1. Introduction
Wound healing complications after trauma, surgery, acute illness, or chronic disease conditions, and subsequent appearance of chronic wounds, affect millions of people worldwide each year. Delayed healing is often caused by dysregulation of the response to wounding resulting from inflammation, angiogenesis, matrix deposition, and/or cell recruitment. The inability to restore the function and architecture of injured tissues is typical of chronic skin wounds resulting from thermal, chemical, or radiation burns of a large area, as well as civilization diseases such as diabetes or atherosclerosis. The need for clinical strategies that might activate natural repair mechanisms leading to scarless wound healing and tissue repair is still growing [
1]. One of the possible strategies is pharmacological stimulation of natural regenerative processes in the body. Peptides, which are biocompatible, biodegradable, bioactive, and relatively amenable to large-scale production, are currently gaining interest as potential agents in regenerative medicine. Among many biologically active molecules is the RDKVYR peptide, which is known as thymohexin and marketed as Imunofan (IM). IM is a hydrophilic hexapeptide which was designed based on the sequence at positions 32–37 of the thymopoietin hormone (RKDVYV) (information from the manufacturer’s website). Thymopoietin is produced by the thymus and its function is to increase lymphopoiesis and to selectively induce differentiation of prothymocytes and thymocytes [
2]. Studies have shown that certain thymus hormones stimulate regeneration of hematopoietic progenitor cells damaged by ionizing radiation [
3].
IM has been reported to demonstrate antioxidative and immunomodulatory properties. The IM peptide, in combination with L-arginine, exerts protective activity in epinephrine- and indomethacin-induced ulcers and gastric lesions in rats [
4], where it decreases the activity of inducible nitric oxide synthase (iNOS) as well as the lipoperoxidation process in gastric mucosa. After subcutaneous injection in patients, IM showed immunomodulatory effects, as manifested by decreasing levels of interleukin-6 (IL-6) and tumor necrosis factor (TNF) in response to diphtheria toxoid vaccination [
5].
IM causes ultrastructural changes of thyrotropic cells of the adenohypophysis, mainly by increasing their size [
6]. It has been proven that application of IM in combination with ozone therapy and surgical treatment can cause improvements in healing processes in orthopedic patients affected by puruloseptic complications [
7]. It is also known that stimulation of the immune system by IM, in combination with local electrolyte detoxification, promotes endometrium regeneration in puerperal endometritis. In these studies, migration of lymphoblasts and phagocytes was detected at the endometrial level, thus illustrating endometrium regeneration (Dorina Muntean, PhD thesis “Local electrochemical detoxication and immunomodulation in prophylaxis and treatment of puerperal endometritis”, 2006). In another in vivo experiment, IM displayed a positive effect on cell profiles in the thymus (“Immunomodulator Imunofan affects cell profile of morphofunctional zones of rat thymus and delays its age-related involution“—data published in Russian). The obtained results indicated a delay in thymic involution. The summarized data from the literature show that the IM peptide stimulates immune system activity and it possesses ability to activate the antioxidative system of the human body.
The described above data about pro-regenerative therapies with IM as a supplement and reported immunomodulatory activity of IM, added to the lack of observed side effects [
5], encouraged us to investigate whether the peptide has the potential to promote tissue repair and skin wound healing using in vitro and in vivo models. In addition, we performed conformational studies of the IM peptide, in order to search for the structural properties associated with its biological activity.
3. Discussion
Chronic wounds are one of the problems associated with civilization diseases, which creates a demand for finding new therapeutics for promoting skin wound healing. Diabetes, cancers, congenital defects, or postoperative wounds create a mortal threat. Regenerative medicine is trying to develop solutions to these challenges. Therapies are still being sought which could accelerate normal physical responses to injury which might be limited or non-existent in patients with these conditions. Due to the complexity of regeneration processes, multidisciplinary approaches combining cell and molecular biology, genetics, biomaterial sciences, chemistry, bioengineering, and tissue engineering are needed. Peptides appear to display properties that could be useful in promoting skin regeneration processes. They may stimulate fibroblasts to proliferate and to produce elastin and collagen. A good example of a pro-regenerative peptide is a thrombin derived peptide, TP508, also known as Chrysalin (OrthoLogic, Tempe, Arizona—23 amino acid residues), which has been reported to accelerate wound healing [
14,
15]. Another example is the GHK tripeptide, which participates in the activation of processes related to skin remodeling, and also in activation of fibroblasts leading to the synthesis of collagen and elastin in the human body [
16,
17]. Further examples of peptides reported to display pro-regenerative properties are Tylotoin [
18] and the AH90 peptide [
19,
20].
In this study a small, hexapeptide, with sequence RDKVYR known as Imunofan, has been tested for potential regenerative properties. IM has been reported to exert an immunomodulatory activity [
5] that encouraged us to consider it as an agent for promoting wound healing and tissue repair. For this purpose, we investigated the physicochemical and structural properties of this peptide and characterized its effects on processes related to wound healing, such as proliferation and migration in cultured skin cells. In addition, we performed immunogenicity and cytotoxicity tests on IM and analyzed transcriptional responses to IM stimulation. In order to assess whether IM can promote tissue repair, we examined the results of subcutaneous IM administration in the ear pinna injury model in mice.
Our studies confirm that this peptide is stable in water. We also demonstrated that IM rapidly disappeared in plasma where it was probably bound to albumin, as indicated by affinity tests we carried out. Albumin is a serum protein known to transport large amounts of hormones (e.g., cortisol) [
21], drugs (e.g., antibiotics) [
22], as well as vitamins [
23], fatty acids [
24], and lipids [
25]. However, the fact that IM is rapidly bound by albumin, is not a proof that IM can be transported to the wound area. To verify this, additional experiments are needed beyond the scope of this manuscript.
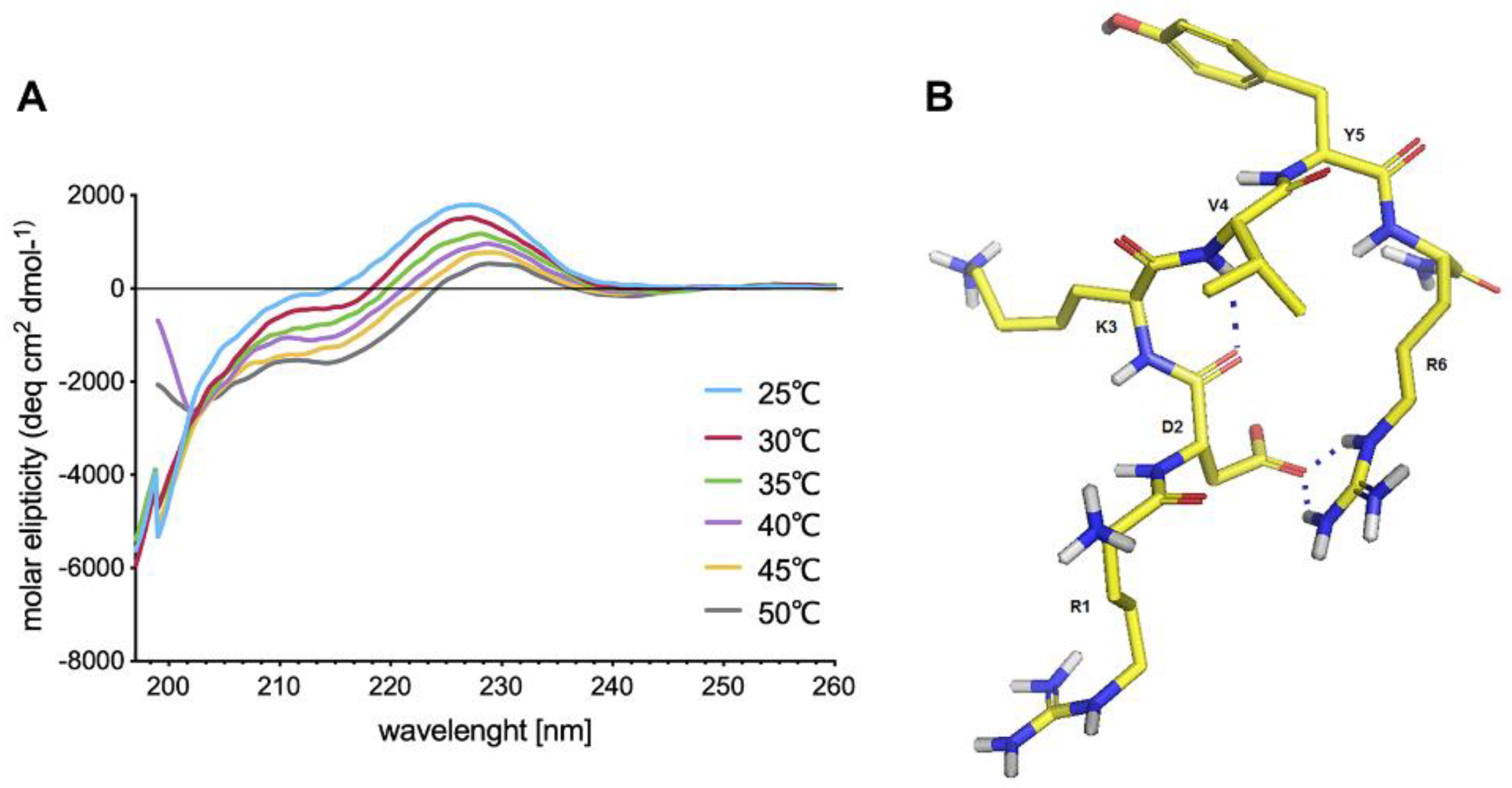
Cell receptors for IM have not been identified yet but examination of its spatial structure may provide clues about the nature of possible IM interactions with target proteins. Despite IM being a relatively short peptide shows a tendency to adopt a bent structure stabilized by a strong salt bridge and a hydrogen bond. Nevertheless, IM shows high conformational flexibility, and thus it may be adaptable to multiple different receptors hence spatial structure yields no clues.
