Synthesis, Characterization, Photoluminescence, Molecular Docking and Bioactivity of Zinc (II) Compounds Based on Different Substituents
Abstract
1. Introduction
2. Results and Discussion
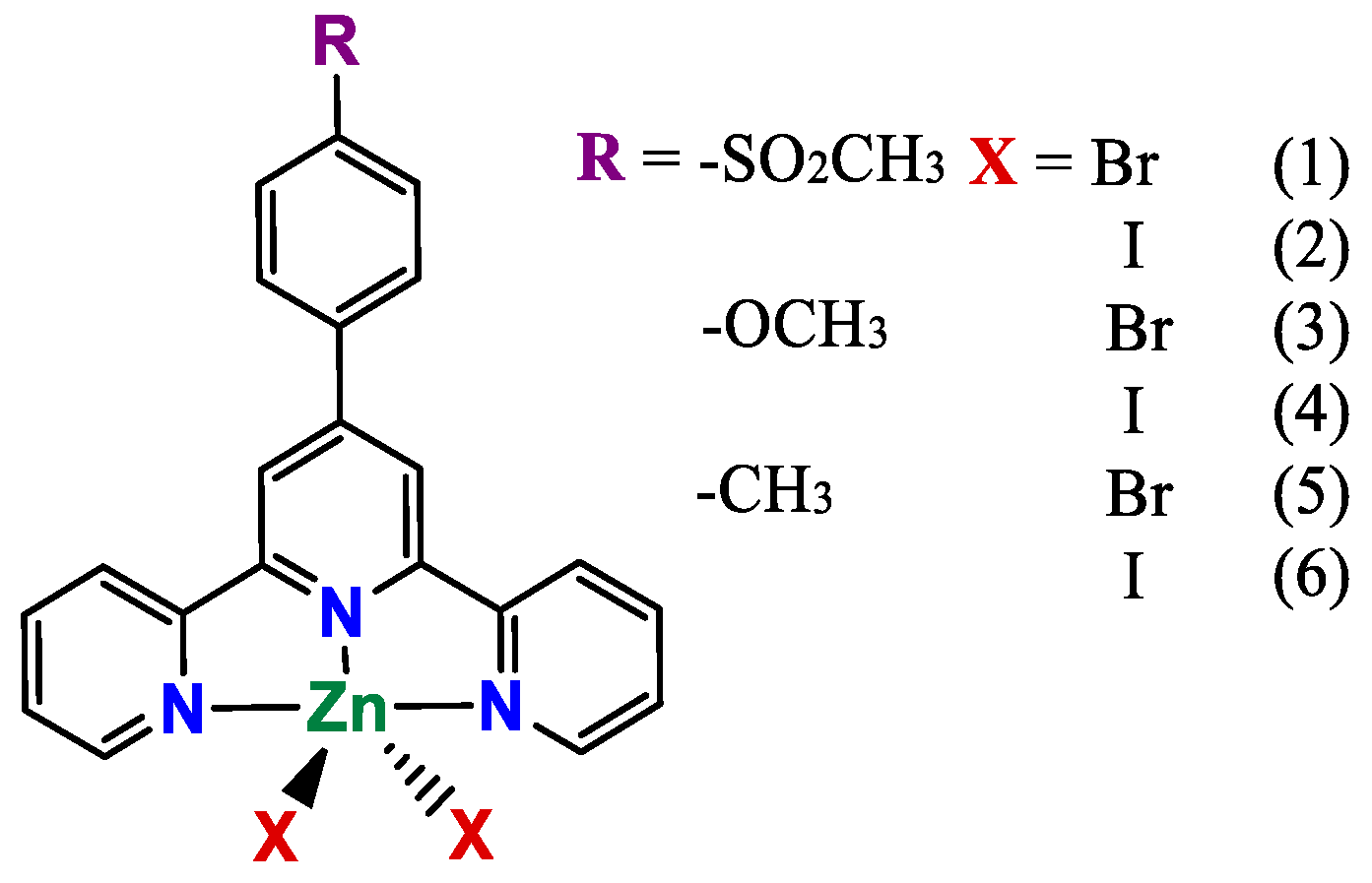
2.1. Syntheses and Characterization
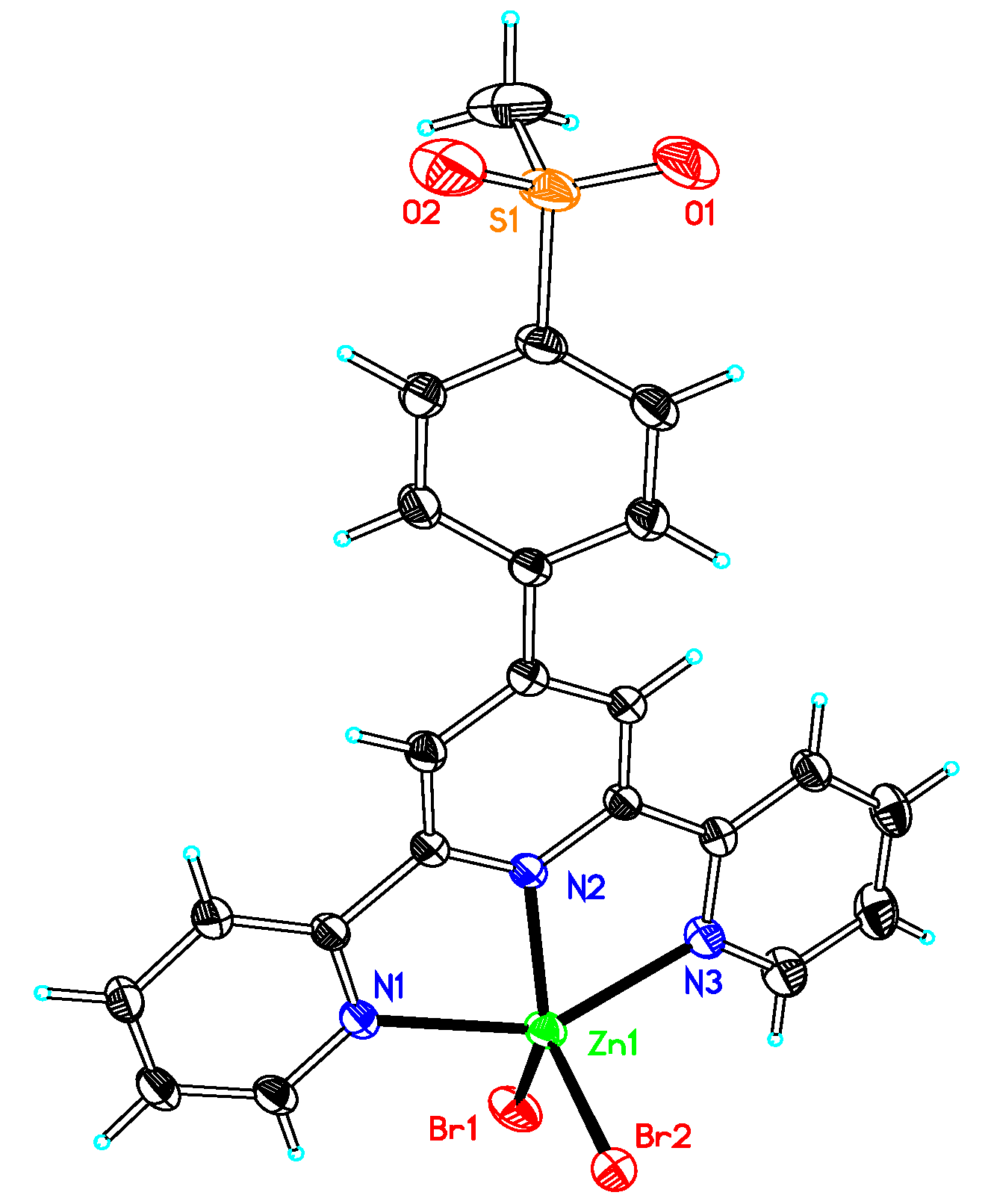
2.2. Elucidation of the Structures of the Compounds
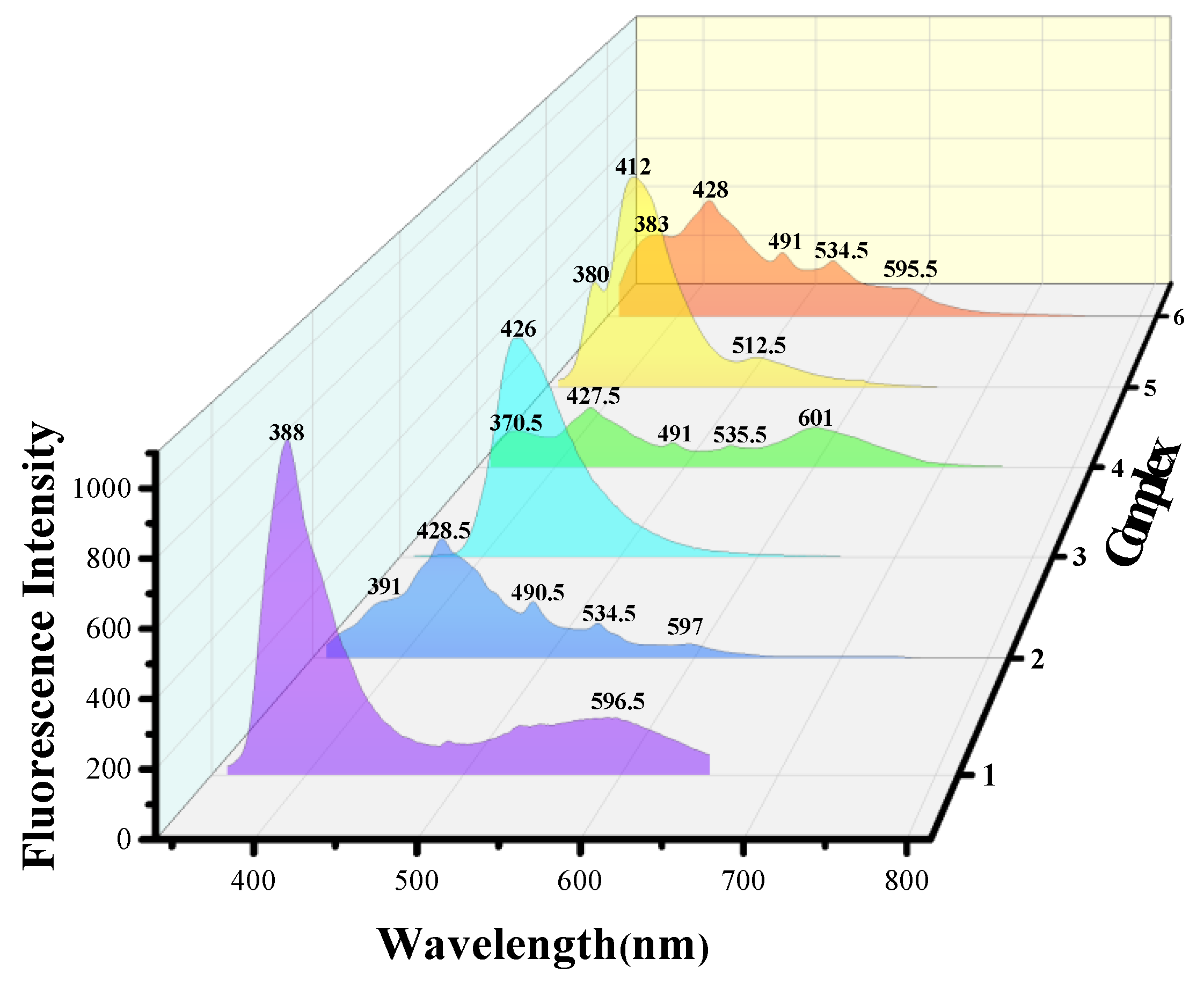
2.3. Photoluminescent Properties
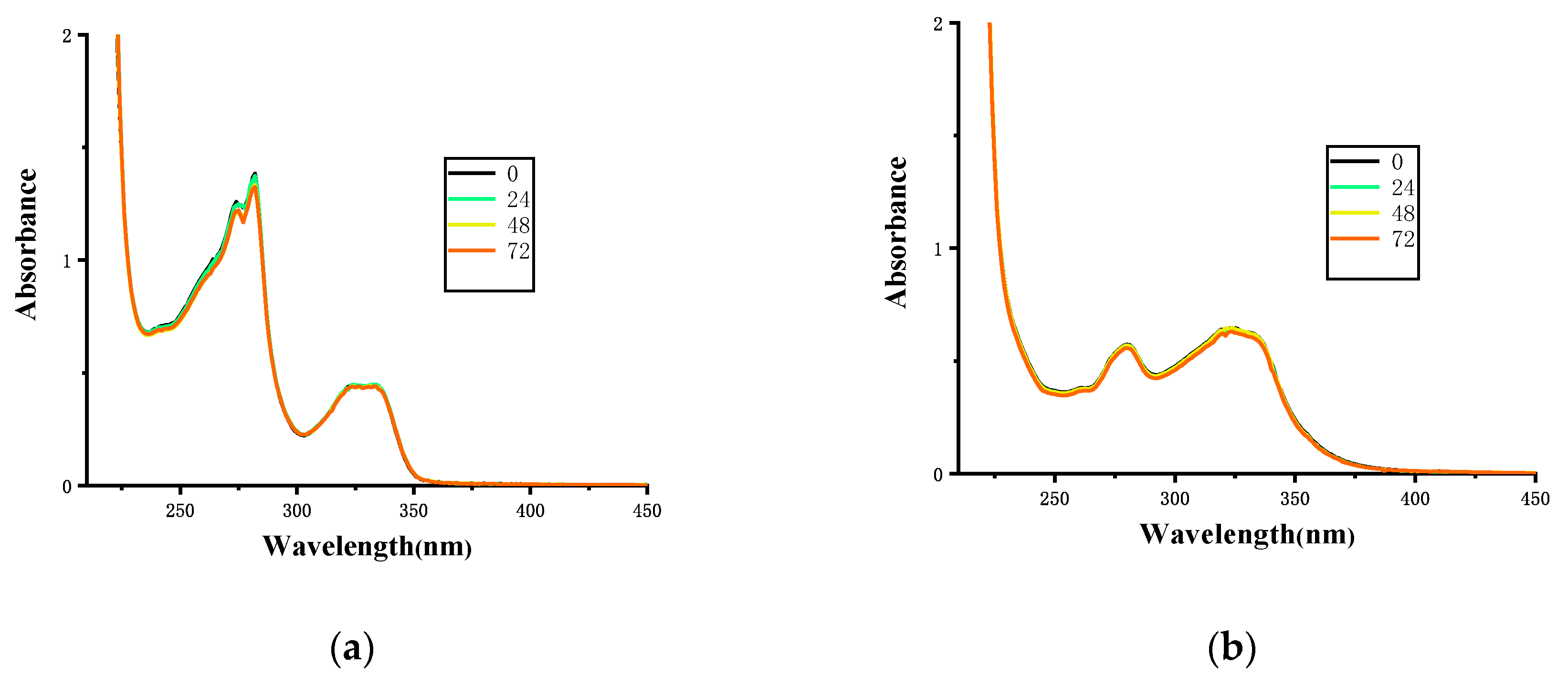
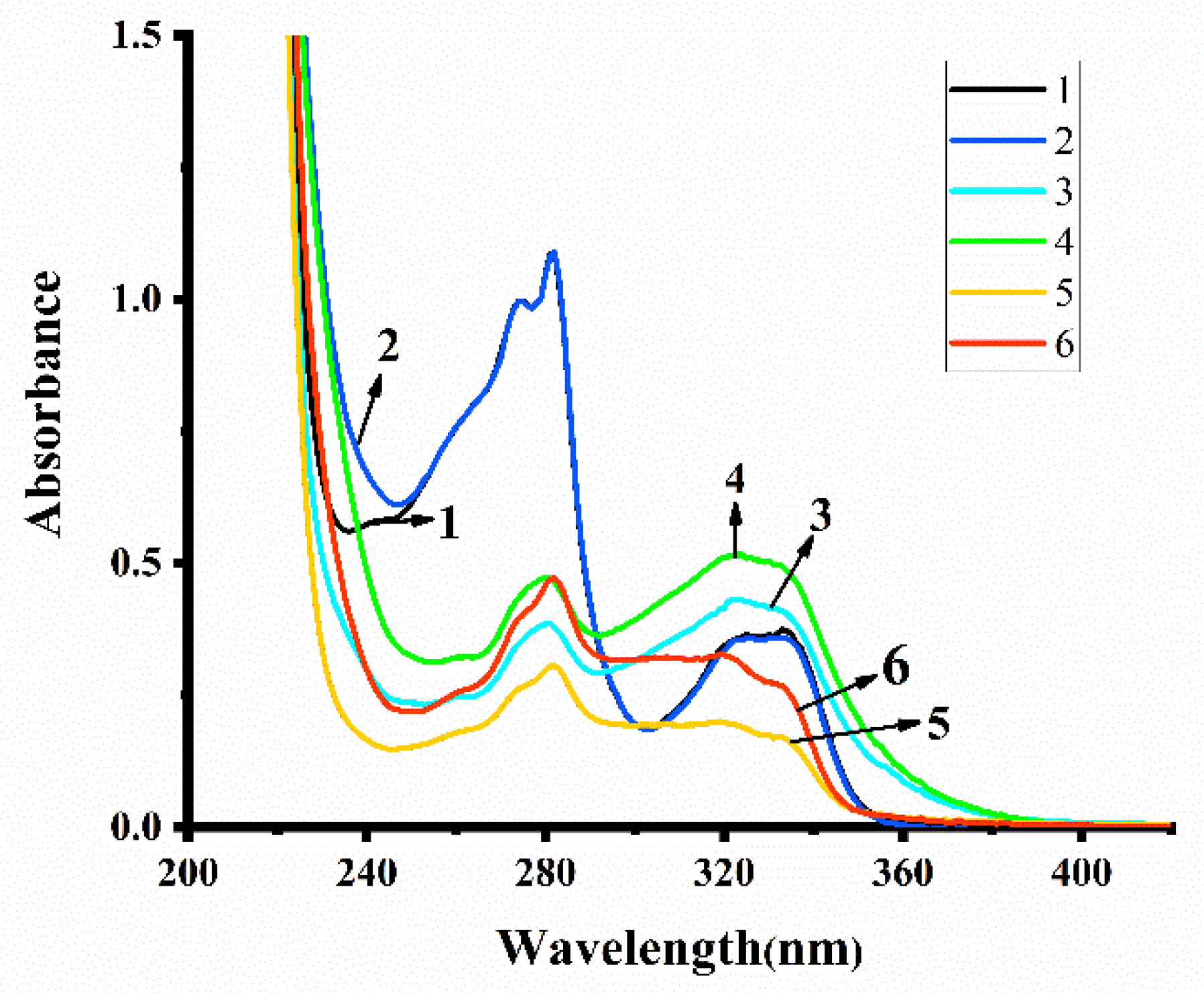
2.4. Solution Stability
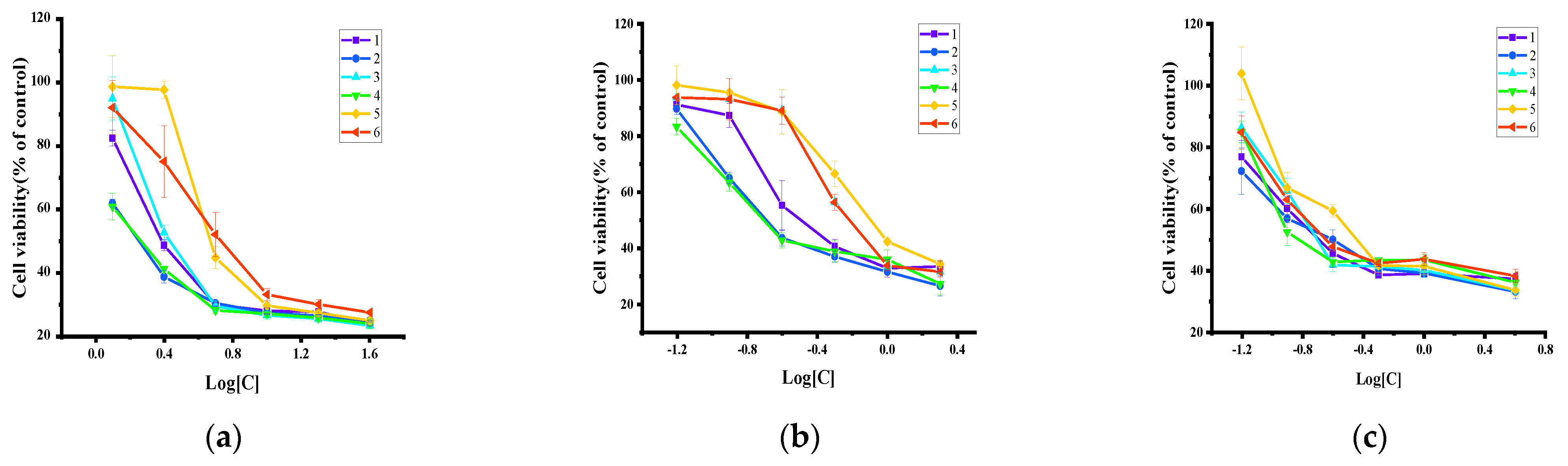
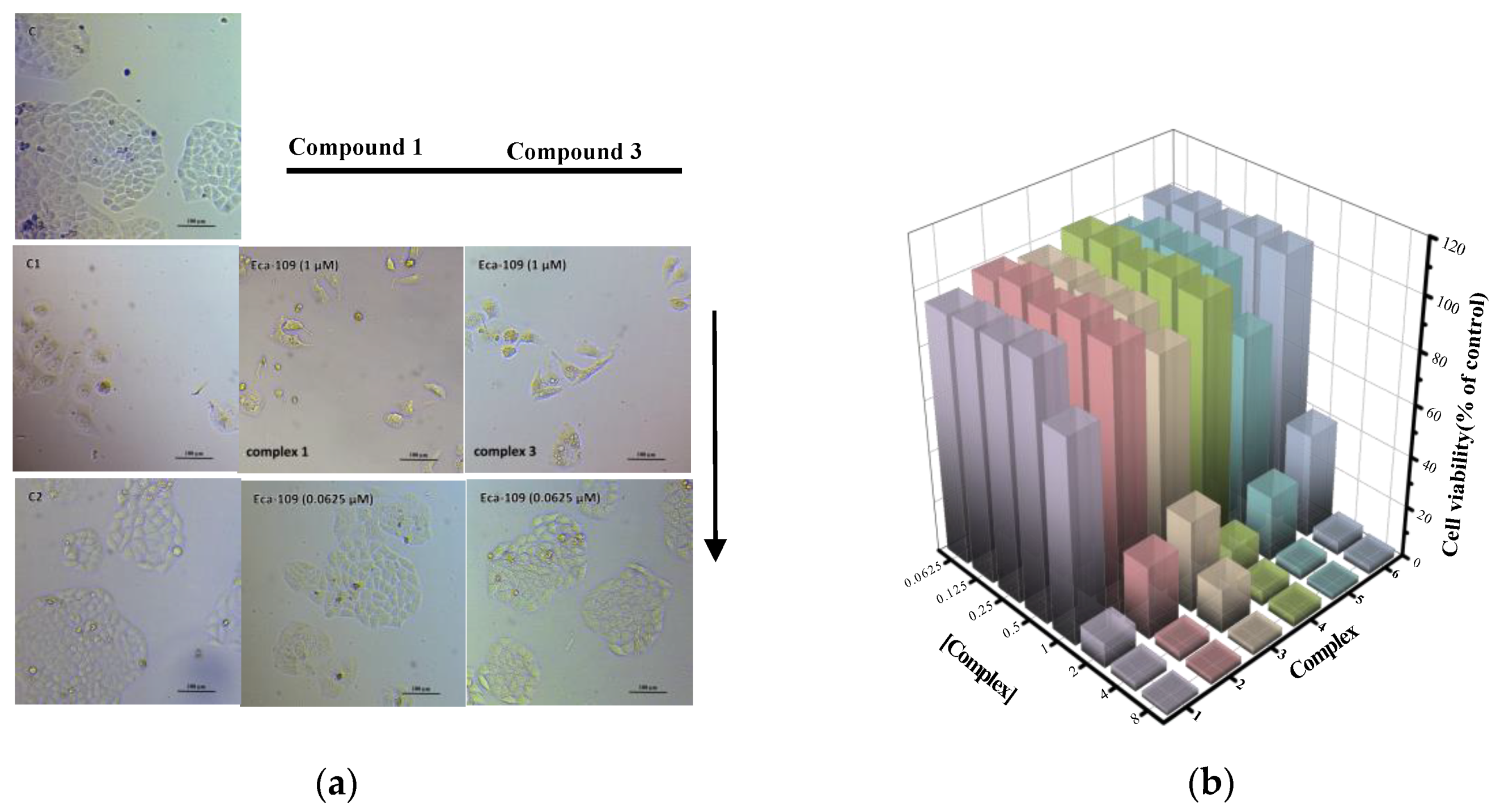
2.5. Antiproliferative Activity and Toxicity Studies
2.6. Protein Binding Analysis
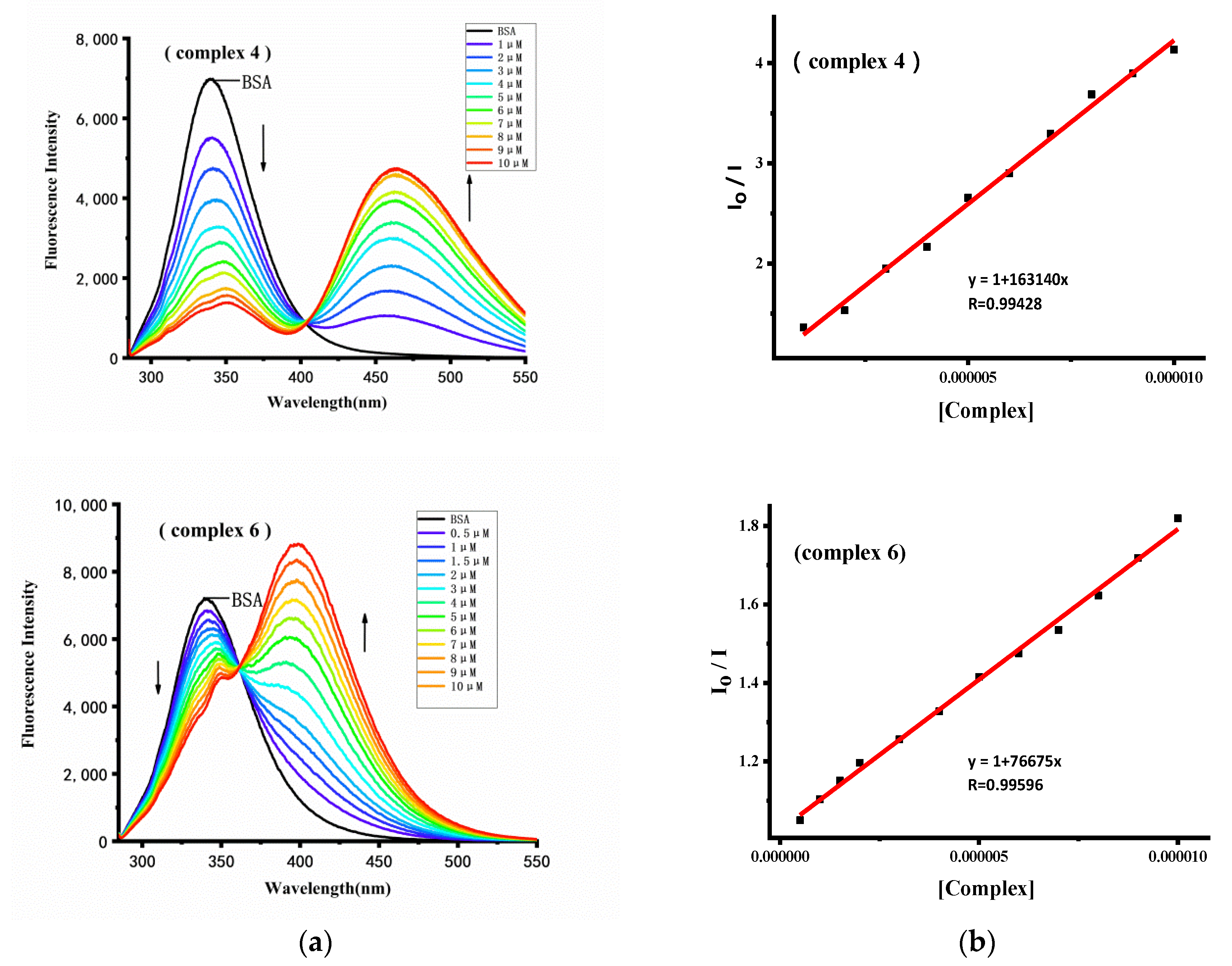
2.6.1. Fluorescence Quenching Analysis
2.6.2. Binding Constant and Site Analysis
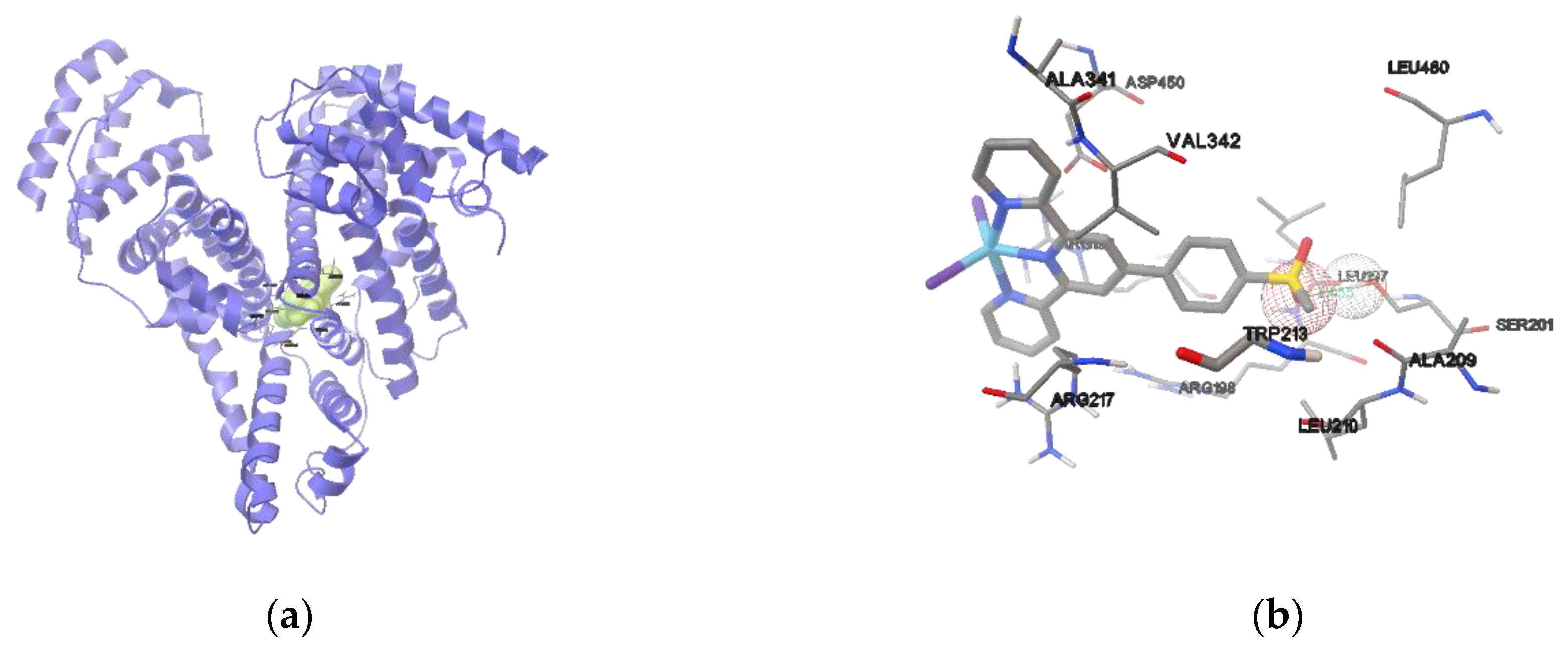
2.6.3. Protein Docking
2.7. DNA Binding Studies
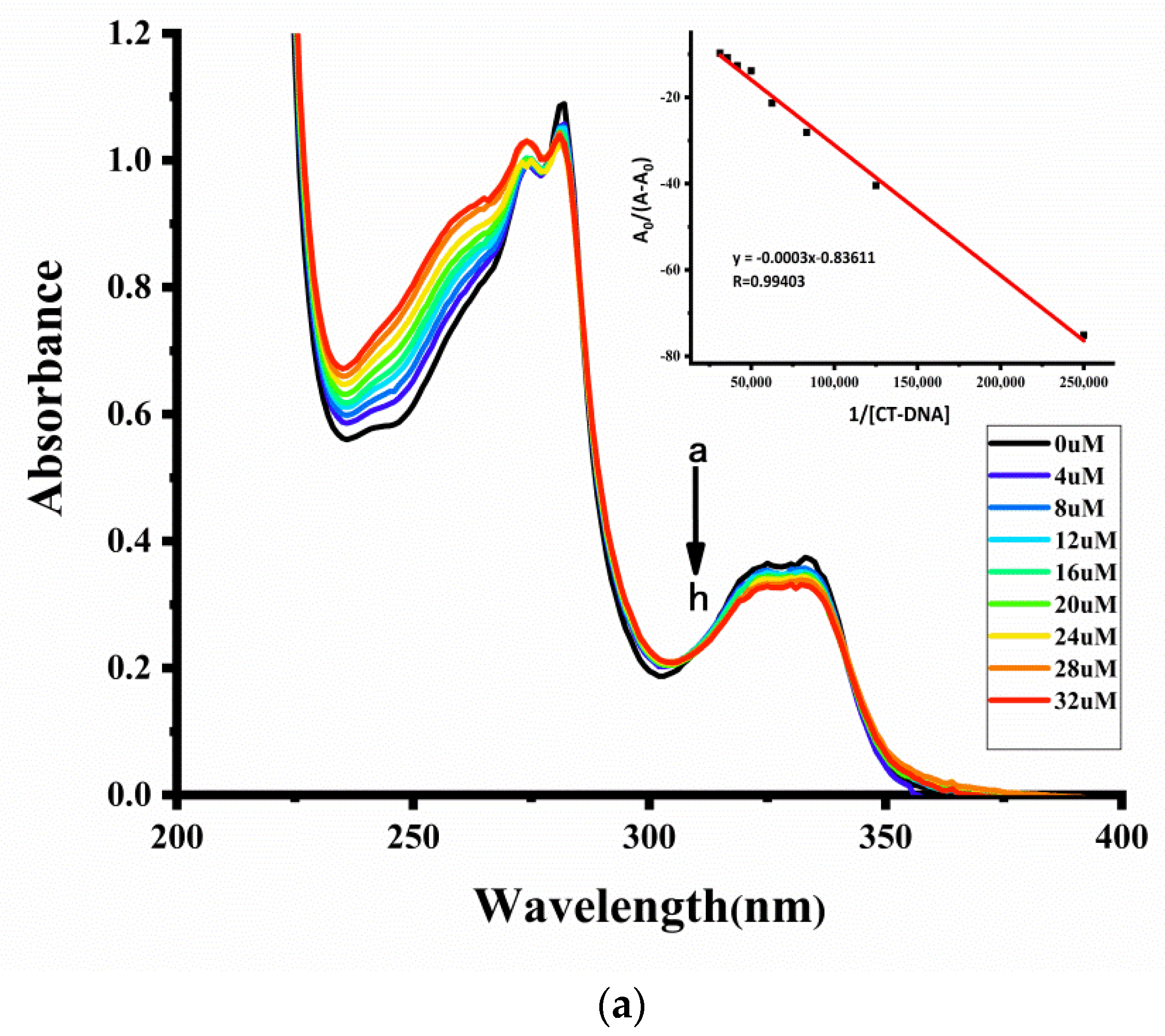
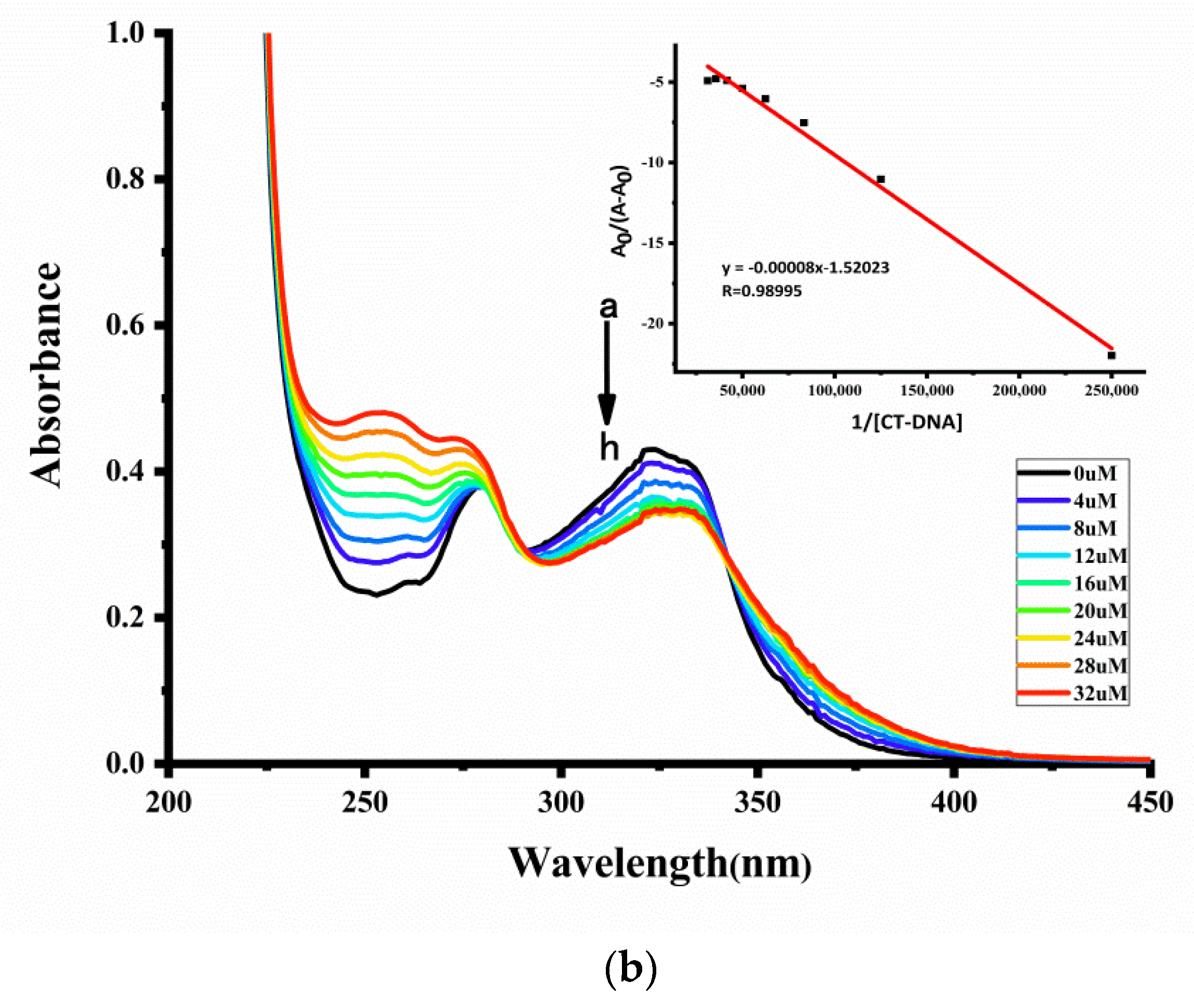
2.7.1. UV-Vis Spectroscopy
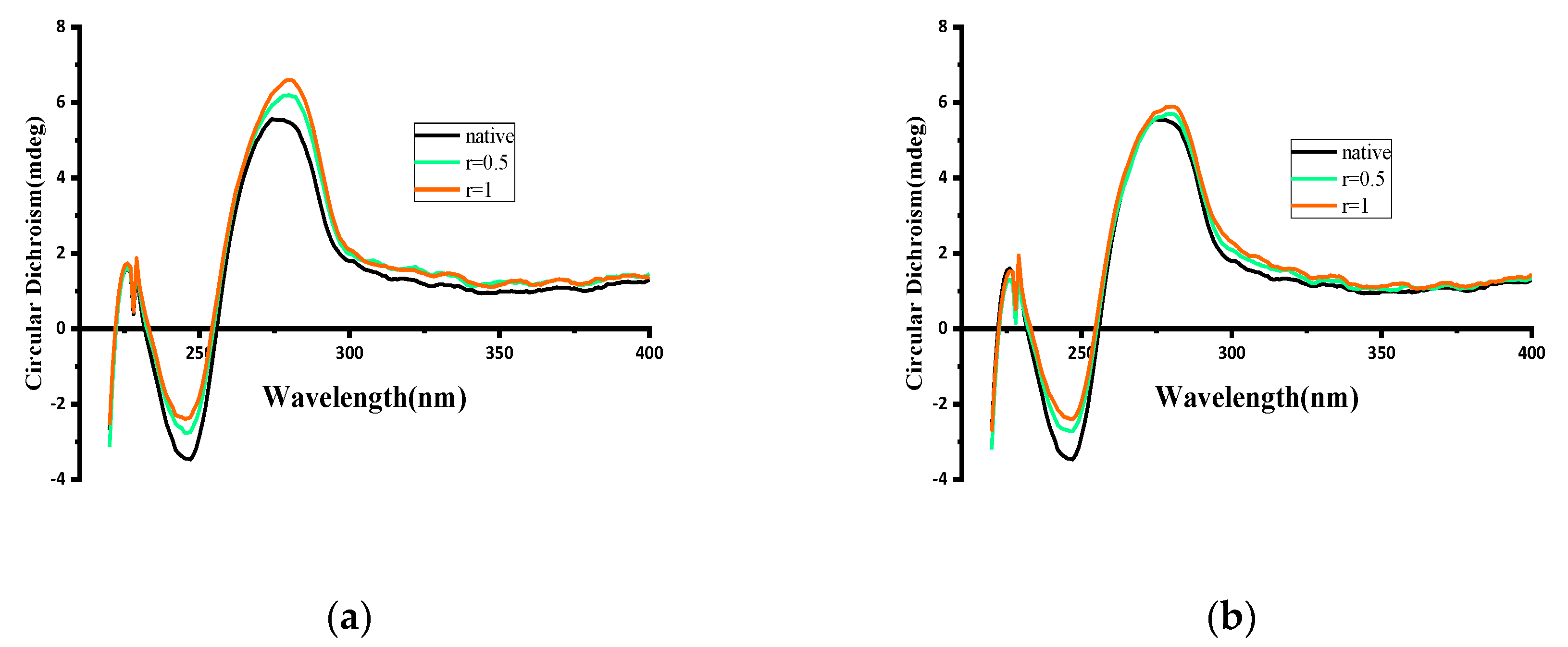
2.7.2. Circular Dichroism Analysis
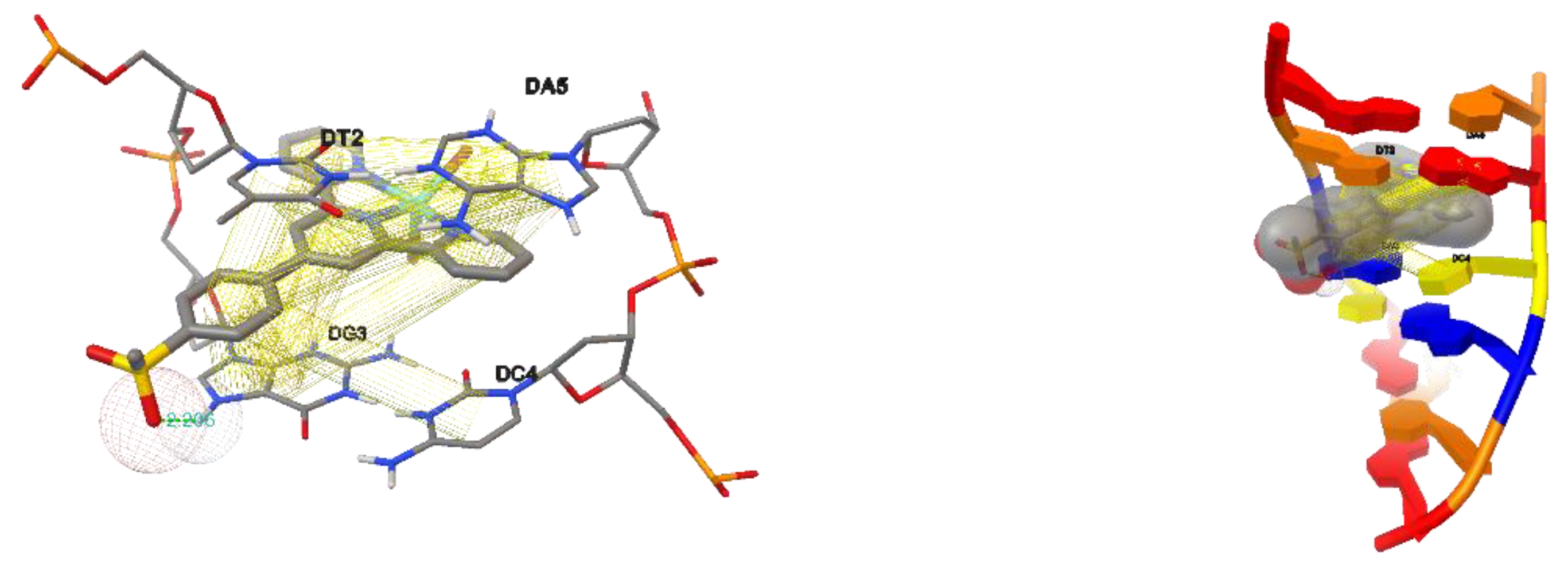
2.7.3. Molecular Docking Studies with CT-DNA
3. Materials and Methods
3.1. Synthesis
3.1.1. [ZnBr2L1] (1)
3.1.2. [ZnI2L1] (2)
3.1.3. [ZnBr2L2] (3)
3.1.4. [ZnI2L2] (4)
3.1.5. [ZnBr2L3] (5)
3.1.6. [ZnI2L3] (6)
3.2. X-ray Determinations
3.3. Solution Chemistry
3.4. Cell Study
3.5. Protein Binding Studies
3.6. DNA Binding Studies
3.6.1. UV-Vis Spectra Titration
3.6.2. Circular Dichroism (CD) Measurements
3.7. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Baig, M.H.; Adil, M.; Khan, R.; Dhadi, S.; Ahmad, K.; Rabbani, G.; Bashir, T.; Imran, M.A.; Husain, F.M.; Lee, E.J.; et al. Enzyme targeting strategies for prevention and treatment of cancer: Implications for cancer therapy. Semin. Cancer Boil. 2019, 56, 1–11. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2018, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2011, 31, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Qu, Y.; Li, C.; Huang, A.; Tong, S.; Wu, C.; Fan, K. Deregulation of lncRNA-AC078883.3 and microRNA-19a is involved in the development of chemoresistance to cisplatin via modulating signaling pathway of PTEN/AKT. J. Cell. Physiol. 2019, 234, 22657–22665. [Google Scholar] [CrossRef]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef]
- Karges, J.; Blacque, O.; Jakubaszek, M.; Goud, B.; Goldner, P.; Gasser, G. Systematic investigation of the antiproliferative activity of a series of ruthenium terpyridine complexes. J. Inorg. Biochem. 2019, 198, 110752. [Google Scholar] [CrossRef]
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorganic Chem. 2019, 88, 102925. [Google Scholar] [CrossRef]
