Dipeptidyl Peptidase-IV Inhibitory Activity and Related Molecular Mechanism of Bovine α-Lactalbumin-Derived Peptides
Abstract
1. Introduction
2. Results
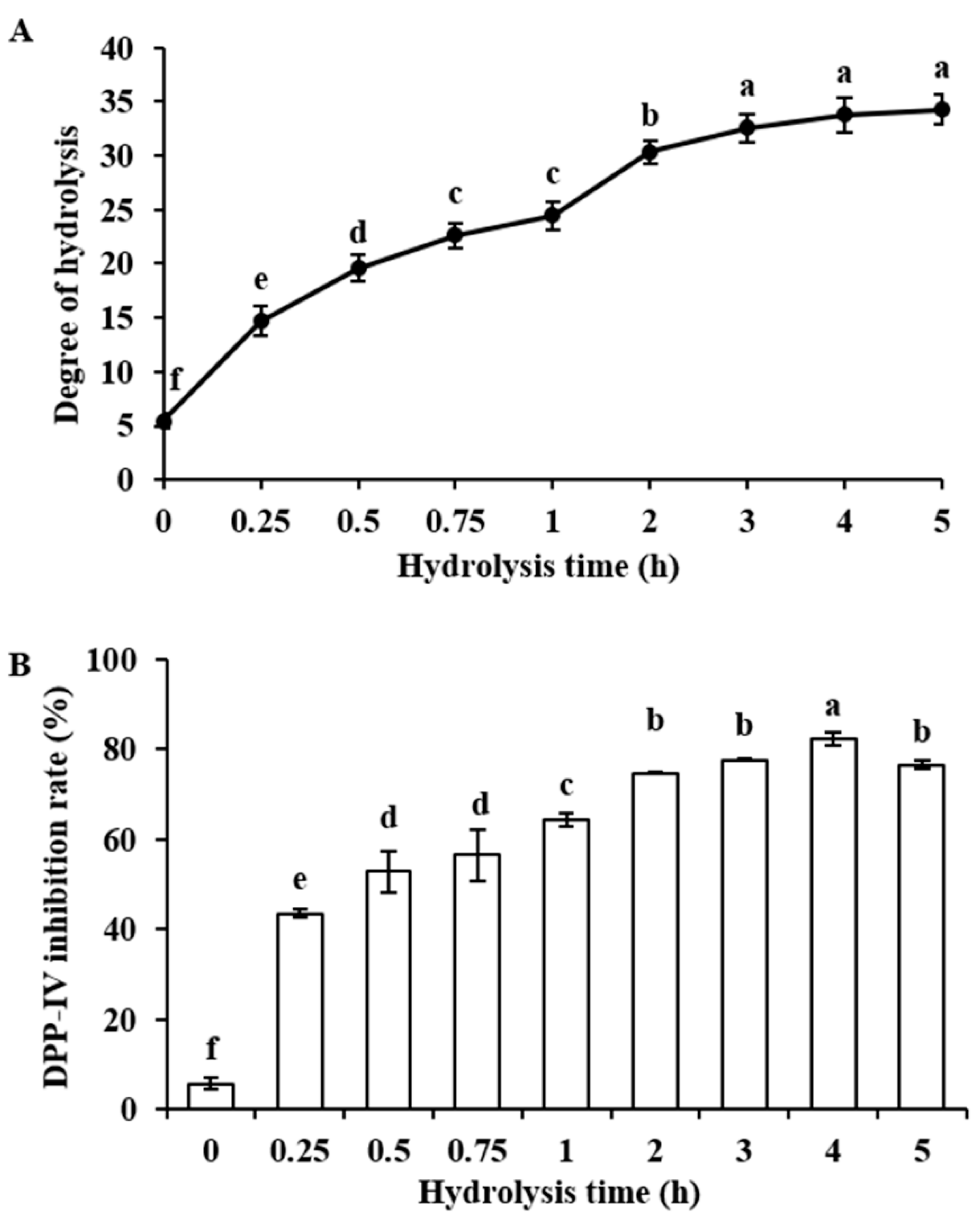
2.1. Degree of Hydrolysis and DPP-IV Inhibitory Activity of Bovine α-Lactalbumin Hydrolysates
2.2. DPP-IV Inhibitory Activity of Bovine α-Lactalbumin Hydrolysates Fractionated by Sephadex G-25 Gel Filtration Chromatography
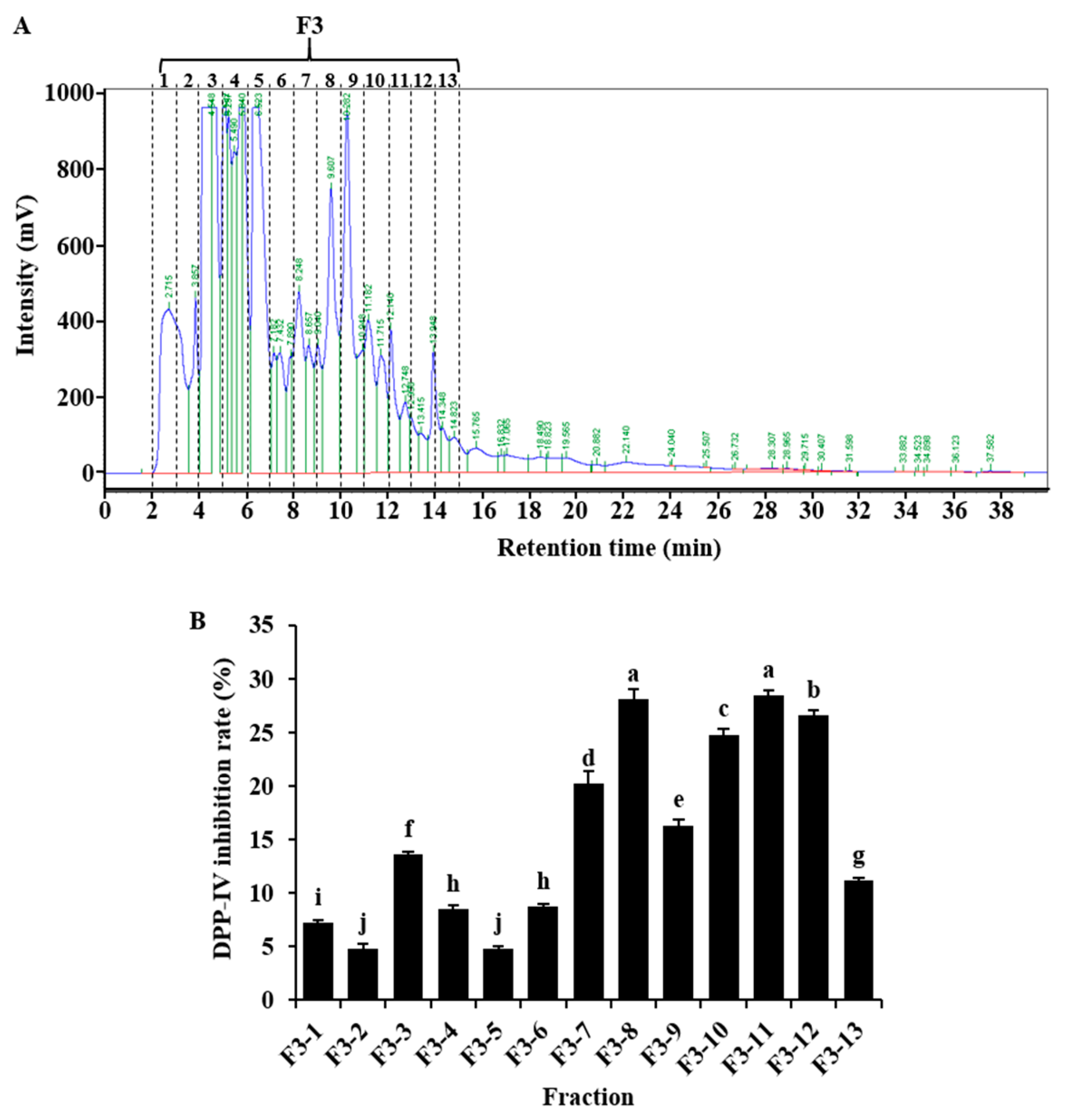
2.3. DPP-IV Inhibitory Activity of F3 Fractionated by RP-HPLC
2.4. Identification of DPP-IV Inhibitory Peptides
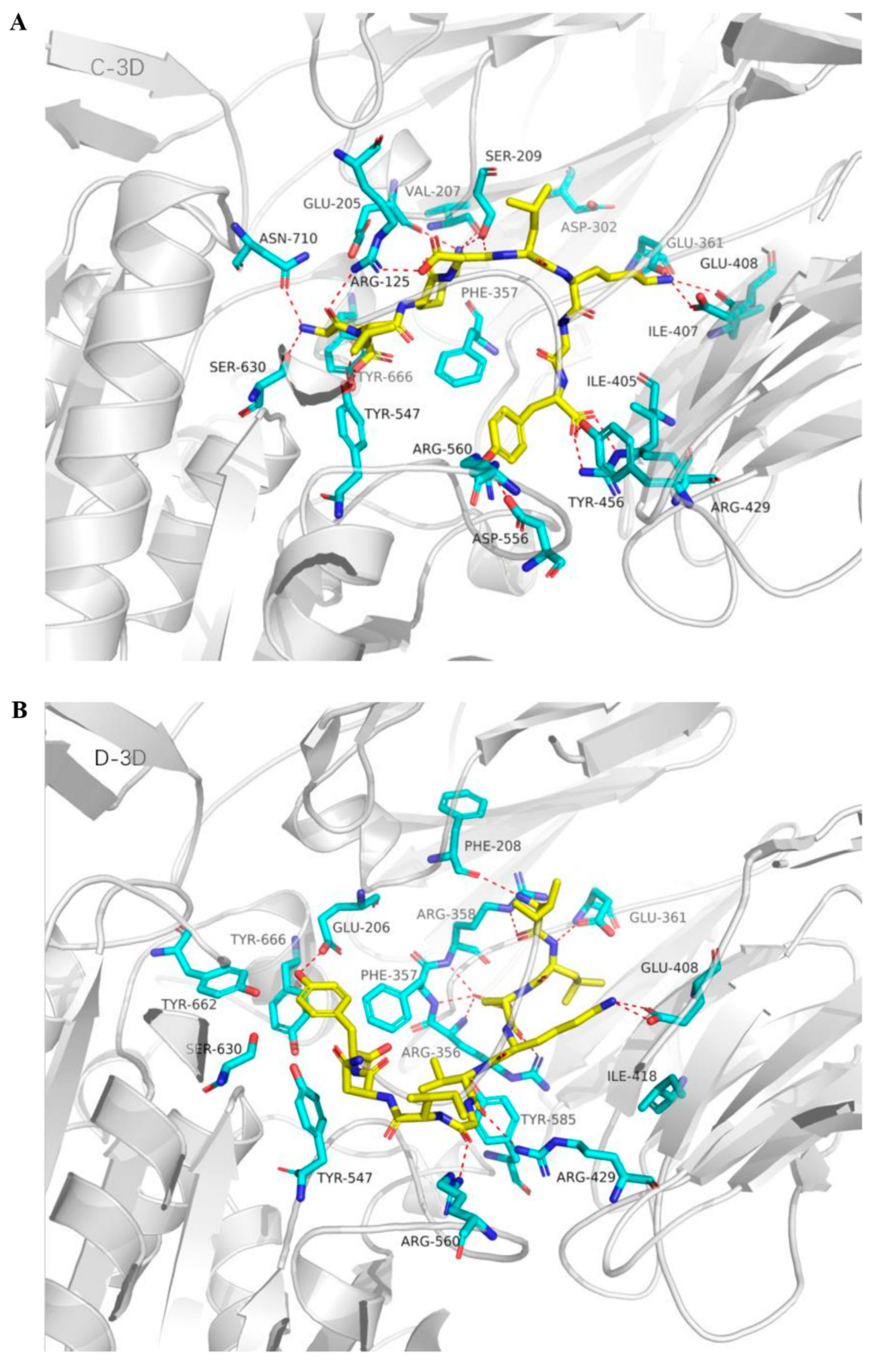
2.5. Molecular Docking Analysis of Bovine α-Lactalbumin Derived Peptides to DPP-IV
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of Bovine α-Lactalbumin Hydrolysates
4.3. Determination of Degree of Hydrolysis
4.4. Determination of DPP-IV Inhibitory Activity
4.5. Gel Filtration Chromatography
4.6. Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC)
4.7. Identification of Peptide Sequence by Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry (LC-ESI MS/MS)
4.8. Molecular Docking
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- International Diabetes Federation. IDF Diabetes Atlas, 9th ed.; International Diabetes Federation: Brussels, Belgium, 2019. [Google Scholar]
- Chatterjee, S.; Khunti, K.; Davies, M.J. Type 2 diabetes. Lancet 2017, 389, 2239–2251. [Google Scholar] [CrossRef]
- Lima, R.D.L.; Berg, R.S.; Ronning, S.B.; Afseth, N.K.; Knutsen, S.H.; Staerk, D.; Wubshet, S.G. Peptides from chicken processing by-product inhibit DPP-IV and promote cellular glucose uptake: Potential ingredients for T2D management. Food Funct. 2019, 10, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- Juillerat-Jeanneret, L. Dipeptidyl peptidase IV and its inhibitors: Therapeutics for type 2 diabetes and what else? J. Med. Chem. 2014, 57, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Andersen, E.S.; Deacon, C.F.; Holst, J.J. Do we know the true mechanism of action of the DPP-4 inhibitors? Diabetes Obes. Meta. 2018, 20, 34–41. [Google Scholar] [CrossRef]