In our previous work [
26], we optimized a procedure to use fibroblasts, keratinocytes, and ASCs as models in skin regeneration studies. We used serum free culture media, because addition of FBS, which contains various growth factors, which stimulate cell proliferation or migration, could mask IM’s effect. Moreover, IM could be captured by albumins present in FBS, which probably would minimize its effect. We found that IM is not cytotoxic to human skin cells (fibroblasts and keratinocytes). It also does not influence the viability of neural cells (
Supplementary Materials Figure S6) and proliferation of two different breast cancer cell lines (see
Supplementary Materials Figures S11 and S12). The lack of cytotoxicity of IM to different cells is a great asset, indicating that it can be used without the risk of damaging cells and tissues. The low cytotoxicity of IM demonstrated in this research corroborates the results of other studies [
4,
27].
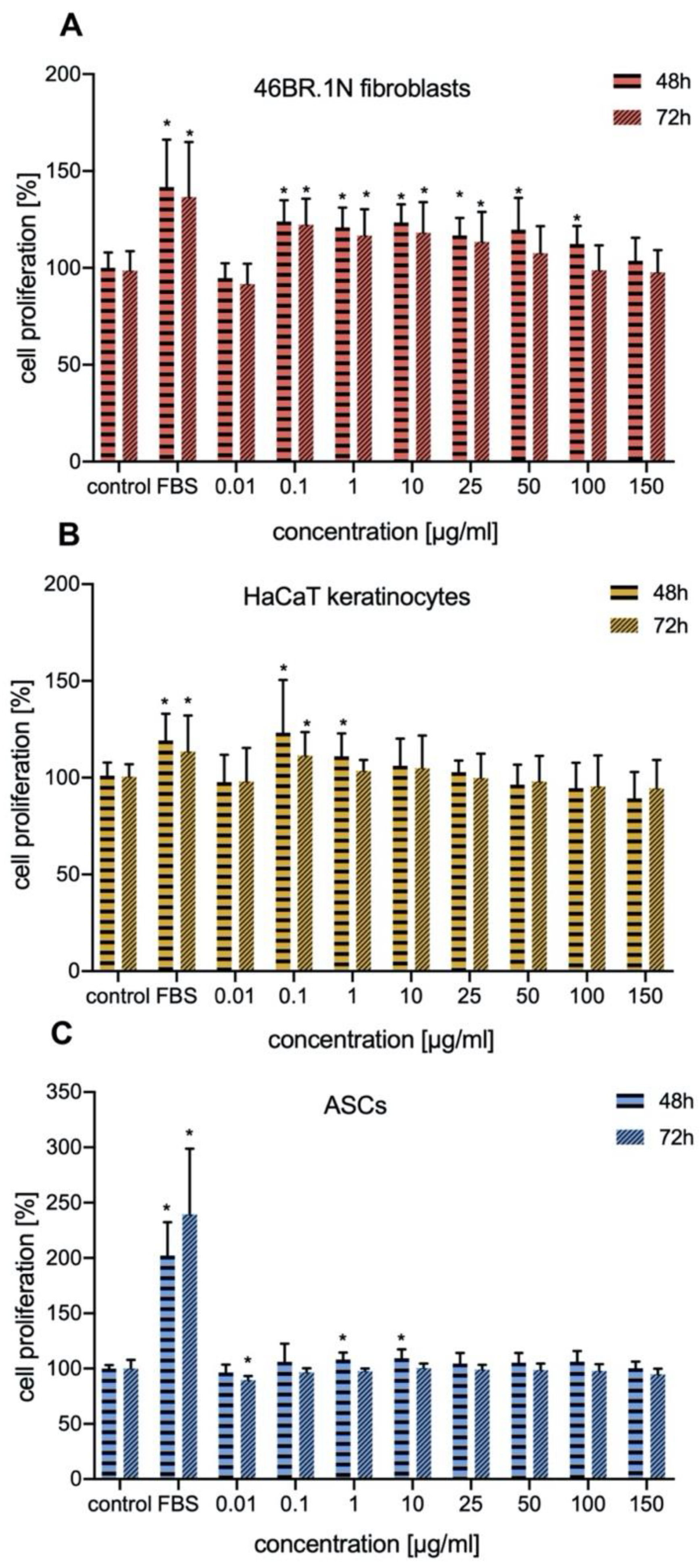
In our work we investigated the effect of IM on proliferation of human fibroblasts and keratinocytes. The proliferation of these cells is crucial for proper skin regeneration processes. Our studies indicated that IM stimulated proliferation of human skin cells (fibroblasts and keratinocytes), mainly in concentrations of 0.1–25 μg/mL, with similar effect on both types of cells. It also slightly stimulate proliferation of human ASCs. Similar studies have been conducted for TP5, a peptide which is very similar to IM. TP5 has no inhibitory effect on proliferation of healthy human peripheral lymphocytes and peripheral blood mononuclear cells (PBMCs) [
28,
29], and at the same time it induces proliferation of spleen cells [
30].
Migration and chemotaxis of fibroblasts and keratinocytes, as well as paracrine factors are essential for skin wound healing. In the present study, IM was not found to stimulate migration and chemotaxis of skin cells at any of the tested concentrations, which indicated that its biological actions are not related to these activities. Cells undergoing intensified processes of proliferation usually have limited migration and chemotactic properties [
31].
