibata, U.; Inouye, S. Amicoumacin-A, a new antibiotic with strong antiinflammatory and antiulcer activity. J. Antibiot. (Tokyo) 1981, 34, 611–613. [Google Scholar] [CrossRef] [PubMed]
- Azumi, M.; Ogawa, K.; Fujita, T.; Takeshita, M.; Yoshida, R.; Furumai, T.; Igarashi, Y. Bacilosarcins A and B, novel bioactive isocoumarins with unusual heterocyclic cores from the marine-derived bacterium Bacillus subtilis. Tetrahedron 2008, 64, 6420–6425. [Google Scholar] [CrossRef]
- Han, X.; Liu, S.; Wang, F.; Liu, J.; Hu, X.; You, X.; Shang, G.; Zhang, Y.; Sun, C. Research progress of amicoumacin group antibiotic. World Notes Antibiot. 2013, 34, 106–115. [Google Scholar]
- Polikanov, Y.S.; Osterman, I.A.; Szal, T.; Tashlitsky, V.N.; Serebryakova, M.V.; Kusochek, P.; Bulkley, D.; Malanicheva, I.A.; Efimenko, T.A.; Efremenkova, O.V.; et al. Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome. Mol. Cell 2014, 56, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Prokhorova, I.V.; Akulich, K.A.; Makeeva, D.S.; Osterman, I.A.; Skvortsov, D.A.; Sergiev, P.V.; Dontsova, O.A.; Yusupova, G.; Yusupov, M.M.; Dmitriev, S.E. Amicoumacin a induces cancer cell death by targeting the eukaryotic ribosome. Sci. Rep.-UK 2016, 6, 27720. [Google Scholar] [CrossRef]
- Wu, G.; Liu, S.; Wang, T.; Jiang, Z.; Lv, K.; Wang, Y.; Sun, C. Total synthesis of originally proposed and revised structure of hetiamacin A. Org. Lett. 2018, 20, 3566–3569. [Google Scholar] [CrossRef]
- Bai, J.; Liu, D.; Yu, S.; Proksch, P.; Lin, W. Amicoumacins from the marine-derived bacterium Bacillus sp. with the inhibition of NO production. Tetrahedron Lett. 2014, 55, 6286–6291. [Google Scholar] [CrossRef]
- Osterman, I.A.; Komarova, E.S.; Shiryaev, D.I.; Korniltsev, I.A.; Khven, I.M.; Lukyanov, D.A.; Tashlitsky, V.N.; Serebryakova, M.V.; Efremenkova, O.V.; Ivanenkov, Y.A.; et al. Sorting out antibiotics’ mechanisms of action: A double fluorescent protein reporter for high throughput screening of ribosome and DNA biosynthesis inhibitors. Antimicrob. Agents Chemother. 2016, 2116–2117. [Google Scholar] [CrossRef] [PubMed]
- Mondol, M.A.; Shin, H.J.; Islam, M.T. Diversity of secondary metabolites from marine Bacillus species: Chemistry and biological activity. Mar. Drugs 2013, 11, 2846–2872. [Google Scholar] [CrossRef]