- Hamers, F.; Gispen, W.; Neijt, J. Neurotoxic side-effects of cisplatin. Eur. J. Cancer Clin. Oncol. 1991, 27, 372–376. [Google Scholar] [CrossRef]
- Munteanu, A.-C.; Notaro, A.; Jakubaszek, M.; Cowell, J.; Tharaud, M.; Goud, B.; Uivarosi, V.; Gasser, G. Synthesis, Characterization, Cytotoxic Activity, and Metabolic Studies of Ruthenium(II) Polypyridyl Complexes Containing Flavonoid Ligands. Inorg. Chem. 2020, 59, 4424–4434. [Google Scholar] [CrossRef]
- Huang, W.; Liu, Y.; Wang, J.; Yuan, X.; Jin, H.-W.; Zhang, L.-R.; Zhang, J.-T.; Liu, Z.-M.; Cui, J.-R. Small-molecule compounds targeting the STAT3 DNA-binding domain suppress survival of cisplatin-resistant human ovarian cancer cells by inducing apoptosis. Eur. J. Med. Chem. 2018, 157, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Bruijnincx, P.C.A.; Sadler, P.J. New trends for metal complexes with anticancer activity. Curr. Opin. Chem. Boil. 2008, 12, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.X.; Lippard, S.J. New metal complexes as potential therapeutics. Curr. Opin. Chem. Boil. 2003, 7, 481–489. [Google Scholar] [CrossRef]
- Gezici, S.; Șekeroğlu, N. Current Perspectives in the Application of Medicinal Plants Against Cancer: Novel Therapeutic Agents. Anti-Cancer Agents Med. Chem. 2019, 19, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Barabas, K.; Milner, R.J.; Lurie, D.; Adin, C. Cisplatin: A review of toxicities and therapeutic applications. Vet. Comp. Oncol. 2008, 6, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Cleare, M. Transition metal complexes in cancer chemotherapy. Co-Ord. Chem. Rev. 1974, 12, 349–405. [Google Scholar] [CrossRef]
- Frezza, M.; Hindo, S.; Chen, D.; Davenport, A.; Schmitt, S.; Tomco, D.; Dou, Q.P. Novel metals and metal complexes as platforms for cancer therapy. Curr. Pharm. Des. 2010, 16, 1813–1825. [Google Scholar] [CrossRef]
- Farrell, N. Transition Metal Complexes as Drugs and Chemotherapeutic Agents; Springer Science and Business Media LLC: Berlin, Germany; Kluwer Academic: Dordrecht, The Netherlands, 2012; Volume 11. [Google Scholar]
- Liang, J.-X.; Zhong, H.-J.; Yang, G.; Vellaisamy, K.; Ma, D.-L.; Leung, C.-H. Recent development of transition metal complexes with in vivo antitumor activity. J. Inorg. Biochem. 2017, 177, 276–286. [Google Scholar] [CrossRef]
- Al-Fartusie, F.S.; Mohssan, S.N. Essential trace elements and their vital roles in human body. Indian J. Adv. Chem. Sci. 2017, 5, 127–136. [Google Scholar] [CrossRef]
- Stefanidou, M.E.; Maravelias, C.; Donà, A.; Spiliopoulou, C. Zinc: A multipurpose trace element. Arch. Toxicol. 2005, 80, 1–9. [Google Scholar] [CrossRef]