Peptides play an important role in immunology, often through being epitopes for adaptive immune systems [
32,
33]. Besides, they take part in mobilizing immune cells. For example, cationic host defense peptides, which possess antimicrobial properties, have the ability to control infections and inflammation because of their influence on innate and adaptive immunity [
34]. Unfortunately, application of synthetic peptides can lead to unwanted immune activation or allergies and immunogenicity or immune-related adverse effects can limit the effectiveness of peptide-based therapies [
35,
36,
37]. IM has been reported to be a compound with in vivo immunoregulatory properties, for example it has been reported to increase the production of IgM, IgG, and IgA in human lymphocyte cultures and to suppress the synthesis of IgE. However, IM has been found to induce the production of immunoglobulins following stimulation with pokeweed mitogen but no influence has been observed without pre-treatment of lymphocytes with the mitogen [
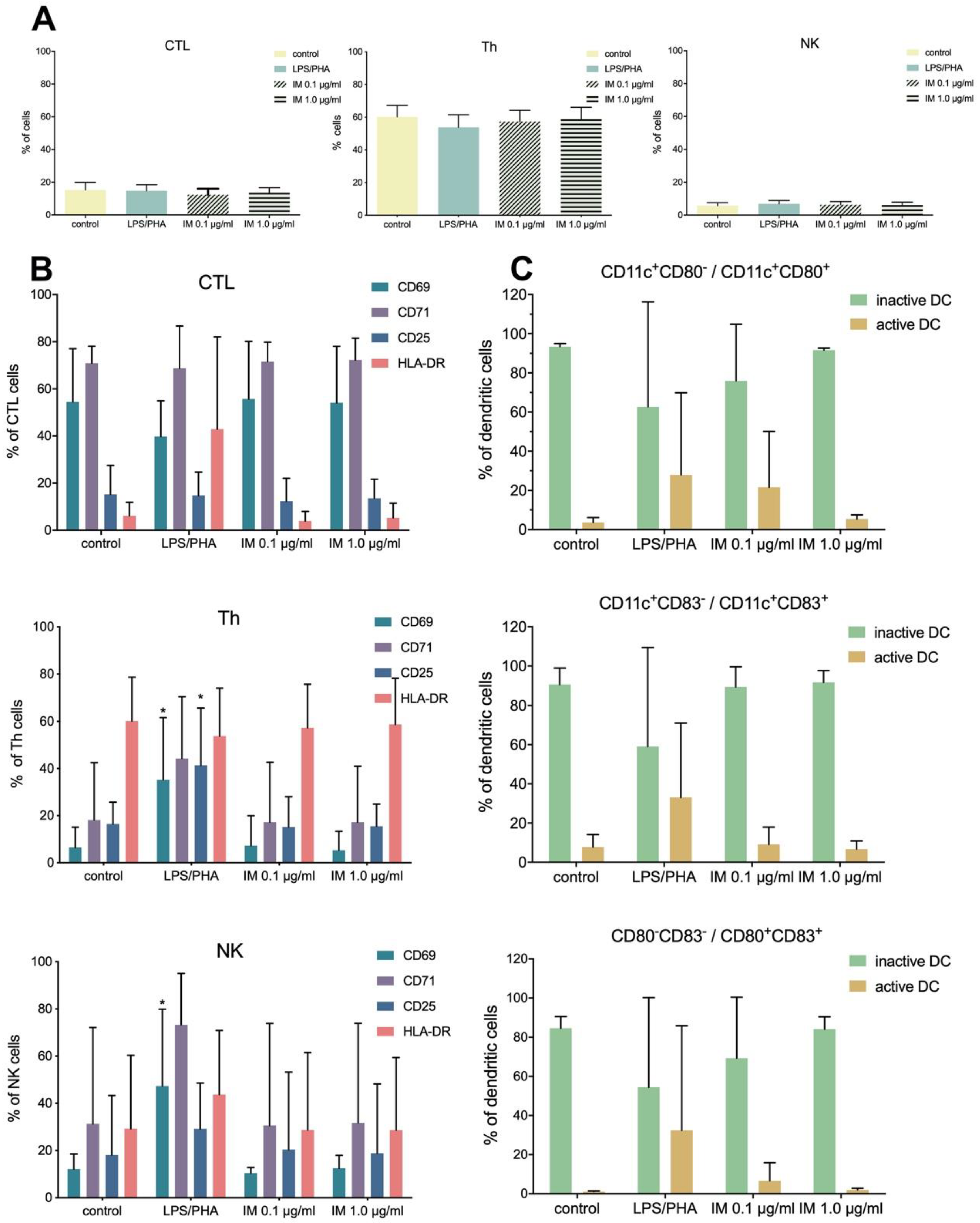
38]. Our studies were conducted in vitro on isolated PBMCs and indicated that IM did not activate the immune system (no activation of CTL, Th, and NK lymphocytes), thus suggesting IM is safe for use.
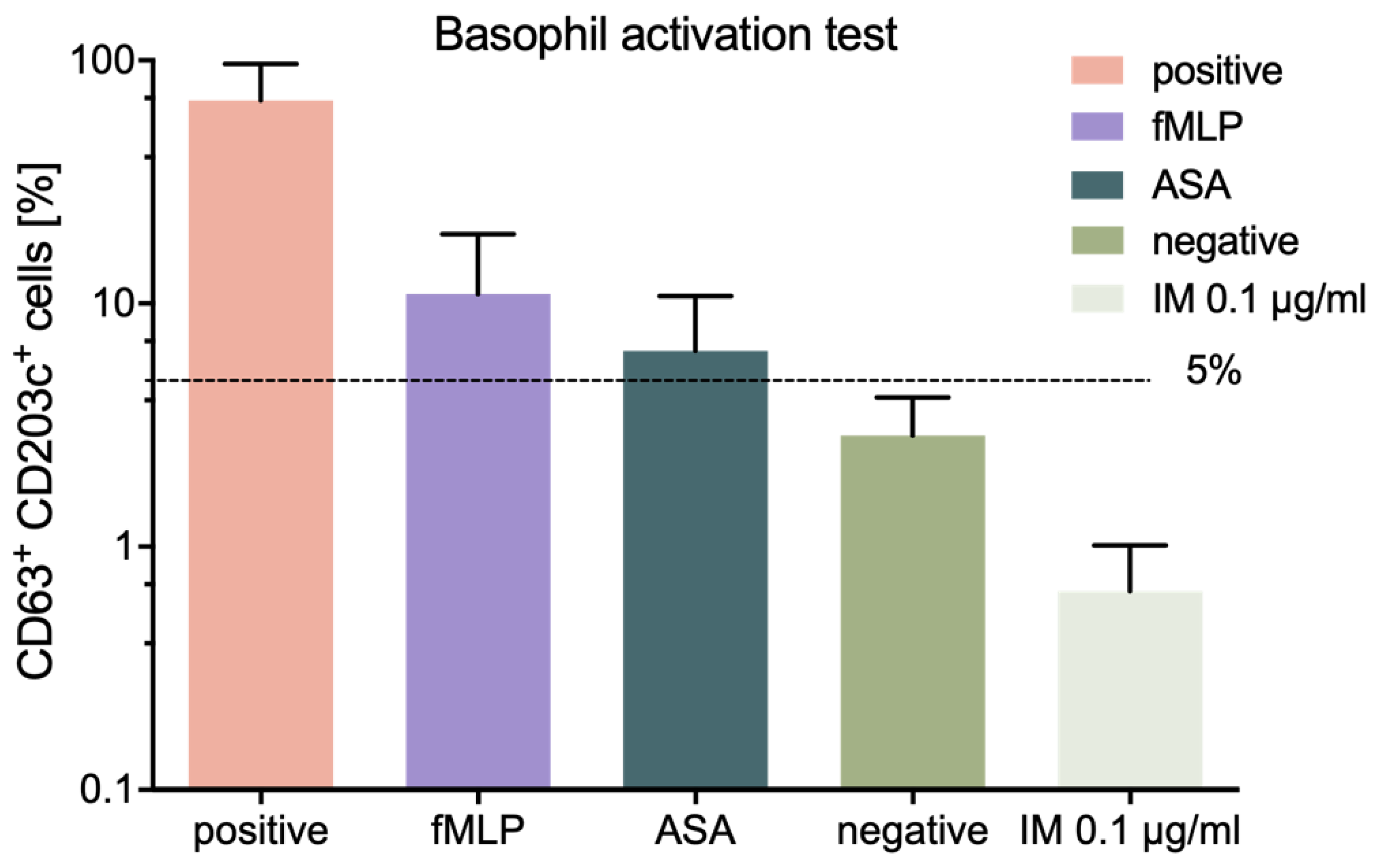
We also assessed the allergic potential of IM peptide, using an in vitro method, namely the BAT described previously [
26]. The conducted tests showed low allergic potential of IM peptide, thus confirming the immunological safety of this peptide.