- Poucher, S.M.; Cheetham, S.; Francis, J.; Zinker, B.; Kirby, M.; Vickers, S.P. Effects of sitagliptin on glycaemic control and pancreatic beta-cell mass in a streptozotocin-induced mouse model of type 2 diabetes. Diabetes Obes. Meta. 2012, 14, 918–926. [Google Scholar] [CrossRef]
- Li, N.; Wang, L.J.; Jiang, B.; Li, X.Q.; Guo, C.L.; Guo, S.J.; Shi, D.Y. Recent progress of the development of dipeptidyl peptidase-4 inhibitors for the treatment of type 2 diabetes mellitus. Eur. J. Med. Chem. 2018, 151, 145–157. [Google Scholar] [CrossRef]
- Akarte, A.S.; Srinivasan, B.P.; Gandhi, S. Vildagliptin selectively ameliorates GLP-1, GLUT4, SREBP-1c mRNA levels and stimulates beta-Cell proliferation resulting in improved glucose homeostasis in rats with streptozotocin-induced diabetes. J. Diabetes Complicat. 2012, 26, 266–274. [Google Scholar] [CrossRef]
- Hirukawa, H.; Kaneto, H.; Shimoda, M.; Kimura, T.; Okauchi, S.; Obata, A.; Kohara, K.; Hamamoto, S.; Tawaramoto, K.; Hashiramoto, M.; et al. Combination of DPP-4 inhibitor and PPAR gamma agonist exerts protective effects on pancreatic beta-cells in diabetic db/db mice through the augmentation of IRS-2 expression. Mol. Cell. Endocrinol. 2015, 413, 49–60. [Google Scholar] [CrossRef]
- Kern, M.; Kloting, N.; Niessen, H.G.; Thomas, L.; Stiller, D.; Mark, M.; Klein, T.; Bluher, M. Linagliptin improves insulin sensitivity and hepatic steatosis in diet-induced obesity. PLoS ONE 2012, 7, e38744. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Z.; Huang, Y.; Wang, J. Effects of chronic administration of alogliptin on the development of diabetes and β-cell function in high fat diet/streptozotocin diabetic mice. Diabetes Obes. Metab. 2011, 13, 337–347. [Google Scholar] [CrossRef]