- Liang, X.; Jiang, J.; Xue, X.; Huang, L.; Ding, X.; Nong, D.; Chen, H.; Pan, L.; Ma, Z. Synthesis, characterization, photoluminescence, anti-tumor activity, DFT calculations and molecular docking with proteins of zinc(ii) halogen substituted terpyridine compounds. Dalton Trans. 2019, 48, 10488–10504. [Google Scholar] [CrossRef]
- Huang, L.; Liu, R.; Li, J.; Liang, X.; Lan, Q.; Shi, X.; Pan, L.; Chen, H.; Ma, Z. Synthesis, characterization, anti-tumor activity, photo-luminescence and BHb/HHb/Hsp90 molecular docking of zinc(II) hydroxyl-terpyridine complexes. J. Inorg. Biochem. 2019, 201, 110790. [Google Scholar] [CrossRef] [PubMed]
- Lazarevic, T.; Rilak, A.; Bugarčić, Ž.D. Platinum, palladium, gold and ruthenium complexes as anticancer agents: Current clinical uses, cytotoxicity studies and future perspectives. Eur. J. Med. Chem. 2017, 142, 8–31. [Google Scholar] [CrossRef] [PubMed]
- Belicchi-Ferrari, M.; Bisceglie, F.; Buschini, A.; Franzoni, S.; Pelosi, G.; Pinelli, S.; Tarasconi, P.; Tavone, M. Synthesis, structural characterization and antiproliferative and toxic bio-activities of copper(II) and nickel(II) citronellal N4-ethylmorpholine thiosemicarbazonates. J. Inorg. Biochem. 2010, 104, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Adhireksan, Z.; Davey, G.E.; Campomanes, P.; Groessl, M.; Clavel, C.M.; Yu, H.; Nazarov, A.A.; Yeo, C.H.F.; Ang, W.H.; Dröge, P.; et al. Ligand substitutions between ruthenium–cymene compounds can control protein versus DNA targeting and anticancer activity. Nat. Commun. 2014, 5, 3462. [Google Scholar] [CrossRef]
- Jain, S.; Khan, T.A.; Patil, Y.P.; Pagariya, D.; Kishore, N.; Tapryal, S.; Naik, A.D.; Naik, S.G. Bio-affinity of copper(II) complexes with nitrogen and oxygen donor ligands: Synthesis, structural studies and in vitro DNA and HSA interaction of copper(II) complexes. J. Photochem. Photobiol. B Boil. 2017, 174, 35–43. [Google Scholar] [CrossRef]
- Farooqi, S.I.; Arshad, N.; Channar, P.A.; Perveen, F.; Saeed, A.; Larik, F.A.; Javeed, A. Synthesis, theoretical, spectroscopic and electrochemical DNA binding investigations of 1, 3, 4-thiadiazole derivatives of ibuprofen and ciprofloxacin: Cancer cell line studies. J. Photochem. Photobiol. B Boil. 2018, 189, 104–118. [Google Scholar] [CrossRef]
- Liedtke, K.R.; Freund, E.; Hermes, M.; Oswald, S.; Heidecke, C.D.; Partecke, L.I.; Bekeschus, S. Gas plasma-conditioned ringer’s lactate enhances the cytotoxic activity of cisplatin and gemcitabine in pancreatic cancer in vitro and in ovo. Cancers 2020, 12, 123. [Google Scholar] [CrossRef]
- Boulikas, T.; Vougiouka, M. Cisplatin and platinum drugs at the molecular level. (Review). Oncol. Rep. 2003, 10, 1663–1682. [Google Scholar] [CrossRef]