In our research, we also examined the effect of IM on the secretion of specific growth factors and cytokines by fibroblasts, keratinocytes, and ASCs. For this purpose, we applied Luminex technology (Luminex xMAP), which is a trusted, widely used platform for biomarker screening and protein analysis. Our results showed that IM exerted little impact on the secretion pattern of cells.
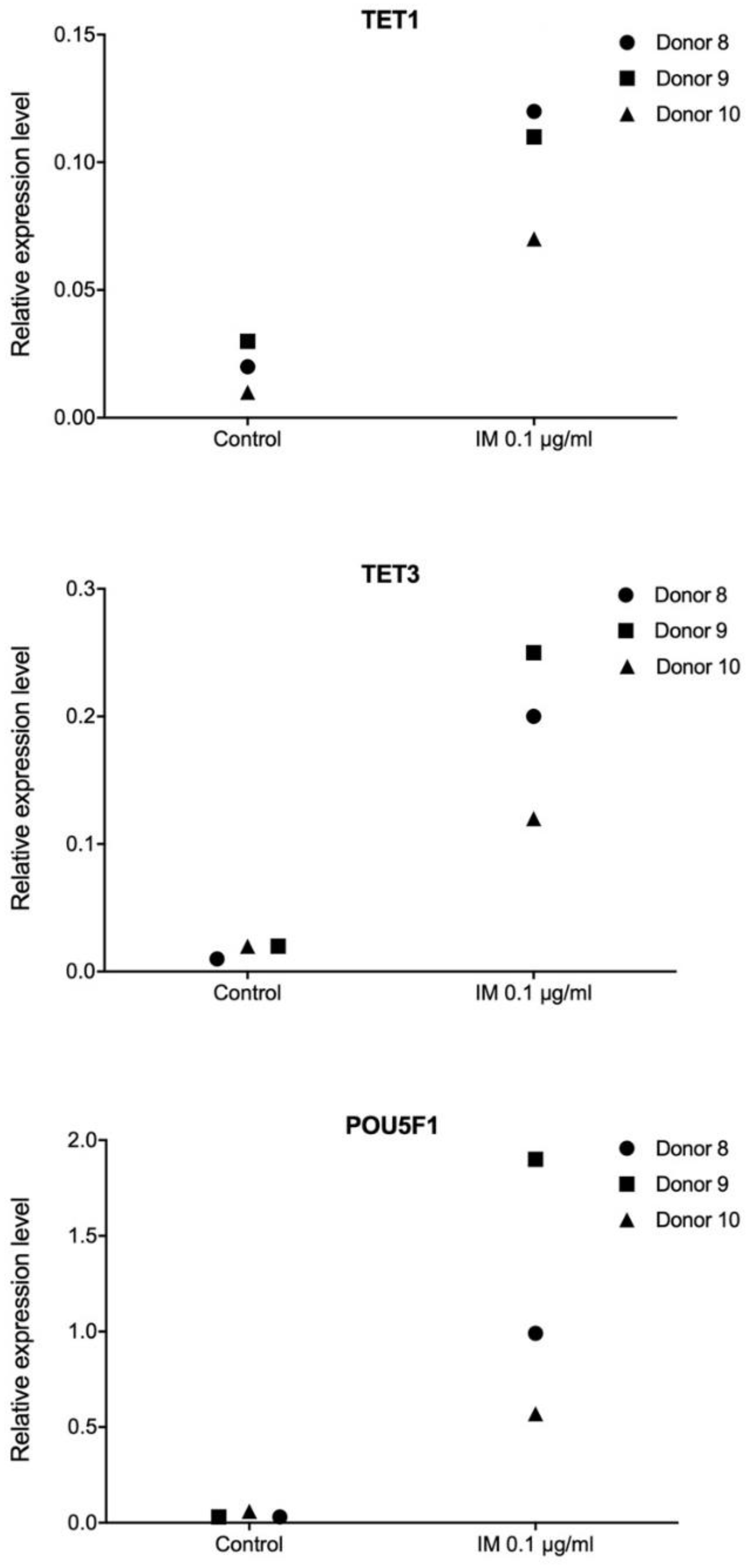
Pluripotency is defined as the ability of a cell to differentiate into any type of somatic cell, apart from trophoblasts. Establishment and maintenance of a pluripotent state depends on a group of transcription factors. Therefore, in our work, we performed transcriptome profiling in primary human fibroblasts, keratinocytes, and ASCs, in order to observe extensive alterations following stimulation with IM peptide. Transcriptional responses to IM of genes controlling cell proliferation potential and DNA methylation were evaluated. The analysis revealed no consistent transcriptional pattern changes following IM treatment, however, we did observe some individual responses. We found very strong induction of
POU5F1 encoding
OCT4, a key pluripotency factor, as well as of the
TET1 and
TET3 genes involved in active DNA demethylation. A recent study suggests that
TET enzymes play roles in controlling the expression of genes regulating regenerative processes in the skin [
39].
OCT4 is the main factor responsible for pluripotency induction [
40], and it retains high activity in pluripotent cells and is quenched after differentiation [
41]. Interestingly, as described by Costa et al. [
42],
TET dioxygenases and
OCT4 are likely to induce local transcriptional changes in shared target genes that regulate pluripotency.
Transcriptome profiling is a useful tool for evaluation of potential drugs. It provides an insight into the activity of investigated molecules. High-throughput next generation sequencing of RNA which can reveal mechanisms of action of tested pharmacological agents, thanks to its potential to characterize and quantify genome-wide gene expression [
43]. Utilization of this technology for investigating the effect of PDGF2 peptide administration on skin cells is presented in our previous research [
26]. In this work, we also performed RNA sequencing analysis for ASCs stimulated with IM. Although the response to IM varied between individual donors, we were able to observe the activation of processes related to wound healing: Activation of cells, proliferation and cell cycle progression, a pathway for synthesis of nitric oxide, and slight anti-inflammatory activity.