- Chen, X.W.; He, Z.X.; Zhou, Z.W.; Yang, T.X.; Zhang, X.J.; Yang, Y.X.; Duan, W.; Zhou, S.F. An update on the clinical pharmacology of the dipeptidyl peptidase 4 inhibitor alogliptin used for the treatment of type 2 diabetes mellitus. Clin. Exp. Pharmacol. Physiol. 2015, 42, 1225–1238. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.H.; Wang, T.Y.; Huang, C.C.; Jao, C.L.; Hsieh, Y.L.; Wu, S.X.; Hsu, K.C. In silico, in vitro and in vivo analyses of dipeptidyl peptidase IV inhibitory activity and the antidiabetic effect of sodium caseinate hydrolysate. Food Funct. 2016, 7, 1122–1128. [Google Scholar] [CrossRef] [PubMed]
- Harnedy, P.A.; O’Keeffe, M.B.; FitzGerald, R.J. Purification and identification of dipeptidyl peptidase (DPP) IV inhibitory peptides from the macroalga Palmaria palmata. Food Chem. 2015, 172, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.Y.; Hsieh, C.H.; Hung, C.C.; Jao, C.L.; Lin, P.Y.; Hsieh, Y.L.; Hsu, K.C. A study to evaluate the potential of an in silico approach for predicting dipeptidyl peptidase-IV inhibitory activity in vitro of protein hydrolysates. Food Chem. 2017, 234, 431–438. [Google Scholar] [CrossRef]
- Mudgil, P.; Jobe, B.; Kamal, H.; Alameri, M.; Al Ahbabi, N.; Maqsood, S. Dipeptidyl peptidase-IV, alpha-amylase, and angiotensin I converting enzyme inhibitory properties of novel camel skin gelatin hydrolysates. LWT-Food Sci. Technol. 2019, 101, 251–258. [Google Scholar] [CrossRef]
- Lin, Y.S.; Han, C.H.; Lin, S.Y.; Hou, W.C. Synthesized peptides from yam dioscorin hydrolysis in silico exhibit dipeptidyl peptidase-IV inhibitory activities and oral glucose tolerance improvements in normal mice. J. Agric. Food Chem. 2016, 64, 6451–6458. [Google Scholar] [CrossRef]
- Lamas, E.M.; Barros, R.M.; Balcao, V.M.; Malcata, F.X. Hydrolysis of whey proteins by proteases extracted from Cynara cardunculus and immobilized onto highly activated supports. Enzyme Microb. Tech. 2001, 28, 642–652. [Google Scholar] [CrossRef]
