Abstract
Flavonoids and triterpenoids were revealed to be the potential inhibitors on soluble epoxide hydrolase (sEH). The aim of this study is to reveal sEH inhibitors from Fuji apples. A flavonoid and three triterpenoids derived from the fruit of Malus domestica were identified as quercetin-3-O-arabinoside (1), ursolic acid (2), corosolic acid (3), and 2-oxopomolic acid (4). They had half-maximal inhibitory concentration of the inhibitors (IC50) values of 39.3 ± 3.4, 84.5 ± 9.5, 51.3 ± 4.9, and 11.4 ± 2.7 μM, respectively, on sEH. The inhibitors bound to allosteric sites of enzymes in mixed (1) and noncompetitive modes (2–4). Molecular simulations were carried out for inhibitors 1 and 4 to calculate the binding force of ligands to receptors. The inhibitors bound to the left (1) and right (4) pockets next to the enzyme’s active site. Based on analyses of their molecular docking and dynamics, it was shown that inhibitors 1 and 4 can stably bind sEH at 1 bar and 300 K. Finally, inhibitors 1 and 4 are promising candidates for further studies using cell-based assays and in vivo cardiovascular tests.
1. Introduction
Arachidonic acid is converted to epoxyeicosatrienoic acids (EETs) by cytochrome P450 epoxygenase [1]. EETs exist as four regioisomeric metabolites: 5,6-, 8,9-, 11,12-, and 14,15-EET [1]. Soluble epoxide hydrolase (sEH, E.C.3.3.2.10) biosynthesizes dihydroxyeicosatrienic acids (DHETs) from EETs by hydrolyzing their epoxide rings into a diol [2]. EETs were first revealed to be endothelium-derived hyperpolarizing factors in 1996 and have since been found to have various biological activities, including anti-inflammatory, antihypertensive, kidney-protective, and cardiac-protective effects [3]. 11,12-EET was reported to inhibit nuclear factor kappa B signaling pathways and the cytokine-induced expression of cellular adhesion molecules [4]. 11,12-/14,15-EETs stimulate the large-conductance Ca2+-activated K+ channel and expand blood vessels [3]. Thus, sEH, which is encoded in the EPHX2 gene located at chromosomal region 8p21-p12, has gained attention as a therapeutic target for the treatment of cardiovascular disease [5]. 12-(3-adamantan -1-yl-ureido)dodecanoic acid (AUDA) is an effective sEH inhibitor [6]. Owing to its low solubility, AUDA is dissolved in dimethyl sulfoxide for in vitro experiments and formulated with large amounts of 2-hydroxylpropyl β-cyclodextrin for in vivo use [3]. Therefore, the aim of this study is to find sEH inhibitors that have higher solubility than that of AUDA.
The apple is one of the most widely consumed fruits worldwide [7]. The Fuji apple (Malus domestica Brokh.), which originated in Japan in 1962 [8], is the most consumed apple variety in Korea [9]. This fruit has been reported to contain phenolic compounds such as caffeoylquinic acid, epicatechin, and procyanidins, which provide health benefits such as decreased risk of cardiovascular disease and cancer [10]. Recently, triterpenoids from Cimicifuga dahurica Maxim. [11], flavonoids from Tetrastigma hemsleyanum [12], and flavonoid glycosides [13] have been reported as noncompetitive agents blocking the catalytic activity of sEH [12,13]. In the present study, flavonoid and triterpenoid compounds were isolated from apples, and their structures were identified using spectroscopic methods. Isolated compounds were evaluated to assess their inhibitory activity against sEH and beneficial effects for cardiovascular disease.
2. Results and Discussion
2.1. Isolation, Identification, and Enzyme Assay
In recent studies, we revealed that flavonoids and triterpenoids have inhibitory activity against sEH [11,12,13]. A flavonoid glycoside (1) and triterpenoids (2–4) were purified from the fruit of M. domestica. Their chemical structures were identified as quercetin 3-O-arabinofuranoside (1) (Figure S1) [14], ursolic acid (2) (Figure S2) [15], corosolic acid (3) (Figure S3) [16], and 2-oxopomolic acid (4) (Figure S4) [17] based on nuclear magnetic resonance (NMR) analysis and comparisons with the reported data (Figure 1).