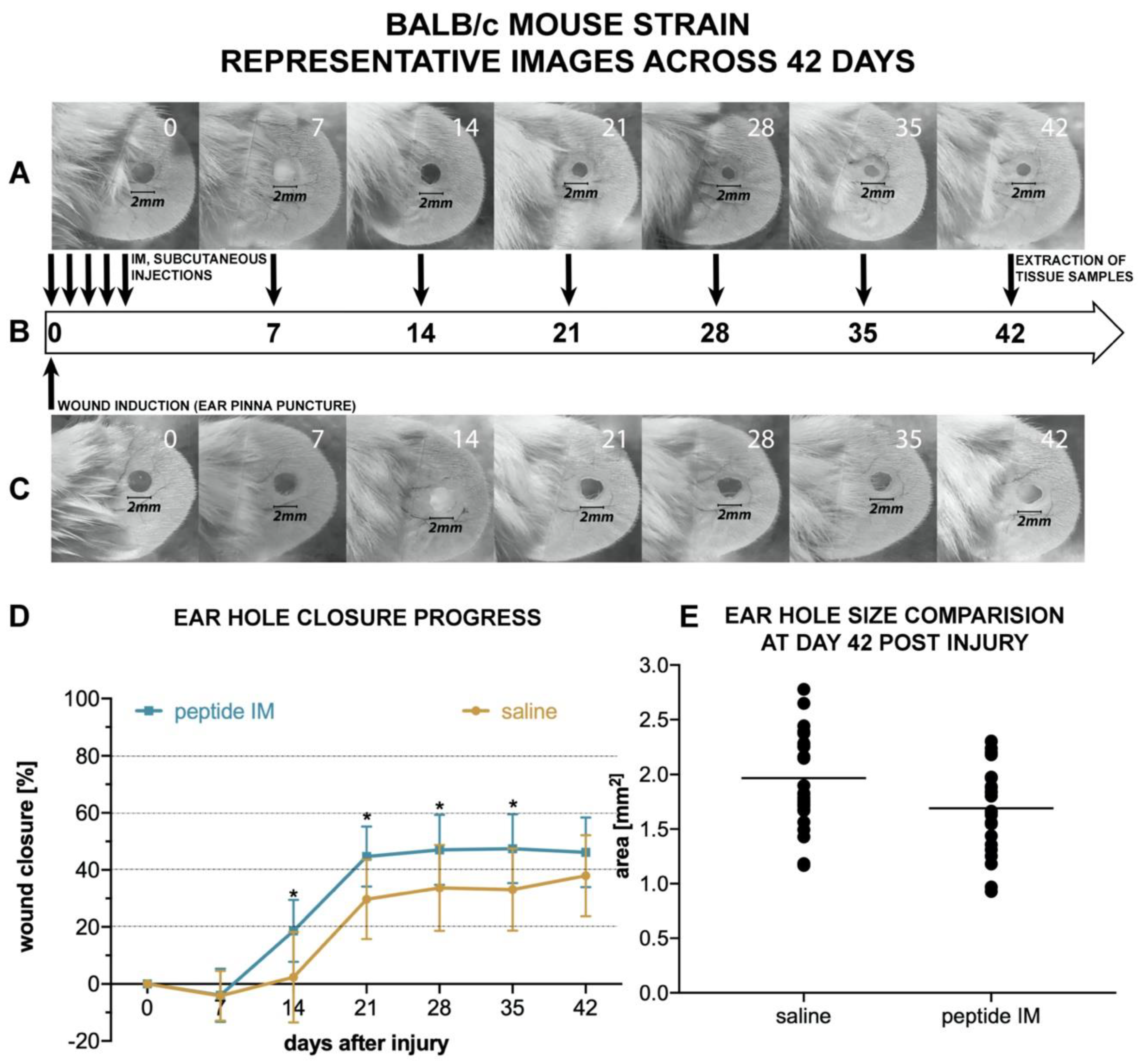
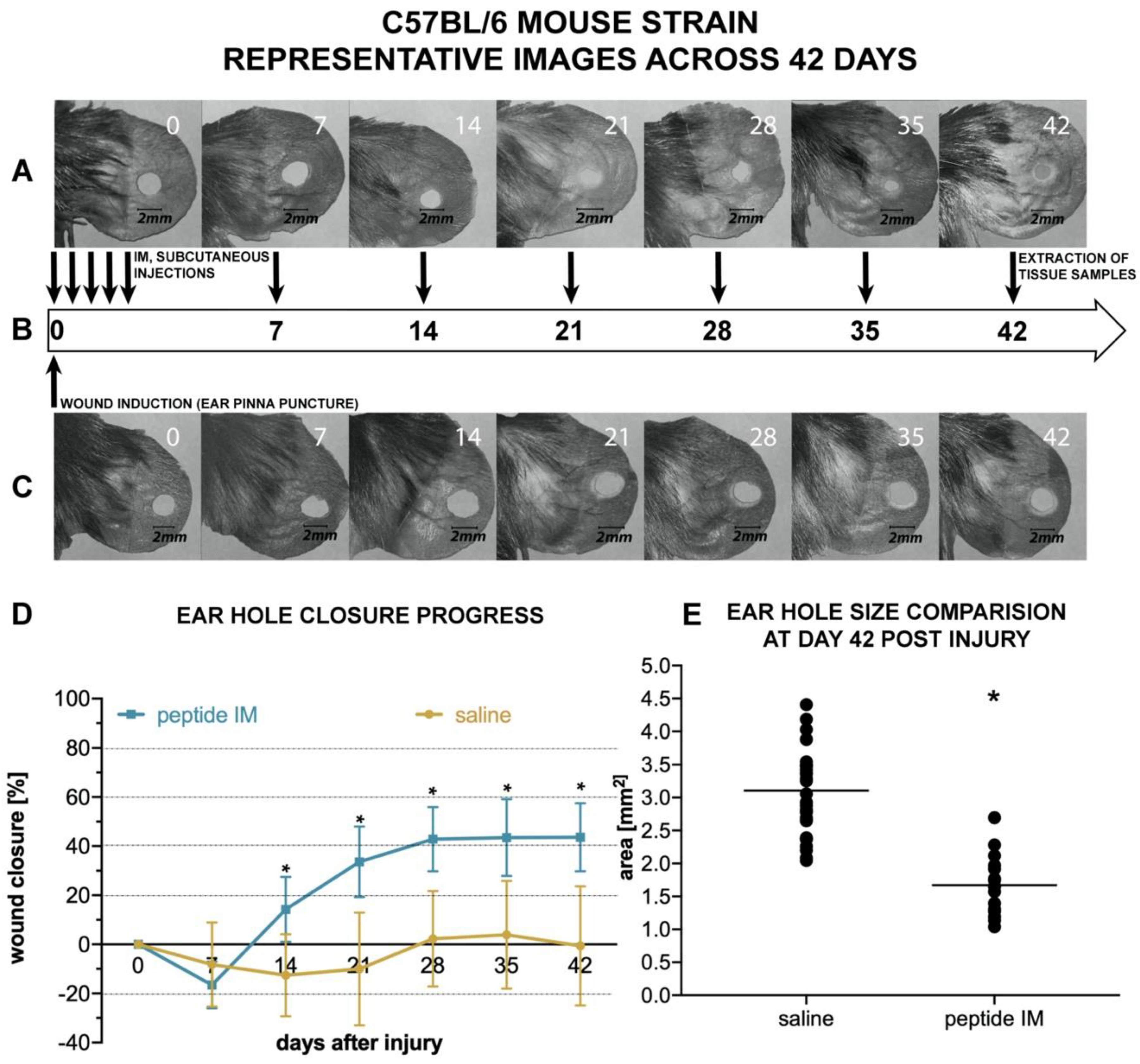
There are few animal models used for assessing the potential pro-regenerative activity of chemical compounds. The ear punch wound model in mice, though simple to perform, is a powerful model of complex tissue regeneration in mammals [
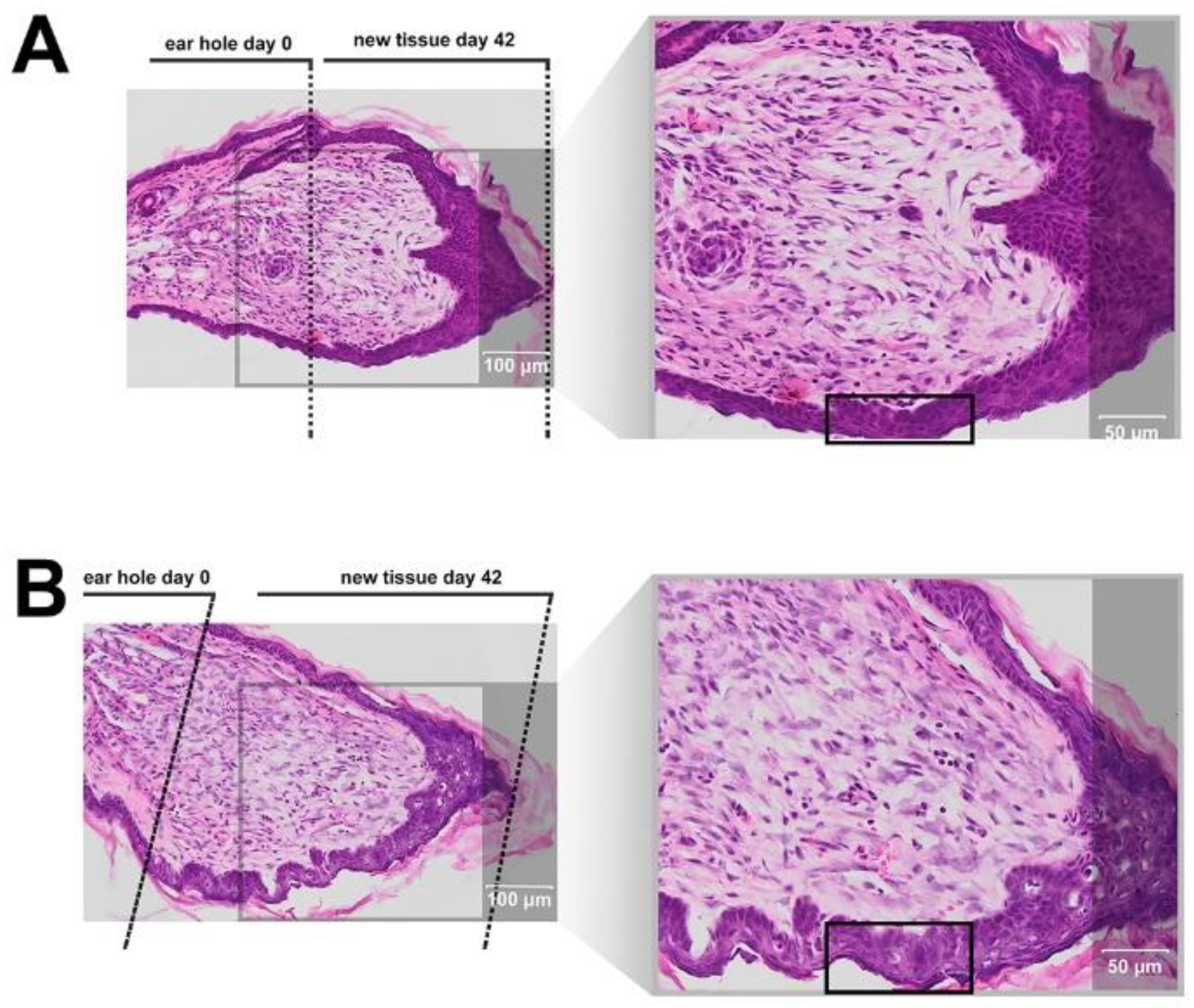
9]. The experiment involves making a 2 mm hole in the center of ear pinna in normal laboratory mice, such as BALB/c ad C57BL/6J. Quantitation of ear pinna hole closure in response to pharmacological treatment is used to assess the pro-regenerative activities of tested chemical compounds. Histological examination is also performed to evaluate if tissue architecture is restored within the re-grown region, which indicates a true regenerative response. In the presented research, we carried out tests of IM healing effects using this model. While limited ear hole closure was determined in mice administered physiological saline (38% in BALB/c and 8% in C57BL/6), restoration of lost tissue was observed in the IM-treated animals (46% in BALB/c and 44% in C57BL/6) 42 days after injury. Subcutaneous injection of IM activated moderate tissue repair in both mouse strains, however a much stronger effect was observed in C57BL/6 mice. Histological examination showed that the restoration in ear pinnae involved not only the formation of connective tissue but also the presence of numerous fibroblasts and epithelial cells. However, the presented work did not fully investigate the mechanism of IM action. Hence, further studies are required to determine molecular and cellular mechanism of IM action. A more detailed analysis of IM effect on immune cells is also necessary.