- Annaraj, B.; Neelakantan, M.A. Synthesis, crystal structure, spectral characterization and biological exploration of water soluble Cu(II) complexes of vitamin B6 derivative. Eur. J. Med. Chem. 2015, 102, 1–8. [Google Scholar] [CrossRef]
- Ma, Z.; Cao, Y.; Li, Q.; Da Silva, M.F.C.G.; Da Silva, J.J.F.; Pombeiro, A.J. Synthesis, characterization, solid-state photo-luminescence and anti-tumor activity of zinc (II) 4′-phenyl-terpyridine compounds. J. Inorg. Biochem. 2010, 104, 704–711. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Li, J.; Liu, C.; Liu, R.; Liang, X.; Zhou, Y.; Pan, L.; Chen, H.; Ma, Z. Study on the substitution effects of zinc benzoate terpyridine complexes on photoluminescence, antiproliferative potential and DNA binding properties. JBIC J. Boil. Inorg. Chem. 2020, 25, 311–324. [Google Scholar] [CrossRef]
- Li, J.; Liu, R.; Jiang, J.; Liang, X.; Huang, L.; Huang, G.; Chen, H.; Pan, L.; Ma, Z. Zinc(II) Terpyridine Complexes: Substituent Effect on Photoluminescence, Antiproliferative Activity, and DNA Interaction. Molecules 2019, 24, 4519. [Google Scholar] [CrossRef] [PubMed]
- Addison, A.W.; Rao, T.N.; Reedijk, J.; Van Rijn, J.; Verschoor, G.C. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen-sulphur donor ligands; The crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2’-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 11, 1349–1356. [Google Scholar] [CrossRef]
- Sathyadevi, P.; Krishnamoorthy, P.; Bhuvanesh, N.S.; Kalaiselvi, P.; Padma, V.V.; Dharmaraj, N. Organometallic ruthenium(II) complexes: Synthesis, structure and influence of substitution at azomethine carbon towards DNA/BSA binding, radical scavenging and cytotoxicity. Eur. J. Med. Chem. 2012, 55, 420–431. [Google Scholar] [CrossRef]
- Senthil Raja, D.; Bhuvanesh, N.S.P.; Natarajan, K. Effect of N(4)-Phenyl Substitution in 2-Oxo-1,2-dihydroquinoline-3-carbaldehyde Semicarbazones on the Structure, DNA/Protein Interaction, and Antioxidative and Cytotoxic Activity of Cu(II) Complexes. Inorg. Chem. 2011, 50, 12852–12866. [Google Scholar] [CrossRef]
- Wani, T.A.; Bakheit, A.H.; Zargar, S.; Bhat, M.A.; Al-Majed, A.-R.A. Molecular docking and experimental investigation of new indole derivative cyclooxygenase inhibitor to probe its binding mechanism with bovine serum albumin. Bioorg. Chem. 2019, 89, 103010. [Google Scholar] [CrossRef]
- Moghaddam, M.M.; Pirouzi, M.; Saberi, M.R.; Chamani, J. Comparison of the binding behavior of FCCP with HSA and HTF as determined by spectroscopic and molecular modeling techniques. Luminescence 2013, 29, 314–331. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, S.; Tian, Z.; Xu, Z.; Zhang, Y.; Xia, X.; Chen, X.; Liu, Z. Potential anticancer agent for selective damage to mitochondria or lysosomes: Naphthalimide-modified fluorescent biomarker half-sandwich iridium (III) and ruthenium (II) complexes. Eur. J. Med. Chem. 2019, 181, 111599. [Google Scholar] [CrossRef]