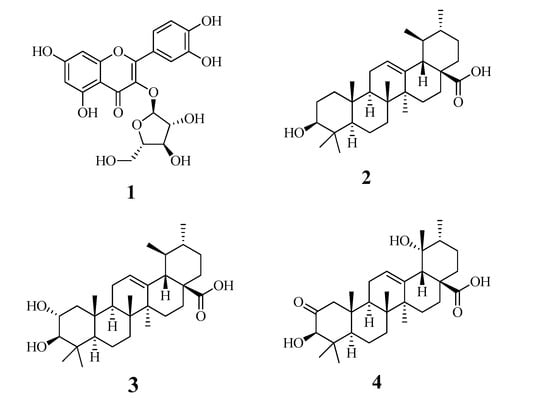
Figure 1.
The structures of compounds 1–4 isolated from the fruits of Malus domestica.
Purified compounds 1–4 (Figure S5) were evaluated to assess their sEH-inhibitory activity in vitro. The inhibitory activity of each concentration of compound was calculated based on the difference in enzyme products with and without the presence of the compound using Equation (1). Figure 2A shows that the compounds had sEH inhibition activity in a dose-dependent manner at 3.1–100 μM concentrations. To calculate the half-maximal inhibitory concentration of the inhibitors (IC50), inhibition rate values were fitted using Equation (2). Inhibitors 1–4 were found to have IC50 values of 39.3 ± 3.4, 84.5 ± 9.5, 51.3 ± 4.9, and 11.4 ± 2.7 μM, respectively (Table 1). AUDA was used as the positive control (1.5 ± 1.2 nM).
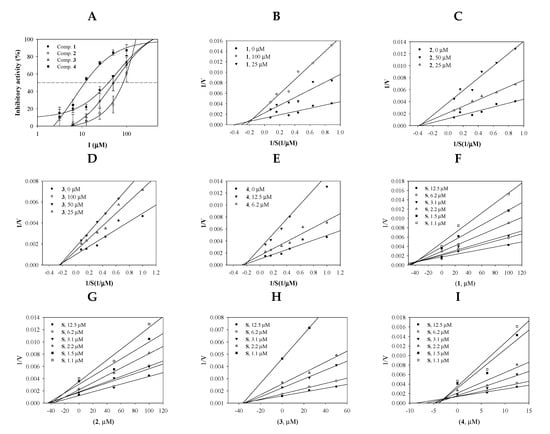
Figure 2.
Inhibitory activities of the inhibitors 1–4 at a variety of concentrations toward soluble epoxide hydrolase (sEH) (A). Lineweaver-Burk plots (B–F) and Dixon plots (G–I) for the inhibition of sEH by compounds 1–4.

Table 1.
The soluble epoxide hydrolase (sEH) inhibitory activities of compounds 1–4.
The enzyme kinetic study was performed to find the binding position of the inhibitor into sEH. The initial velocity (v0) for the three different concentrations of inhibitors was calculated relative to the substrate concentrations (1.1, 1.5, 2.2, 3.1, 6.2, and 12.5 μM). The inverse of these concentrations and v0 values were used to construct Lineweaver-Burk plots. As indicated in Figure 2B–E, flavonoid 1 and triterpenoids 2–4 exhibited mixed inhibitions (different −1/Km and 1/Vmax values) and noncompetitive inhibitions (a −1/Km value and different 1/Vmax values) (Table 1).