4. Materials and Methods
4.1. Peptide Synthesis and Purification
The IM peptide (H-Arg-Asp-Lys-Val-Tyr-Arg-OH) was synthesized by the standard solid phase method using
N-Fmoc protected amino acids in a Liberty BlueTM automated microwave synthesizer (CEM Corporation, Matthews, NC, USA). Cl-TCP(Cl) ProTide (CEM Corporation, Matthews, NC, USA) resin was used as a solid support with a capacity of 0.4 mmol/g. During the synthesis, a 20% piperidine solution in DMF was used as a remover of the Fmoc amino group, a 0.5 M DIC solution in DMF was used as the coupling reagent, a 1 M solution of Oxyma pure was used as a suppressor of racemization, and 0.1 M DIPEA in DMF was used as a scavenger of free radicals. The peptide was cleaved from the solid support, along with removal of protecting groups from the amino acid side chains, using the mixture TFA/phenol/TIPSI/H
2O (88:5:5:2 v/v/v/v). The IM peptide was purified by preparative high-performance liquid chromatography on a reversed phase (RP-HPLC), at room temperature, using an appropriate water/acetonitrile gradient with TFA. The crude product of the synthesis was purified using a Jupiter
® Proteo C12 semipreparative column (Phenomenex, Torrance, CA, USA) with dimensions 21.2 mm by 250 mm, 4 μm, 90 Å. The pure product was analyzed using analytical RP-HPLC and mass spectrometry (BIFLEX III MALDI-TOF, Bruker, Germany, and ESI LCMS IT TOF, Shimadzu, Japan) (see
Supplementary Data Figure S13). The purified peptide was subjected to exchange of the trifluoroacetate counter ions to acetate using 0.1 M aqueous ammonium acetate.
4.2. Stability in Water and Plasma
A stock of IM peptide in sterile water (1 mg/mL) was prepared and divided into 200 µL portions. The sample tubes were stored in an incubator at 37 °C and analysis was performed at various time points (0, 1, 2, 3, 6, and 24 h). The analyses were performed by HPLC using a Luna C18(2) (5 µm, 100 Å 4.6 by 250 mm) column (Phenomenex, Torrance, CA, USA) and monitored by a PDA detector. Then, 0.01% TFA in H2O (A) and 0.01% TFA in 80% ACN in water (B) were used as solvents. A linear gradient of 5–100% B over 60 min and a flow rate of 1 mL/min was applied.
A stability test in plasma was performed as previously described [
44]. Blood was collected from healthy anonymous donors and EDTA was then used as an anticoagulant. The plasma was aliquoted and incubated with IM peptide for 1, 2, 3, 6, and 24 h at 37 °C, at a final concentration of 300 µM. The analyses were performed by HPLC applying the conditions specified above.
For calculations, peak areas were compared to a calibration curve calculated by LC solution Software (Shimadzu, Kyoto, Japan).
4.3. CD and NMR Studies
CD studies of IM were performed in PBS (pH 7.4). The CD spectra were recorded for 0.25 mg/mL peptide, over the range 185–260 nm, with a Jasco J-815 spectropolarimeter (Jasco, Victoria, BC, Canada). For IM peptide, the CD spectra were recorded three times, over a temperature range of 25–50 °C and using a 1-mm cell, and depicted as the mean residue molar ellipticity (MRME, deg × cm2 × dmol−1) against the wavelength λ (nm).
NMR experiments were performed in water (90%:10% H
2O:D
2O). The peptide concentration was 4.0 mM. All NMR datasets were acquired at 303 K on a Bruker Avance III NMR System 700 NMR spectrometer (Brucker, Billerica, MA, USA). Proton chemical shifts (ppm) of IM peptide are listed in
Supplementary Table S2. Sequential backbone resonance assignments were obtained according to a standard procedure based on analysis of the 2D NMR spectra [
45]. In our study, the analyzed NMR datasets included the following: (i) double-quantum filtered correlation spectroscopy (DQF-COSY) [
46]; (ii) total chemical shift correlation spectroscopy (TOCSY) [
47], acquired at a mixing time of 80 ms; (iii) nuclear Overhauser effect spectroscopy (NOESY) [
48], recorded with mixing times of 200 ms; and (iv) rotating-frame Overhauser enhancement spectroscopy (ROESY) [
49], collected with mixing times of 200 ms. The acquired datasets were processed with NMRPipe software [
50] and analyzed with the Sparky [
51] program.
4.4. Structure Calculations
The interproton distances were calculated on the basis of the NOE intensities extracted from the NOESY spectra at a mixing time of 200 ms. Here, 98 distance restraints were provided for IM. The initial structure of the peptide was calculated by a simulated annealing algorithm, which included 10,000 steps of MD in the torsion angle space (CYANA software, Frankfurt, Germany) [
52]. As a result, the calculation protocol initialized with 200 structures created randomly chosen torsion angles. The 3D structures of IM, evaluated with CYANA, were used as the starting structure in the MD calculations in water. The entire system was subjected to energy minimization (steepest descent method). The system was then subjected to NTP MD for 10 ns at 305 K, with time steps of 2 fs. During the MD stimulation, a non-bonded cutoff of 9 Å was chosen. The MD studies in water were performed with AMBER software [
53], using a protocol including time-averaged NMR restraints (TAV) derived from NMR spectroscopy, with the force constant for interproton connectivity equal to 50 kcal/(mol × Å
2). No torsion angle restraints were used in the calculations. The geometry of the peptide groups was kept all trans, with the force constant of f = 50 kcal/mol × rad
2, according to the NMR data. The coordinates were collected every 2,000 steps. The shape of the IM structure was evaluated using the PyMOL program [
54].