- Anjomshoa, M.; Fatemi, S.J.; Torkzadeh-Mahani, M.; Hadadzadeh, H. DNA- and BSA-binding studies and anticancer activity against human breast cancer cells (MCF-7) of the zinc(II) complex coordinated by 5,6-diphenyl-3-(2-pyridyl)-1,2,4-triazine. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 127, 511–520. [Google Scholar] [CrossRef]
- Zhou, P.; Wu, S.; Hegazy, M.; Li, H.; Xu, X.; Lu, H.; Huang, X. Engineered borate ester conjugated protein-polymer nanoconjugates for pH-responsive drug delivery. Mater. Sci. Eng. C 2019, 104, 109914. [Google Scholar] [CrossRef] [PubMed]
- Behnamfar, M.T.; Hadadzadeh, H.; Simpson, J.; Darabi, F.; Shahpiri, A.; Khayamian, T.; Ebrahimi, M.; Rudbari, H.A.; Salimi, M. Experimental and molecular modeling studies of the interaction of the polypyridyl Fe(II) and Fe(III) complexes with DNA and BSA. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 134, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Heydari, A.; Mansouri-Torshizi, H. Design, synthesis, characterization, cytotoxicity, molecular docking and analysis of binding interactions of novel acetylacetonatopalladium(ii) alanine and valine complexes with CT-DNA and BSA. RSC Adv. 2016, 6, 96121–96137. [Google Scholar] [CrossRef]
- Ali, M.S.; Al-Lohedan, H.A. Deciphering the interaction of procaine with bovine serum albumin and elucidation of binding site: A multi spectroscopic and molecular docking study. J. Mol. Liq. 2017, 236, 232–240. [Google Scholar] [CrossRef]
- Shi, J.-H.; Pan, D.-Q.; Jiang, M.; Liu, T.-T.; Wang, Q. Binding interaction of ramipril with bovine serum albumin (BSA): Insights from multi-spectroscopy and molecular docking methods. J. Photochem. Photobiol. B Boil. 2016, 164, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Khan, I.A.; Banipal, P.K.; Banipal, T.S. Deciphering the complexation process of a fluoroquinolone antibiotic, levofloxacin, with bovine serum albumin in the presence of additives. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 191, 259–270. [Google Scholar] [CrossRef]
- Alsaeedi, M.S.; Babgi, B.A.; Hussien, M.; Abdellattif, M.H.; Humphrey, M.G. DNA-Binding and Anticancer Activity of Binuclear Gold(I) Alkynyl Complexes with a Phenanthrenyl Bridging Ligand. Molecules 2020, 25, 1033. [Google Scholar] [CrossRef]