2.2. Molecular Docking
In 1894, the “lock-and-key” model was first suggested by Emil Fisher to describe the interactions of enzymes (locks) with substrates (keys) [18]. Molecular docking is a virtual technology that simulates these interactions, and such computer technologies have been used for drug development by Merck Inc. since the 1980s [19]. We carried out molecular docking analyses using the Autodock 4.2 program to gain insight into the inhibitor-sEH interactions. Based on the enzyme kinetic results, blind docking was performed to identify an allosteric site to which inhibitors 1 and 4 with IC50 values below 50 μM could bind. Two compounds had the highest-scoring poses, with binding energies of −9.03 and −10.18 kcal/mol, respectively (Figure 3A,B and Table 2). As shown in Figure 3C and Table 2, flavonoid glycoside (1) had three hydrogen bonds between its sugar group and Lys495 (at a 2.62-Å distance), Asp496 (at a 2.56-Å distance), and Phe497 (at a 2.82-Å distance). Triterpenoid (4) formed two hydrogen bonds: between its carboxyl group and Asn366 at a distance of 3.21 Å and between its hydroxyl and ILe363 at a distance of 2.77 Å (Figure 3D and Table 2). This hydroxyl unit is a functional group only in compound 4 among triterpenoids. It can be confirmed that the sEH inhibitory activity increases as this functional group binds to the enzyme. Through a molecular docking study, the allosteric sites were identified as the left (1) and right (4) pockets next to the catalytic site, respectively.
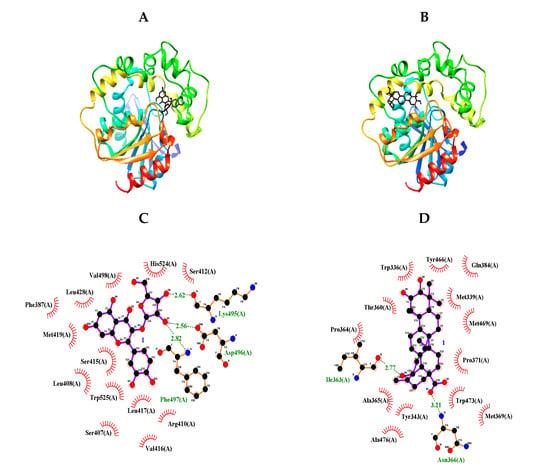
Figure 3.
Predicted docking poses between compounds 1 and 4 and sEH (A,B). Hydrogen bonds of inhibitors 1 and 4 of the catalytic site, respectively (C,D).

Table 2.
Interaction and binding energy of sEH with inhibitors 1 and 4.
2.3. Molecular Dynamics
Molecular dynamics (MD) simulation was used to obtain computational information on the bovine pancreatic trypsin inhibitor under vacuum conditions by McCammon in 1977 [19]. MD is a state-of-the-art computational technique for investigating interactions between ligands and receptors [19]. The “induced fit” model of enzyme-inhibitor binding was proposed by Koshland, as inhibitors can induce enzyme conformation changes during binding [19]. NMR and X-ray crystallography have provided limited insight into the static binding pose of ligands and enzymes [20]. In the present study, MD simulation using the Gromacs 4.6 program was carried out to assess the interactions between the inhibitors (1 and 4) and sEH. As shown in Figure 4A,B, inhibitors 1 and 4 bound with sEH, reaching a steady state with ~9.1 × 155 kJ/mol potential energy (Figure 4C). The protein root mean square deviation (RMSD) values of compounds 1 and 4 were ~2.8 and ~2.5 Å, respectively (Figure 4D). Amino acids that bonded with the two compounds exhibited root mean square fluctuation (RMSF) values within ~3.5 Å (Figure 4E). Inhibitors 1 and 4 maintained mainly 1–5 and 1–3 hydrogen bonds with sEH residues for 10 ns (Figure 4F,G). Hydrogen bonds in sEH-inhibitor complexes were analyzed at 1-ns intervals during their trajectory (Table 3). As a result, inhibitor 1 consistently formed hydrogen bonds with four residues: Ser415, Val416, Met419, and Lys495. Inhibitor 4 was found to form hydrogen bonds with Tyr343 and Ser374 continuously. These results were different from the ligand-receptor hydrogen bond in molecular docking. These facts suggest that the two inhibitors affect the enzymatic structure. Accordingly, inhibitors 1 and 4 were assigned to the “induced fit” in “lock-and-key” models. In silico, it was confirmed that the hydrogen bonds of the inhibitors were different in stationary (docking) and fluid (dynamic) enzymes. These results suggest that the two compounds bind to the enzyme by changing the enzyme structure as the “induced fit” theory.