- Layman, D.K.; Lonnerdal, B.; Fernstrom, J.D. Applications for alpha-lactalbumin in human nutrition. Nutr. Rev. 2018, 76, 444–460. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Evaluation of the potential of dietary proteins as precursors of dipeptidyl peptidase (DPP)- IV inhibitors by an in silico approach. J. Funct. Foods 2012, 4, 403–422. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Inhibition of dipeptidyl peptidase (DPP)- IV and alpha-glucosidase activities by pepsin-treated whey proteins. J. Agric. Food Chem. 2013, 61, 7500–7506. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Isolation and characterization of peptides with dipeptidyl peptidase-IV inhibitory activity from pepsin-treated bovine whey proteins. Peptides 2014, 54, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; FitzGerald, R.J. An in silico model to predict the potential of dietary proteins as sources of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem. 2014, 165, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; Le Maux, S.; Hamayon, J.; FitzGerald, R.J. Strategies for the release of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides in an enzymatic hydrolyzate of alpha-lactalbumin. Food Funct. 2016, 7, 3437–3443. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Song, J.J.; Du, M.; Mao, X.Y. Bovine alpha-lactalbumin hydrolysates (alpha-LAH) ameliorate adipose insulin resistance and inflammation in high-fat diet-fed C57BL/6J mice. Nutrients 2018, 10, 242. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Song, J.J.; Du, M.; Mao, X.Y. Bovine alpha-lactalbumin hydrolysates (alpha-LAH) attenuate high-fat diet induced nonalcoholic fatty liver disease by modulating hepatic lipid metabolism in C57BL/6J mice. J. Funct. Foods 2019, 54, 254–262. [Google Scholar] [CrossRef]
- Hwang, H.J.; Jung, T.W.; Kim, B.H.; Hong, H.C.; Seo, J.A.; Kim, S.G.; Kim, N.H.; Choi, K.M.; Choi, D.S.; Baik, S.H.; et al. A dipeptidyl peptidase-IV inhibitor improves hepatic steatosis and insulin resistance by AMPK-dependent and JNK-dependent inhibition of LECT2 expression. Biochem. Pharmacol. 2015, 98, 157–166. [Google Scholar] [CrossRef]