- Esfahani, N.H.; Salami, F.; Saberi, Z.; Karami, K.; Lighvan, Z.M.; Ramezani, M.; Alibolandi, M.; Farzad, S.A.; Khayamian, T. DNA G-quadruplexes binding and antitumor activity of palladium aryl oxime ligand complexes encapsulated in either albumin or algal cellulose nanoparticles. Colloids Surf. B Biointerfaces 2019, 176, 70–79. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug–DNA interactions and their study by UV–Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Boil. 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Kellett, A.; Molphy, Z.; Slator, C.; McKee, V.; Farrell, N.P. Molecular methods for assessment of non-covalent metallodrug–DNA interactions. Chem. Soc. Rev. 2019, 48, 971–988. [Google Scholar] [CrossRef]
- Draksharapu, A.; Boersma, A.J.; Leising, M.; Meetsma, A.; Browne, W.R.; Roelfes, G. Binding of copper(ii) polypyridyl complexes to DNA and consequences for DNA-based asymmetric catalysis. Dalton Trans. 2015, 44, 3647–3655. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.G.; De Almeida, S.M.V.; De Oliveira, J.F.; Souza, T.R.C.D.L.; Dos Santos, K.L.; Albuquerque, A.P.D.B.; Nogueira, M.C.D.B.L.; Junior, L.B.D.C.; De Moura, R.O.; Da Silva, A.C.; et al. Novel 4-quinoline-thiosemicarbazone derivatives: Synthesis, antiproliferative activity, in vitro and in silico biomacromolecule interaction studies and topoisomerase inhibition. Eur. J. Med. Chem. 2019, 182, 111592. [Google Scholar] [CrossRef] [PubMed]
- Meenongwa, A.; Brissos, R.F.; Soikum, C.; Chaveerach, P.; Gamez, P.; Trongpanich, Y.; Chaveerach, U. Effects of N, N-heterocyclic ligands on the in vitro cytotoxicity and DNA interactions of copper (II) chloride complexes from amidino-O-methylurea ligands. New. J. Chem. 2016, 40, 5861–5876. [Google Scholar] [CrossRef]
- Pagadala, N.S.; Syed, K.; Tuszynski, J.A. Software for molecular docking: A review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Mariappan, M.; Alagarsamy, R.; Panneerselvam, A.P.; Anbazhagan, V.; Rajendran, S.; Arunachalam, J. Synthesis, solvatochromism, photochemistry, DNA binding, photocleavage, cytotoxicity and molecular docking studies of a ruthenium(II) complex bearing photoactive subunit. J. Photochem. Photobiol. A Chem. 2018, 356, 617–626. [Google Scholar] [CrossRef]
- Ibezim, A.; Debnath, B.; Ntie-Kang, F.; Mbah, C.J.; Nwodo, N.J. Binding of anti-Trypanosoma natural products from African flora against selected drug targets: A docking study. Med. Chem. Res. 2017, 26, 562–579. [Google Scholar] [CrossRef]
- CrysAlis PRO. Agilent; Agilent Technologies: Yarnton, UK, 2012. [Google Scholar]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2007, 64, 112–122. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1–6 are available from the authors. |
| Compound | Eca-109 | A549 | Bel-7042 | |||
|---|---|---|---|---|---|---|
| IC50 (μM) | 95% Confidence | IC50 (μM) | 95% Confidence | IC50 (μM) | 95% Confidence | |
| Intervals (μM) | Intervals (μM) | Intervals (μM) | ||||