- Cochrane, S.A.; Vederas, J.C. Lipopeptides from Bacillus and Paenibacillus spp.: A gold mine of antibiotic candidates. Med. Res. Rev. 2016, 36, 4–31. [Google Scholar] [CrossRef]
- Tyurin, A.P.; Efimenko, T.A.; Prokhorenko, I.A.; Rogozhin, E.A.; Malanicheva, I.A.; Zenkova, V.A.; Efremenkova, O.V.; Korshun, V.A. Amicoumacins and related compounds: Chemistry and biology. In Studies in Natural Products Chemistry; Atta, R., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 385–441. [Google Scholar]
- Shankarrao, K.O.; Angadrao, K.T.; Vasudha, D.K. Partial identification of antimicrobial compound produced by thermotolerant Bacillus subtilis KFSB5 isolated from compost soil. Res. J. Biotechnol. 2014, 92, 23–28. [Google Scholar]
- Pinchuk, I.V.; Bressollier, P.; Verneuil, B.; Fenet, B.; Sorokulova, I.B.; Mégraud, F.; Urdaci, M.C. In vitro anti-Helicobacter pylori activity of the probiotic strain Bacillus subtilis 3 is due to secretion of antibiotics. Antimicrob. Agents Ch. 2001, 45, 3156–3161. [Google Scholar] [CrossRef] [PubMed]
- Kaspar, F.; Neubauer, P.; Gimpel, M. Bioactive secondary metabolites from Bacillus subtilis: A comprehensive review. J. Nat. Prod. 2019, 82, 2038–2053. [Google Scholar] [CrossRef]
- Hegazi, N.M.; Radwan, R.A.; Bakry, S.M.; Saad, H.H. Molecular networking aided metabolomic profiling of beet leaves using three extraction solvents and in relation to its anti-obesity effects. J. Adv. Res. 2020, 24, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Oh, J.H.; Lee, M.Y.; Lee, S.G.; Ha, I.J. Molecular network-guided alkaloid profiling of aerial parts of Papaver nudicaule L. Using LC-HRMS. Molecules 2020, 25, 2636. [Google Scholar] [CrossRef] [PubMed]
- Buedenbender, L.; Astone, F.A.; Tasdemir, D. Bioactive molecular networking for mapping the antimicrobial constituents of the baltic brown alga Fucus vesiculosus. Mar. Drugs 2020, 18, 311. [Google Scholar] [CrossRef]
- Lu, Q.; Ye, J.; Huang, Y.; Liu, D.; Liu, L.; Dong, K.; Razumova, E.A.; Osterman, I.A.; Sergiev, P.V.; Dontsova, O.A.; et al. Exploitation of potentially new antibiotics from mangrove actinobacteria in maowei sea by combination of multiple discovery strategies. Antibiotics (Baselswitzerland) 2019, 8, 236. [Google Scholar] [CrossRef]