- Nakamura, K.; Fukunishi, S.; Yokohama, K.; Ohama, H.; Tsuchimoto, Y.; Asai, A.; Tsuda, Y.; Higuchi, K. A long-lasting dipeptidyl peptidase-4 inhibitor, teneligliptin, as a preventive drug for the development of hepatic steatosis in high-fructose diet-fed ob/ob mice. Int. J. Mol. Med. 2017, 39, 969–983. [Google Scholar] [CrossRef]
- Nauck, M. Incretin therapies: Highlighting common features and differences in the modes of action of glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors. Diabetes Obes. Metab. 2016, 18, 203–216. [Google Scholar] [CrossRef]
- Silveira, S.T.; Martinez-Maqueda, D.; Recio, I.; Hernandez-Ledesma, B. Dipeptidyl peptidase-IV inhibitory peptides generated by tryptic hydrolysis of a whey protein concentrate rich in beta-lactoglobulin. Food Chem. 2013, 141, 1072–1077. [Google Scholar] [CrossRef]
- Hutch, C.R.; Roelofs, K.; Haller, A.; Sorrell, J.; Leix, K.; D’Alessio, D.D.; Augustin, R.; Seeley, R.J.; Klein, T.; Sandoval, D.A. The role of GIP and pancreatic GLP-1 in the glucoregulatory effect of DPP-4 inhibition in mice. Diabetologia 2019, 62, 1928–1937. [Google Scholar] [CrossRef]
- Xie, C.; Wang, X.Y.; Jones, K.L.; Horowitz, M.; Sun, Z.L.; Littele, T.J.; Rayner, C.K.; Wu, T.Z. Role of endogenous glucagon-like peptide-1 enhanced by vildagliptin in the glycaemic and energy expenditure responses to intraduodenal fat infusion in type 2 diabetes. Diabetes Obes. Metab. 2019, 22, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Li-Chan, E.C.Y.; Hunag, S.L.; Jao, C.L.; Ho, K.P.; Hsu, K.C. Peptides derived from atlantic salmon skin gelatin as dipeptidyl-peptidase IV inhibitors. J. Agric. Food Chem. 2012, 60, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Zhang, C.; Ji, H. Purification, identification and molecular mechanism of two dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from Antarctic krill (Euphausia superba) protein hydrolysate. J. Chromatogr. B. 2017, 1064, 56–61. [Google Scholar] [CrossRef]