| 1 | 0.079 | 0.0533–0.1188 | 0.309 | 0.6419–0.8715 | 0.358 | 0.3468–0.7850 |
| 2 | 0.066 | 0.0464–0.0946 | 0.290 | 0.3822–0.6072 | 0.331 | 0.2434–0.5271 |
| 3 | 0.169 | 0.1246–0.2311 | 0.482 | 0.2465–0.3824 | 0.414 | 0.2817–0.6089 |
| 4 | 0.075 | 0.0503–0.1144 | 0.307 | 0.2501–0.3819 | 0.397 | 0.2408–0.6553 |
| 5 | 0.197 | 0.1449–0.2693 | 0.748 | 0.2072–0.4068 | 0.616 | 0.3681–1.032 |
| 6 | 0.146 | 0.1208–0.1780 | 0.945 | 0.6751–0.9521 | 0.522 | 0.2533–0.4325 |
| cisplatin | 5.465 | 4.982–5.994 | 11.99 | 10.79–13.32 | 3.088 | 2.856–3.340 |
| Compound | Ksv (M−1) | Kq (M−1) | Kb (M−1) | n | Ra |
|---|---|---|---|---|---|
| 1 | 1.76 × 105 | 1.76 × 1013 | 4.47 × 103 | 0.72 | 0.9783 |
| 2 | 1.76 × 105 | 1.76 × 1013 | 1.80 × 104 | 0.83 | 0.9989 |
| 3 | 1.15 × 105 | 1.15 × 1013 | 2.95 × 105 | 1.07 | 0.9919 |
| 4 | 1.63 × 105 | 1.63 × 1013 | 3.80 × 105 | 1.08 | 0.9943 |
| 5 | 4.63 × 104 | 4.63 × 1012 | 1.07 × 104 | 0.87 | 0.9946 |
| 6 | 7.67 × 104 | 7.67 × 1012 | 2.24 × 104 | 0.89 | 0.9960 |
| Compound | BSA n | DNA(4JD8) |
|---|---|---|
| 1 | −7.69 | −7.90 |
| 2 | −8.28 | −8.65 |
| 3 | −7.00 | −8.42 |
| 4 | −6.99 | −8.76 |
| 5 | −7.26 | −8.51 |
| 6 | −7.79 | −8.78 |
| Compound | Kb (M−1) n | Ra |
|---|---|---|
| 1 | 2.79 × 103 | 0.9940 |
| 2 | 2.47 × 103 | 0.9906 |
| 3 | 1.90 × 104 | 0.9900 |
| 4 | 2.15 × 104 | 0.9924 |
| 5 | 6.63 × 104 | 0.9931 |
| 6 | 6.69 × 104 | 0.9974 |
| Compound | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Empirical formula | C44H38Br4N6O6S2Zn2 | C24H23I2N3O3S2Zn | C22H17Br2N3OZn | C22H17I2N3OZn | C22H17Br2N3Zn | C22H17I2N3Zn |
| Formula weight | 1261.3 | 784.74 | 564.58 | 658.56 | 548.58 | 642.56 |
| Temperature | 293(2) K | 293(2) K | 293(2) K | 571(2) K | 571(2) K | 571(2) K |
| Crystal system | Monoclinic | Orthorhombic | Monoclinic | Monoclinic | Monoclinic | Monoclinic |
| space group | C2/c | P212121 | P21/c | P21/n | P21/c | P21/c |
| a (Ǻ) | 13.2530(5) | 11.748(2) | 8.2757(2) | 8.8978(2) | 8.23930(10) | 8.1062(2) |
| b (Ǻ) | 14.4141(5) | 20.209(3) | 14.8951(5) | 15.9409(3) | 14.9859(2) | 15.5081(3) |
| c (Ǻ) | 24.6133(9) | 23.075(4) | 17.1200(6) | 15.8601(3) | 16.7323(3) | 17.3032(4) |
| α (°) | 90 | 90 | 90 | 90 | 90 | 90 |
| β (°) | 95.604(3) | 90 | 98.939(2) | 105.1540(10) | 97.2180(10) | 96.9060(10) |
| γ (°) | 90 | 90 | 90 | 90 | 90 | 90 |
| Volume (Ǻ3) | 4679.4(3) | 5478.7(16) | 2084.71(11) | 2171.35(8) | 2049.62(5) | 2159.43(8) |
| Z | 4 | 8 | 4 | 4 | 4 | 4 |
| Calculated density (Mg/m3) | 1.79 | 1.903 | 1.799 | 2.015 | 1.778 | 1.976 |
| Absorption coefficient (mm−1) | 4.584 | 3.334 | 5.03 | 3.993 | 5.11 | 4.009 |
| F (000) | 2496 | 3040 | 1112 | 1256 | 1080 | 1224 |