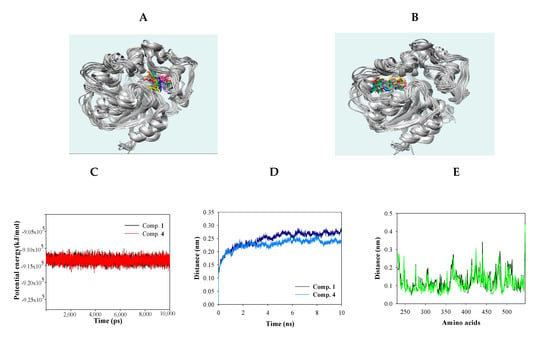
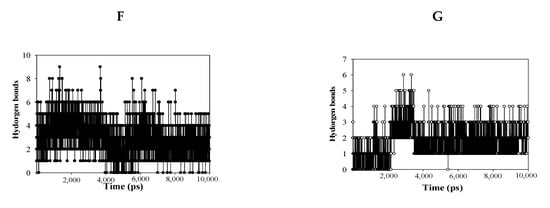
Figure 4.
Superpositions of sEH with compounds 1 and 4 for simulation trajectory (0 ns: red, 1: orange, 2: yellow, 3: green, 4: forest green, 5: cyan, 6: blue, 7: cornflower blue, 8: purple, 9: hot pink, and 10: magenta) (A,B). The potential energy (C), root mean square deviation (RMSD) (D), root mean square fluctuation (RMSF) (E), and hydrogen bonds (F,G) of the simulation calculated with 1 and 4 during 10 ns.

Table 3.
Hydrogen bond analysis of the inhibitor with sEH at 1-ns intervals for 10 ns.
The sEH-inhibitor complexes that were analyzed by the Dictionary of Secondary Structure protein (DSSP) of 1 depended on the Ser415-His420, Lys495-Val500, and Cys522-Trp525 loops in the left pocket (Figure 5A), as well as 4 anchored to the Thr360-Pro371 loop when binding to the right pocket (Figure 5B). As shown in Figure 5A,B, this loop maintained a coil and band structure without any major structural changes. This finding confirms that the inhibitor is the correct size for the loop pocket (Figure 3A). The key amino acids-containing loop (Ser415, Val416, and Met419) in sEH was partially transformed into various secondary structures in the form of a bend, turn, and α-helix by compound 1, whereas the Lys495 loop showed a fixed bend and turn structure (Figure 5A). As indicated in Figure 5B, the Thr360-Pro371 loop was retained as a fixed secondary structure despite binding to inhibitor 4. These facts confirmed that, after binding of the sEH and inhibitors, the secondary structure of the loop containing key amino acids was not affected.
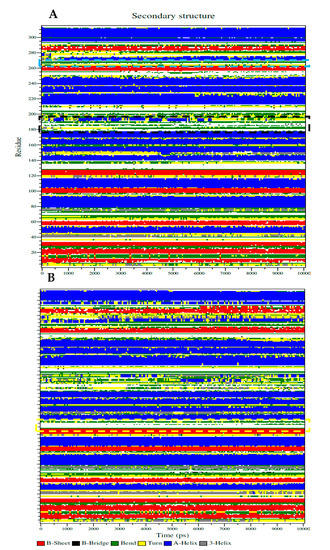
Figure 5.
The Dictionary of Secondary Structure protein (DSSP) represented with secondary structure changes of sEH with inhibitor 1 (A) or 4 (B) (yellow square: Thr360-Pro371, black square: Ser415-Glu424, and blue square: Glu494-Pro501).
3. Materials and Methods
3.1. General Experimental Procedures
Column chromatography were performed using silica gel (Kieselgel 60, 70–230, and 230–400 mesh, Merck, Darmstadt, Germany); Sephadex LH-20 (GE Healthcare, Uppsala, Sweden); and C-18 (ODS-A 12 nm S-150 and S-75 μm; YMC Co., Dinslaken, Germany) resins. Thin-layer chromatography (TLC) was performed using precoated silica gel 60 F254 and RP-18 F254S plates (both 0.25 mm, Merck). Spots in TLC were visualized by spraying with 10% aqueous H2SO4 solution, followed by heating at 300 °C dried air. Nuclear magnetic resonance (NMR) spectra were recorded using the JEOL ECA 500 spectrometer (1H, 500 MHz and 13C, 125 MHz) (Tokyo, Japan). AUDA (10007927), soluble epoxide hydrolase (10011669), and 3-phenyl-cyano(6-methoxy-2-naphthalenyl) methyl ester-2-oxiraneacetic acid (PHOME) (10009134) were purchased from Cayman (Cayman, Ann Arbor, MI, USA).