- Wang, T.; Li, F.; Lu, Q.; Wu, G.; Jiang, Z.; Liu, S.; Habden, X.; Razumova, E.A.; Osterman, I.A.; Sergiev, P.V.; et al. Studies on diversity, novelty, antimicrobial activity, and new antibiotics of cultivable endophytic actinobacteria isolated from psammophytes collected in Taklamakan Desert. J. Pharm. Anal. 2020. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1 and 2 are available from the authors. |
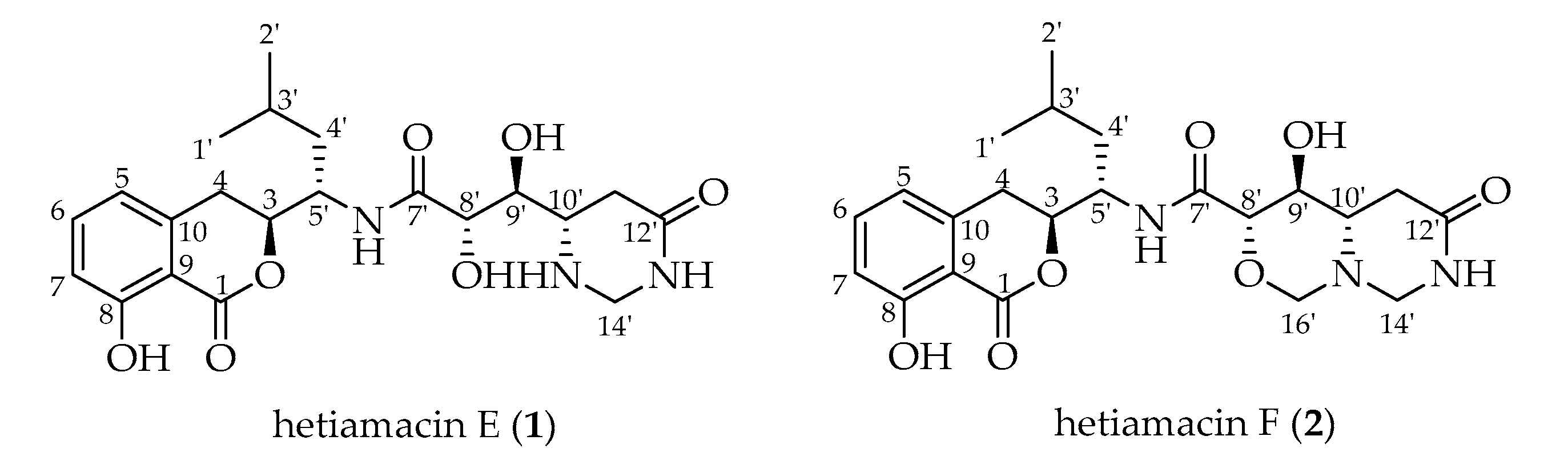
| No. | Hetiamacin E (1) a | Hetiamacin F (2) b | ||||
|---|---|---|---|---|---|---|
| δC | δH, Mult (J in Hz) | HMBC | δC | δH, Mult (J in Hz) | HMBC | |
| 1 | 169.1, C | 7 | 169.1, C | |||
| 3 | 81.1, CH | 4.70, d (12.5) | 4,4′ | 81.2, CH | 4.69, dt (12.8, 2.7) | 4 |
| 4 | 29.1, CH2 | 3.08–3.02, m; 2.90–2.82, m | 5 | 29.2, CH2 | 3.00, dd (16.3, 13.0); 2.82, dd (16.7, 2.8) | 5 |
| 5 | 118.5, CH | 6.83, d (7.3) | 7,4 | 118.6, CH | 6.82, d (7.5) | 7,4 |
| 6 | 136.3, CH | 7.49, t (7.9) | 5 | 136.3, CH | 7.51–7.47, m | |
| 7 | 115.2, CH | 6.85, d (8.4) | 5 | 115.3, CH | 6.85, d (8.4) | 5 |
| 8 | 160.8, C | 6,7 | 160.9, C | 6 | ||
| 8–OH | 10.78, s | 10.82, s | ||||
| 9 | 108.3, C | 6,5,7,4 | 108.3, C | 7,5,4 | ||
| 10 | 140.7, C | 6,5,3,4 | 140.7, C | 6,4 | ||
| 1′ | 21.5, CH3 | 0.85, d (6.2) | 4′,2′ | 21.5, CH3 | 0.86, d (6.5) | 2′,4′ |
| 2′ | 23.3, CH3 | 0.89, d (6.4) | 1′ | 23.3, CH3 | 0.91, d (6.6) | 4′,1′ |
| 3′ | 23.9, CH | 1.67–1.59, m | 4′,1′,2′ | 24.1, CH | 1.67–1.58, m | |
| 4′ | 40.0, CH2 | 1.73–1.67, m; 1.36–1.28, m | 3, 1′, 2′ | 40.6, CH2 | 1.75–1.67, m; 1.38–1.31, m | 1′,2′ |
| 5′ | 47.9, CH | 4.24–4.14, m | 6′–NH,4,4′ | 48.2, CH | 4.24–4.18, m | 4,4′ |
| 6′–NH | 7.64, d (9.5) | 8.12, d (9.2) | ||||
| 7′ | 172.7, C | 6′–NH,8′ | 168.9, C | 6′-NH,8′ | ||
| 8′ | 72.3, CH | 3.90, d (6.6) | 8′–OH | 81.0, CH | 3.72, d (9.2) | 16′ |
| 8′–OH | 5.57, s | |||||
| 9′ | 73.7, CH | 3.68, d (5.7) | 8′,10′,11′ | 65.9, CH | 3.50–3.44, m | 8′,10′,11′ |
| 9′–OH | 4.99, d (5.6) | 5.29, d (6.2) | ||||
| 10′ | 52.6, CH | 3.02–2.95, m | 14′,8′,9′,11′ | 59.2, CH | 2.63–2.57, m | 16′,14′,11′ |
| 11′ | 32.8, CH2 | 2.13–2.07, m | 9′,13′–NH | 32.9, CH2 | 2.43, dd (17.7, 6.2); 2.21, dd (17.7, 6.2) | 13′-NH |