| Crystal size (mm−1) | 0.36 × 0.30 × 0.22 | 0.49 × 0.11 × 0.09 | 0.39 × 0.36 × 0.30 | 0.48 × 0.43 × 0.40 | 0.39 × 0.30 × 0.21 | 0.26 × 0.25 × 0.18 |
| θmax, θmin (°) | 2.95, 27.50 | 2.80, 29.69 | 2.73, 27.15 | 1.84, 27.12 | 5.52, 26.63 | 2.37, 27.19 |
| Index range h | −16→17 | −16→17 | −10→10 | −11→11 | −9→10 | −10→10 |
| k | −18→15 | −27→27 | −19→15 | −19→20 | −13→18 | −19→17 |
| l | −31→30 | −31→29 | −14→21 | −20→20 | −20→21 | −22→18 |
| Reflections collected/unique | 13958/5198 | 74225/14185 | 14640/4523 | 29741/4806 | 9325/4150 | 18433/4788 |
| [R(int) = 0.0236] | [R(int) = 0.0297] | [R(int) = 0.0309] | [R(int) = 0.0264] | [R(int) = 0.0541] | [R(int) = 0.0251] | |
| Data/restraints/parameters | 5198/0/290 | 14185/0/631 | 4523/0/262 | 4806/0/262 | 4150/0/253 | 4788/0/253 |
| Goodness-of-fit on F2 | 1.012 | 1.021 | 1.017 | 1.013 | 0.943 | 0.994 |
| Final R indices [I > 2σ(I)] | R1 = 0.0320 | R1 = 0.0317 | R1 = 0.0280 | R1 = 0.0250 | R1 = 0.0344 | R1 = 0.0309 |
| wR2 = 0.0889 | wR2 = 0.0777 | wR2 = 0.0657 | wR2 = 0.0650 | wR2 = 0.0630 | wR2 = 0.0825 | |
| R indices (all data) | R1 = 0.0386 | R1 = 0.0427 | R1 = 0.0445 | R1 = 0.0294 | R1 = 0.0602 | R1 = 0.0409 |
| wR2 = 0.0931 | wR2 = 0.0852 | wR2 = 0.0716 | wR2 = 0.0677 | wR2 = 0.0660 | wR2 = 0.0880 | |
| Largest diff. peak and hole (e Ǻ−3) | 0.939 and −0.562 | 1.580 and −0.806 | 0.703 and −0.448 | 0.395 and −0.920 | 0.965 and −0.565 | 0.807 and −0.822 |
| CCDC number | 1969820 | 1969821 | 1969822 | 1969823 | 1969824 | 1969825 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Yan, H.; Jiang, J.; Li, J.; Liang, X.; Yang, D.; Pan, L.; Xie, T.; Ma, Z. Synthesis, Characterization, Photoluminescence, Molecular Docking and Bioactivity of Zinc (II) Compounds Based on Different Substituents. Molecules 2020, 25, 3459. https://doi.org/10.3390/molecules25153459
Liu R, Yan H, Jiang J, Li J, Liang X, Yang D, Pan L, Xie T, Ma Z. Synthesis, Characterization, Photoluminescence, Molecular Docking and Bioactivity of Zinc (II) Compounds Based on Different Substituents. Molecules. 2020; 25(15):3459. https://doi.org/10.3390/molecules25153459
Chicago/Turabian StyleLiu, Rongping, Hao Yan, Jinzhang Jiang, Jiahe Li, Xing Liang, Dengfeng Yang, Lixia Pan, Tisan Xie, and Zhen Ma. 2020. "Synthesis, Characterization, Photoluminescence, Molecular Docking and Bioactivity of Zinc (II) Compounds Based on Different Substituents" Molecules 25, no. 15: 3459. https://doi.org/10.3390/molecules25153459
APA StyleLiu, R., Yan, H., Jiang, J., Li, J., Liang, X., Yang, D., Pan, L., Xie, T., & Ma, Z. (2020). Synthesis, Characterization, Photoluminescence, Molecular Docking and Bioactivity of Zinc (II) Compounds Based on Different Substituents. Molecules, 25(15), 3459. https://doi.org/10.3390/molecules25153459