3.2. Plant Materials
The fruits of M. domestica were collected in October 2017 at Jansu-gun, Jeollabuk-do, Republic of Korea and were identified by Prof. Y.H. Kim in the College of Pharmacy, Chungnam National University, Daejeon, Republic of Korea. A voucher specimen (CNU 17003) was deposited at the Herbarium of the College of Pharmacy, Chungnam National University.
3.3. Extraction and Isolation
The flesh (peel and seeds removed) and peel of M. domestica were sliced and dried at room temperature for one week. The dried flesh (1.6 kg) and peel (260.0 g) were extracted three times with methanol by ultrasonic solvent extraction in an ultrasound bath at 35 ± 3 °C. After removal of solvents under reduced pressure, methanol extracts of the flesh (725.0 g) and peel (36.0 g) were obtained. Two methanol extracts were suspended in distilled water and successively partitioned with n-hexane, ethyl acetate, and n-BuOH to yield two n-hexane fractions (FH and PH), two ethyl-acetate fractions (FE and PE), and two n-BuOH fractions (FB and PH), respectively. The PE fraction (16.2 g) was subjected to silica gel column chromatography with a gradient of CH2Cl2-MeOH (30:1→1:1) to yield eight fractions (PE1–PE8). Compound 1 (20.0 mg) was isolated from the PE8 fraction using RP-18 silica gel column chromatography with MeOH-H2O (2:1→1:1) as the gradient elution solvent. The FE fraction (18.1 g) was chromatographed using vacuum liquid column chromatography and eluted with a CH2Cl2-MeOH (50:1→1:1) gradient system to yield six fractions (FE1–FE6). The FE2 fraction was chromatographed using RP-18 silica gel column chromatography and eluted with a MeOH-H2O (3:1) isocratic system to yield compound 4 (16.3 mg). The FE3 fraction was separated using RP-18 silica gel column chromatography with MeOH-H2O (3:1) to yield compound 2 (10.1 mg) and compound 3 (1.5 mg), respectively.
3.4. sEH Assay and Kinetic Analysis
The assays were carried out at 37 °C in 0.025-M bis-tris-HCL buffer, pH 7.0, containing 0.1% Bovine Serum Albumin B. PHOME (~2.6 μM) was used as a substrate. The final volume of tests was 200 μM in ~10 ng/mL sEH. The products were monitored at 330 nm of excitation and 465 nm emission for 40 min.
The inhibition rate was calculated according to the following equations:
where ΔC and ΔS represent the gap of control and inhibitor during reaction, respectively.
where “y0” is the point on the axis of ordinates, “a” denotes the difference between the maximum and minimum values, and “b” refers to the “x” value at 50%.
Inhibition effect rate (%) = [(ΔC − ΔS)/ΔC] × 100
y = y0 + [(a × x)/(b + x)]
3.5. Docking Study of sEH with Inhibitor
The 3D structure of the inhibitor was downloaded from the ChemSpider database and then minimized by MM2. The single bonds were designated as rotational bonds by using AutoDockTools. The 3D structure of sEH was downloaded from the research collaboratory for structural bioinformatics protein data bank coded with 3ANS ID. A chain of that with hydrogen atoms and Gasteiger charges was used for docking. The grid dimensions were set at 126 × 126 × 126 number of points (x × y × z). Docking simulation was performed to dock the inhibitor 25,000,000 times into this grid. The results of this study were represented as a diagram by using Chimera (San Francisco, CA, USA) and Ligplot (Cambridge, UK).