| 12′ | 169.4, C | 11′,13′−NH,14′ | 167.7, C | 14′,10′,11′ | ||
| 13′–NH | 7.61, s | 7.90, s | ||||
| 14′ | 56.7, CH2 | 4.03–3.95, m | 10′,13′–NH | 57.6, CH2 | 4.17, dd (9.0, 2.6); 3.66, dd (9.0, 2.1) | 16′, 10′, 13′ |
| 15′–NH | not detected | |||||
| 16′ | 81.5, CH2 | 4.51, d (9.1); 3.95, d (9.2) | ||||
| Test Organisms | MICs (µg/mL) | ||
|---|---|---|---|
| Hetiamacin E (1) | Hetiamacin F (2) | Levofloxacin | |
| S. epidermidis ATCC 12228 (MSSE) | 2 | 32 | 0.25 |
| S. epidermidis 16-4 (MSSE) | 4 | 32 | 0.25 |
| S. epidermidis 16-5 (MRSE) | 4 | 32 | 8 |
| S. aureus ATCC 29213 (MSSA) | 8 | >32 | 0.25 |
| S. aureus ATCC 33591 (MRSA) | 16 | >32 | 0.25 |
| Enterococcus faecium ATCC 700221 (VRE) | 64 | >32 | 32 |
| Escherichia coli ATCC 25922 | 64 | >32 | ≤0.03 |
| Pseudomonas aeruginosa PAO1 | 64 | 32 | 4 |
| Acinetobacter baumannii ATCC 19606 | 64 | 32 | 0.25 |
| Shigella flexneri ATCC 12022 | 64 | >32 | ≤0.03 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Lu, Q.; Sun, C.; Lukianov, D.; Osterman, I.A.; Sergiev, P.V.; Dontsova, O.A.; Hu, X.; You, X.; Liu, S.; et al. Hetiamacin E and F, New Amicoumacin Antibiotics from Bacillus subtilis PJS Using MS/MS-Based Molecular Networking. Molecules 2020, 25, 4446. https://doi.org/10.3390/molecules25194446
Wang T, Lu Q, Sun C, Lukianov D, Osterman IA, Sergiev PV, Dontsova OA, Hu X, You X, Liu S, et al. Hetiamacin E and F, New Amicoumacin Antibiotics from Bacillus subtilis PJS Using MS/MS-Based Molecular Networking. Molecules. 2020; 25(19):4446. https://doi.org/10.3390/molecules25194446
Chicago/Turabian StyleWang, Ting, Qinpei Lu, Chenghang Sun, Dmitrii Lukianov, Ilya Andreevich Osterman, Petr Vladimirovich Sergiev, Olga Anatolievna Dontsova, Xinxin Hu, Xuefu You, Shaowei Liu, and et al. 2020. "Hetiamacin E and F, New Amicoumacin Antibiotics from Bacillus subtilis PJS Using MS/MS-Based Molecular Networking" Molecules 25, no. 19: 4446. https://doi.org/10.3390/molecules25194446
APA StyleWang, T., Lu, Q., Sun, C., Lukianov, D., Osterman, I. A., Sergiev, P. V., Dontsova, O. A., Hu, X., You, X., Liu, S., & Wu, G. (2020). Hetiamacin E and F, New Amicoumacin Antibiotics from Bacillus subtilis PJS Using MS/MS-Based Molecular Networking. Molecules, 25(19), 4446. https://doi.org/10.3390/molecules25194446