4.5. Affinity Test
Immobilization of protein in a microcolumn, and affinity tests, were performed as described previously by Spodzieja et al. [
55]. The IM peptide was added onto an albumin-Sepharose column equilibrated in ammonium hydrogen carbonate, pH 7.4, and incubated for 2 h at 25 °C with gentle shaking. After 2 h, the column was washed with 100 mL of ammonium hydrogen carbonate, collecting the first and last milliliter of the solution (supernatant and last wash fraction). The albumin-peptide complex was dissociated by incubation for 15 min in 0.1% TFA, with gentle shaking. The procedure was repeated twice, and the elution fraction was collected each time. All fractions were lyophilized and analyzed by mass spectrometry (ESI-IT-TOF, Kyoto, Shimadzu Japan).
4.6. Primary Cell Isolation
Human skin and subcutaneous adipose tissue were sampled from patients of the Plastic Surgery Clinic or Oncologic Surgery Clinic in the Medical University of Gdańsk. The procedure was approved by the Independent Bioethics Commission for Research of the Medical University of Gdańsk (NKBBN/387/2014). Tissue samples for cell isolation were obtained from adult donors (
Table S3). Unchanged skin was removed routinely during surgery, e.g., abdominal surgery or mastectomy and adipose tissue was removed with skin fragments or by liposuction. Donors with other medical conditions that could affect the results of tests, e.g., active infections and autoimmune diseases (e.g., scleroderma, lupus, MS), HBV, HCV, HIV carriers, patients with active allergy, diabetes or atherosclerosis, and advanced cancer as well as subjected to neoadjuvant treatment (chemotherapy, radiotherapy, and/or immunotherapy) were excluded from the study. All tissue donors were informed about the assumptions of the project and signed an informed consent. Isolation of adipose-derived stem cells was based on a standard protocol, followed by enzymatic digestion and erythrocyte lysis [
56,
57]. Confirmation of “stemness” of these cells was performed by flow cytometry and analysis of differentiation potential into adipocytes, osteocytes, and chondrocytes (
Figure S14), as previously described by Mieczkowska et al. [
56], and the protocol is summarized in the Supplementary file. Primary epidermal cells were isolated with the protocol described earlier by Langa et al. [
58] and fibroblasts by culture of explants. To allow cells to grow out from the tissue samples, human dermis was placed in six-well plates containing Dulbecco’s modified Eagle’s medium high glucose (4500 mg/L) medium supplemented with 10% of fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO, USA), 100 U/mL of penicillin, and 100 μg/mL streptomycin (Sigma-Aldrich, MO, USA), then placed under standard conditions in a humidified atmosphere with 5% CO
2 at 37 °C. The medium was changed every 2–3 days.
4.7. Cell Culture Conditions
Five types of cells were used in our study: Immortalized human HaCaT keratinocytes (DKFZ Heidelberg, Germany), human dermal fibroblasts of cell line 46BR.1N, human primary fibroblasts and keratinocytes isolated from skin samples, and ASCs. The fibroblast cell line was originally derived from the skin of an anonymous individual and was transformed with the pSV3neo plasmid [
59]. HaCaT keratinocytes were immortalized cells derived from normal epidermal keratinocytes. They maintain most of the normal keratinocyte functions, including differentiation potential and response to different stimuli [
60,
61]. HaCaT, 46BR.1N cells, and human primary fibroblasts were grown in a DMEM medium (Sigma–Aldrich, St. Louis, MO, USA; including 4500 mg/L of glucose, 584 mg/l of l-glutamine, sodium pyruvate, and sodium bicarbonate) supplemented with 10% FBS and 100 units/mL of penicillin and 100 μg/mL of streptomycin (Sigma–Aldrich, St. Louis, MO, USA). Human primary keratinocytes were grown in Keratinocyte Growth Medium (cat. CC-3103, Lonza-Clonetics, Basel, Switzerland) supplemented with epidermal growth factor, hydrocortisone, transferrin, epinephrine, insulin, and gentamycin (cat. CC-4152, Lonza-Clonetics). ASCs were grown in DMEM (Sigma–Aldrich, St. Louis, MO, USA; with 1000 mg/L of glucose, 584 mg/L of l-glutamine, sodium pyruvate, and sodium bicarbonate) supplemented with 10% FBS and 100 units/mL of penicillin and 100 μg/mL of streptomycin (Sigma–Aldrich, St. Louis, MO, USA). The cells were routinely cultured under a humidified atmosphere, with 5% CO
2 and at 37 °C, in culture flasks (growth surface area 25 cm
2).
4.8. Peptide Cytotoxicity Assay
Cell death was quantified by measurement of lactate dehydrogenase activity in cell culture supernatants (Takara, Japan, cat. No. MK401), following the manufacturer’s instructions. Cells were seeded into 96-well plates (BD, cat.no. 353872), at a density of 5000 cells per well, in medium (DMEM) supplemented with 10% FBS. After 24 h, the medium was changed to serum-free medium containing appropriate concentrations of IM peptide. After 48 h of stimulation supernatants were collected for LDH analysis. Cell death was normalized with respect to untreated control (without IM). Non-IM treated cells were used as negative control (0%) and Triton-X 100 (1%) was used as the positive control for maximum LDH release (maximum cytotoxicity, 100%) [
26,
62]. The assay was performed to IM in concentrations of 50–150 µg/mL. IM did not show signs of toxicity in lower concentrations, so it was not necessary to perform LDH analysis for other concentrations.
4.9. Skin Cell and ASCs Proliferation Assay
XTT assay, which is widely used in the measurement of the effect of bioactive compounds (e.g., cytokines, growth factors, and peptides) on cell proliferation was used for the evaluation of IM effect on skin cells and ASCs proliferation [
63,
64]. Cells were seeded at a density of 5000 cells per well into 96-well plates (BD, cat.no. 353872), in a medium supplemented with 10% of FBS, which after 24 h was exchanged for a serum-free DMEM medium containing appropriate concentrations of the IM peptide. IM solutions used in the experiments were prepared with double distilled water under sterile conditions. The cells were incubated with the peptide for 48 h or 72 h, then XTT reagent was added and the plates were incubated at 37 °C for 4 h in the presence of 5% CO
2, following manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO, USA). Plates were then read using a standard plate reader at OD 490 nm. Cell proliferation was normalized with respect to an untreated control (100%) [
26].
4.10. Migration and Chemotaxis Assay
The effect of IM on cell migration after 24 h was determined using ibidi culture inserts (ibidi, Cat. No. 81176, Gräfelfing, Germany). Cells were seeded in culture inserts, at a density of 30,000 per well, in appropriate medium supplemented with 10% FBS, which was changed to serum-free DMEM after 24 h. Mitomycin C (5 μg/mL) was added for 2 h to block cell proliferation. The medium was then changed, again, and cells were stimulated with an appropriate concentration of IM. After 24 h the cells were fixed with 3.7% paraformaldehyde, stained with 0.05% crystal violet, and the effect was measured with a phase-contrast fluorescent microscope (Zeiss, Oberkochen, Germany) and the surface areas were counted with Imaging Software NIS-Element Basic Research program [
26,
62].
The effect of IM on cell chemotaxis was assessed with ThinCert cell culture inserts (8 μm, Greiner Bio-One, Kremsmünster, Austria). Cells were starved overnight in serum-free high-glucose DMEM and then seeded on inserts placed in a 24-well plate, at a density of 100,000 cells, in serum-free medium. Culture medium with appropriate concentrations of IM was then added to the wells of the plate and incubated for 24 h. The cells were stained with 8 μM Calcein-AM (BD Pharmingen), then migratory cells were detached with typsin-EDTA (Sigma-Aldrich, St. Louis, MO, USA). Detached cells were transferred to another plate and read with a fluorescence plate reader at an excitation wavelength of 485 nm and an emission wavelength of 520 nm [
26].