- Wang, L.Y.; Ding, L.; Du, Z.Y.; Yu, Z.P.; Liu, J.B. Hydrolysis and transport of egg white-derived peptides in Caco-2 cell monolayers and everted rat sacs. J. Agric. Food Chem. 2019, 67, 4839–4848. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhou, L.; Zhang, Y.; Sheng, N.J.; Wang, Z.K.; Wu, T.Z.; Wang, T.Z.; Wu, H. Rapid identification of dipeptidyl peptidase-IV (DPP-IV) inhibitory peptides from Ruditapes philippinarum hydrolysates. Molecules 2017, 22, 1714. [Google Scholar] [CrossRef] [PubMed]
- Utomo, D.H.; Widodo, N.; Rifa’I, M. Identifications small molecules inhibitor of p53-mortalin complex for cancer drug using virtual screening. Bioinformation 2012, 8, 426–429. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Paolella, S.; Mudgil, P.; Maqsood, S.; FitzGerald, R.J. Identification of novel dipeptidyl peptidase IV (DPP-IV) inhibitory peptides in camel milk protein hydrolysates. Food Chem. 2018, 244, 340–348. [Google Scholar] [CrossRef]
- Huang, S.L.; Jao, C.L.; Ho, K.P.; Hsu, K.C. Dipeptidyl-peptidase IV inhibitory activity of peptides derived from tuna cooking juice hydrolysates. Peptides 2012, 35, 114–121. [Google Scholar] [CrossRef]
- Farzaneh, P.; Ehsani, M.R.; Khanahmadi, M.; Sharifan, A. Characterization of bio-peptides purified from Terfezia claveryi hydrolysates and their antibacterial effect on raw milk. LWT-Food Sci. Technol. 2019, 116, 108522. [Google Scholar] [CrossRef]
- Khueychai, S.; Jangpromma, N.; Choowongkomon, K.; Joompang, A.; Daduang, S.; Vesaratchavest, M.; Payoungkiattikun, W.; Tachibana, S.; Klaynongsruang, S. A novel ACE inhibitory peptide derived from alkaline hydrolysis of ostrich (Struthio camelus) egg white ovalbumin. Process. Biochem. 2018, 73, 235–245. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, R.; Ma, H.Q.; Chen, S.W. Isolation and identification of dipeptidyl peptidase IV-inhibitory peptides from trypsin/chymotrypsin-treated goat milk casein hydrolysates by 2D-TLC and LC-MS/MS. J. Agric. Food Chem. 2015, 63, 8819–8828. [Google Scholar] [CrossRef] [PubMed]




| Sequence | Calculated Mass (Da) | Origin |
|---|---|---|
| F3-8 | ||
| ELKDLKGY | 929.19 | α-LA f (30–37) |
| F3-11 | ||
| ILDKVGINY | 1016.77 | α-LA f (114–122) |