3.6. Molecular Dynamic Study
To simulate the molecular dynamics of sEH with the inhibitor, the Gromacs program (version 4.6.5) was used. The Itp and gro files of the inhibitor were created in the GlycoBioChem PRODRG2 server. sEH gro (Gromos96 43a1 force field) and topology files were encoded by gdb2gmx. The complex between the inhibitor and sEH was contained as cubes of water molecules measuring 8.5 × 8.5 × 8.5 containing six chloride ions. The energy minimization of that was lowered by 10 kJ/mol maximal force in the steepest descent method. A stable simulated complex was progressively simulated 300 K and at 1 bar for 100 ps, respectively. Finally, the stable product was subjected to molecular dynamics for 10 ns.
3.7. Data Analysis
Data were expressed as the means ± standard deviation (n = 3). All values were analyzed using SigmaPlot to determine the treatment variations.
4. Conclusions
A flavonoid and three triterpenoids from Fuji apples were indicated to have an inhibitory activity at 100 μM toward sEH. Among them, inhibitors 1 and 4 were calculated as IC50 values with 39.3 ± 3.4 and 11.4 ± 2.7 μM, respectively. These inhibitors were interacted into the position next to the catalytic site of the enzyme. Especially, the findings in the molecular simulation suggest that these two compounds bind to the enzyme by changing the enzyme structure as in the “induced fit” theory. These facts confirmed the possibility that quercetin-3-O-arabinoside (1) and 2-oxopomolic acid (4) contained in the fruits of M. domestica are natural sEH inhibitors in vitro and in silico. Inhibitors 1 and 4 are promising candidates for further studies using cell-based assays and in vivo cardiovascular tests.
Supplementary Materials
Figure S1: 1H/13C-NMR spectra of 1. Figure S2: 1H/13C-NMR spectra of 2. Figure S3: 1H/13C-NMR spectra of 3. Figure S4: 1H/13C-NMR spectra of 4. Figure S5: C-18 TLC results of compounds 1–4.
Author Contributions
I.S.C., X.D.S., Y.L. and J.H.K. performed the experiments. I.S.C. and J.H.K. wrote the manuscript. Y.H.K., I.S.C., J.S.K., and S.Y.Y. designed the experiments and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was carried out with support of the Cooperative Research Program for Agriculture Science & Technology Development, Rural Development Administration, Republic of Korea (Project No. PJ01252402).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Batchu, S.N.; Lee, S.B.; Samokhvalon, V.; Chaudhary, K.R.; EI-Sikhry, H.; Weldon, S.M.; Seubert, J.M. Novel soluble epoxide hydrolase inhibitor protects mitochondrial function following stress. Can. J. Physiol. Pharmacol. 2012, 90, 811–823. [Google Scholar] [CrossRef]
- Manickam, M.; Pillaiyar, T.; Boggu, P.; Venkateswararao, E.; Jalani, H.B.; Kim, N.-D.; Lee, S.K.; Jeon, J.S.; Kim, S.K.; Jung, S.-H. Discovery of enantioselectivity of urea inhibitors of soluble epoxide hydrolase. Eur. J. Med. Chem. 2016, 117, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Imig, J.D.; Hammock, B.D. Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat. Rev. Drug Discov. 2009, 8, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.W.; Morisseau, C.; Hammock, B.D. Epoxide hydrolases: Their roles and interactions with lipid metabolism. Prog. Lipid Res. 2005, 44, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.-T.; Wasti, B.; Xu, D.-Y.; Shen, L.; Du, J.-Q.; Zhao, S.-P. Soluble epoxide hydrolase and ischemic cardiomyopathy. Int. J. Cardiol. 2012, 155, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Pecic, S.; Zeki, A.A.; Xu, X.; Jin, G.Y.; Zhang, S.; Kodani, S.; Halim, M.; Morisseau, C.; Hammock, B.D.; Deng, S.-X. Novel piperidine-derived amide sEH inhibitors as mediators of lipid metabolism with improved stability. Prostag. Lipid Med. 2018, 136, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, F.; Tatsuki, M.; Matsubara, K.; Okazaki, K.; Yoshimura, M.; Kasai, S. Methyl ester generation associated with flesh browning in ‘Fuji’ apples after long storage under repressed ethylene function. Postharvest Biol. Technol. 2018, 145, 53–60. [Google Scholar] [CrossRef]
- Lin, M.-Z.; Chai, W.-M.; Chong, O.-Y.; Huang, Q.; Xu, X.-U.; Peng, Y.-Y. Antityrosinases mechanism of omeprazole and its application on the preservation of fresh-cut Fuji apple. Int. J. Biol. Macromol. 2018, 117, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.P.D.; Cho, I.S.; Kim, K.T.; Yang, S.Y.; Kim, Y.H.; Kang, J.S. Evaluation of phenolic compounds from viroid-free and viroid-infected apples using HPLC-PDA-ESI-MS/MS. Phytochem. Anal. 2019, 30, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Ribas-Agustí, A.; Martín-Belloso, O.; Soliva-fortuny, R.; Elez-Martínez, P. Influence of pulsed electric fields processing on the bioaccessible and non-bioaccessible fractions of apple phenolic compounds. J. Funct. Foods 2019, 59, 206–214. [Google Scholar] [CrossRef]
- Thao, N.P.; Kim, J.H.; Luyen, B.T.T.; Dat, N.T.; Kim, Y.H. In silico investigation of cycloartane triterpene derivatives from Cimicifuga dahurica (Turcz.) Maxim. roots for the development of potent soluble epoxide hydrolase inhibitors. Int. J. Biol. Macromol. 2017, 98, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Thao, N.P.; Luyen, B.T.T.; Kim, J.H.; Jo, A.R.; Dat, N.T.; Kiem, P.V.; Minh, C.V.; Kim, Y.H. Identification, characterization, kinetics, and molecular docking of flavonoid constituents from Archidendron clypearia (Jack.) Nielsen leaves and twigs. Bioorg. Med. Chem. 2016, 24, 3125–3132. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Lee, S.; Jang, H.-J.; Su, X.D.; Wang, H.-S.; Kim, Y.H.; Yang, S.Y. Inhibition potential of phenolic constituents from the aerial parts of Tetrastigma hemsleyanum against soluble epoxide hydrolase and nitric oxide synthase. J. Enzym. Inhib. Med. Chem. 2019, 34, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Nugroho, A.; Rhim, T.-J.; Choi, M.-Y.; Choi, J.S.; Kim, Y.-C.; Kim, M.-S.; Park, H.-J. Simultaneous analysis and peroxynitrite-scavenging activity of galloylated flavonoid glycosides and ellagic acid in Euphorbia supine. Arch. Pharm. Res. 2014, 37, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, S.; Wu, L.; Li, Y.; Feng, L.; Shen, Y.; Tian, J.; Tang, J.; Wang, N.; Liu, Y.; et al. Abiesatrines A-J: Anti-inflammatory and antitumor triterpenoids from Abies georgei Orr. Org. Biomol. Chem. 2010, 8, 2609–2616. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Zhou, W.; Oh, J.S.; Choe, S.G.; Kim, D.W.; Lee, S.H.; Na, M. Rhododendric acid A, a new ursane-type PTP1B inhibitor from the endangered plant Rhododendron brachycarpum G. Don. Bioorg. Med. Chem. Lett. 2012, 22, 6116–6119. [Google Scholar] [CrossRef] [PubMed]
- D’Abrosca, B.; Fiorentino, A.; Monaco, P.; Pacifico, S. Raidcal-scavenging activities of new hydroxylated ursane triterpenes from Annurca Apples cv. Chem. Biodivers. 2015, 2, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Bankaitis, V.A. Molecular docking: From lock and key to combination lock. J. Mol. Clin. Appl. 2017, 2, 1–9. [Google Scholar]
- Salmaso, V.; Moro, S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: An overview. Front Pharmacol. 2018, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Lu, D.; Zhang, H.; Zheng, M.; Yang, H.; Xu, Y.; Luo, C.; Zhu, W.; Yu, K.; Jiang, H. Applying high-performance computing in drug discovery and molecular simulation. Natl. Sci. Rev. 2016, 3, 49–63. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).