| Peptide (Ligand) | Target | Binding Energy Score (kcal/mol) |
|---|---|---|
| ELKDLKGY | DPP-IV | −7.771 |
| ILDKVGINY | DPP-IV | −8.037 |
| Peptide | Peptide Residue | Receptor Residue | Type |
|---|---|---|---|
| ELKDLKGY | E1 | Arg125 | H-bond |
| E1 | Ser630 | H-bond | |
| E1 | Asn710 | H-bond | |
| E1 | Tyr547 | H-bond | |
| E1 | Tyr666 | H-bond | |
| E1 | Tyr666 | Pi-cation | |
| K3 | Ser209 | H-bond | |
| K3 | Val207 | H-bond | |
| K3 | Glu205 | H-bond | |
| D4 | Ser209 | H-bond | |
| D4 | Arg125 | H-bond | |
| D4 | Arg125 | Salt bridge | |
| K6 | Glu361 | H-bond | |
| K6 | Glu408 | H-bond | |
| K6 | Glu361 | Salt bridge | |
| K6 | Glu408 | Salt bridge | |
| Y8 | Arg429 | H-bond | |
| Y8 | Arg429 | Salt bridge | |
| Y8 | Tyr456 | H-bond | |
| Y8 | Asp556 | H-bond | |
| ILDKVGINY | I1 | Arg358 | Pi-cation |
| D3 | Arg356 | H-bond | |
| D3 | Arg357 | H-bond | |
| D3 | Arg358 | H-bond | |
| K4 | Glu408 | H-bond | |
| K4 | Glu408 | Salt bridge | |
| V5 | Arg429 | H-bond | |
| G6 | Arg560 | H-bond | |
| Y9 | Arg125 | Pi-cation |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Gong, H.; Mao, X. Dipeptidyl Peptidase-IV Inhibitory Activity and Related Molecular Mechanism of Bovine α-Lactalbumin-Derived Peptides. Molecules 2020, 25, 3009. https://doi.org/10.3390/molecules25133009
Gao J, Gong H, Mao X. Dipeptidyl Peptidase-IV Inhibitory Activity and Related Molecular Mechanism of Bovine α-Lactalbumin-Derived Peptides. Molecules. 2020; 25(13):3009. https://doi.org/10.3390/molecules25133009
Chicago/Turabian StyleGao, Jing, Han Gong, and Xueying Mao. 2020. "Dipeptidyl Peptidase-IV Inhibitory Activity and Related Molecular Mechanism of Bovine α-Lactalbumin-Derived Peptides" Molecules 25, no. 13: 3009. https://doi.org/10.3390/molecules25133009
APA StyleGao, J., Gong, H., & Mao, X. (2020). Dipeptidyl Peptidase-IV Inhibitory Activity and Related Molecular Mechanism of Bovine α-Lactalbumin-Derived Peptides. Molecules, 25(13), 3009. https://doi.org/10.3390/molecules25133009