4.11. Immunological Studies
Immunological tests were conducted on human peripheral blood mononuclear cells, isolated from “buffy coats” using a ficoll density gradient (Histopaque, Sigma-Aldrich, St. Louis, MO, USA). Buffy coats for were obtained from blood of healthy anonymous donors of the Regional Center of Blood Donation and Blood Therapy in Gdansk, Poland. After washing in PBS, erythrocyte lysis, and cell counting (cell counter, Bio-Rad), the PBMCs were seeded onto a 24-well culture plate at a density of 10
6 cells/1 mL of RPMI 1640 (Penicillin/Streptomycin, 10% FBS) per well. After cell resting for 24 h, cells were stimulated with IM peptide (0.1 µg/mL and 1.0 µg/mL) for the next 48 h under appropriate conditions (37 °C, 5% CO
2). The untreated cells were used as a negative control, whereas cells stimulated with LPS (1 µg/mL) and PHA (2.5 µg/mL) were a positive control. After incubation, the PBMCs were collected, washed with PBS, counted and prepared for flow cytometry analysis. 10
4 cells/100 µl were stained with monoclonal, fluorochrome-conjugated antibodies (anti-CD3, -CD4, -CD8, -CD16, -CD56, -CD25, -CD69, -CD71, -HLA-DR, -CD11c, -CD80, and -CD83) and incubated for 30 min in RT in the dark. Stained cells were analyzed using a flow cytometer (LSRFortessa, BD, USA) [
26,
62]. All data are presented as percentages due to the application of hospital diagnostic laboratory standards for immune cells analyses.
4.12. Basophil Activation Test
The commercially available test Flow CAST
® highsens (Bühlmann Laboratories, Schönenbuch, Switzerland) was used to perform BAT test on blood samples collected from healthy volunteers, who were not suffering from allergy, within 24 h of collection, according to the manufacturer’s protocol. Then, 100 µL of blood was incubated and IM peptide was added to the wells at a final concentration of 0.1 µg/mL. The cells were then stained with fluorochrome-stained monoclonal antibodies (anti-CD63, -CD203c, -CCR3) and incubated for 15 min at 37 °C. Then, erythrocyte lysis was performed, the cells were washed and analyzed by flow cytometry (BD FACSCanto II, San Jose, CA, USA). Each sample was accompanied by its own negative and positive control (anti-FcεRI mAb and fMLP) [
62,
65].
4.13. Analysis of Protein and Cytokine Levels in Culture Supernatants
We used Luminex
® xMAP
® technology for analysis of proteins and cytokines in supernatants collected from primary fibroblasts or keratinocytes and cell cultures of ASCs stimulated with IM peptide at 0.1 µg/mL concentration. This method allows multiplex detection of proteins in single biological samples. For analysis of ASC supernatants we used Human Adipocyte Magnetic Bead Panel (Merck Millipore, Burlington, MA, USA) which enables simultaneous quantification of the following: Adiponectin, HGF, IL-1β, IL-6, IL-8, leptin, MCP-1, NGF, PAI-1 (total), resistin, and TNFα. For supernatants of fibroblasts, keratinocytes, and ASCs we assessed concentrations of 12 human cytokines and growth factors (angiopoietin-2, BMP-9, EGF, endoglin, endothelin-1, FGF-1 (acidic FGF), FGF-2 (basic FGF), follistatin, G-CSF, HB-EGF, HGF, IL-8, leptin, PLGF, VEGF-A, VEGF-C and VEGF-D) with Human Angiogenesis/Growth Factor Magnetic Bead Panel 1 (Merck Millipore, Burlington, MA, USA). The analysis was conducted according to the manufacturer’s instructions with a protocol described before by Deptula et al. [
26]. Analysis was performed with a Luminex MAGPIX
® Analyzer (Merck Millipore, Burlington, MA, USA) and data was analyzed in xPONENT 4.2 software. Results are presented as the concentration of cytokines in units of pg per 1 mL.
4.14. Cells Stimulation for Transcriptome Analysis, for Transcript Level Analysis and for Luminex Xmap Analyses
Cell stimulation was performed as previously described [
26]. Primary fibroblasts after the third passage and ASCs after the second passage were cultured in appropriate DMEM medium supplemented with 10% FBS for 24 h. Then, the medium was changed for DMEM with 5% FBS and 24-h incubation was performed. After another medium change for serum-free, cells were stimulated with IM peptide (0.1 µg/mL) for 48 h. Cell cultures deprived of FBS for 48 h were used as a control. After stimulation, the cells were trypsinized, collected, and centrifuged, and the pellet was kept frozen (−80 °C) for transcriptome analysis and transcript level analysis.
Human epidermal cells were cultured in a 25 cm2 t-flask in keratinocyte growth medium (KGM, Lonza, Basel, Switzerland) supplemented with epidermal growth factor, hydrocortisone, transferrin, epinephrine, insulin, gentamycin, and 5% FBS for 24h. Then, they were grown in serum-free KGM for another 2–5 days. Next, the medium was exchanged for keratinocyte basal medium (KBM, Lonza, Basel, Switzerland) and the cells were stimulated with IM peptide (0.1 or 1.0 µg/mL) for 48 h. The cell pellets were collected and kept frozen at −80 °C until RNA isolation for transcriptome analysis and transcript level analysis.
Cell culture supernatants of ASCs, human primary fibroblasts and human epidermal cells were collected and stored frozen at for further Luminex xMAP analyses.
4.15. Transcript Levels
RNA was extracted using an RNeasy Mini kit (Qiagen, Cat. No. 74104, Hilden, Germany). RNA quality and concentrations were examined with a NanoDrop 2000 spectrophotometer. cDNA synthesis was performed in a reaction mixture containing 200 ng of RNA, 100 pmoles of oligo dT20, 4 µl of 5× reaction buffer (250 mM Tris-HCl, 375 mM KCl, 15 mM MgCl
2, and 50 mM DTT), and 200 units of Maxima Reverse Transcriptase (ThermoScientific Bio. Cat. No. EP0742, Waltham, MA USA), at a final volume of 20 µl. PCR reactions were carried out on a LightCycler LC96 device (Roche, Basel, Switzerland), at a final volume of 10 µl containing 5 µl of FastStart Essential DNA Green Master (Roche, Cat. No. 06402712001, Basel, Switzerland), 1 µl of cDNA, and 0.25 µl each of forward and reverse primer (10 µM). The primer sequences are listed in
Supplementary Table S4. The transcript levels were determined using the 2
−ΔCt method relative to
Actb and
Tbp [
9,
26].
4.16. RNA Isolation and Quality Assessment for RNAseq
For three plastic surgery donors (donor 6, donor 11, and donor 12) ASCs (3–4.5 × 10
5 cells) after passage P2, namely cultures stimulated for 48 h with 0.1 µg/mL IM, or control cultured with FBS-deprived medium, were trypsinized, washed, and snap frozen for transcriptome analysis (see
Supplementary Table S5 for clinical information). The pellets were stored at −80 °C prior to RNA isolation. RNA was isolated using a miRNeasy Mini Kit (Qiagen, Hilden, Germany) with two modifications of the original protocol: (i) chloroform was substituted with 1-bromo-3-chloropropane to prevent foaming and emulsification, and (ii) the elution was done with 40 µl of water followed by another elution with the entire volume of the original eluate. The quality and concentration of RNA was assessed using a Bioanalyzer 2100 Instrument and RNA 6000 Nano Kit (Agilent, Waldbronn, Germany)
. The average RNA yield was 2 µg. Samples with RIN (RNA Integrity Number) >8.0 were used for further transcriptome analysis [
26,
56].
4.17. RNA Sequencing and Bioinformatic Analysis
A ribosome reduction kit (Arraystar Inc, Rockville, MD, USA) and SureSelect Strand Specific mRNA library kit (Agilent, Santa Clara, CA, USA) were used for next generation sequencing (NGS) library preparation according to standard procedures. The cDNA libraries were quantitated using qPCR in a Roche LightCycler 480 with the Kapa Library Quantification Kit for Illumina platforms (Kapa Biosystems, Woburn, MA, USA). Onboard cluster generation and 50 nt paired end RNA sequencing were performed on an Illumina HiSeq2500 device using Rapid Run v2 sequencing chemistry and flow cells as recommended by the manufacturer (Illumina Inc., San Diego, CA, USA). Paired end 50 bp sequencing runs were completed and the data was converted to the FASTQ Sanger format. Trim Galore was used to remove residual adapter sequences from the reads (
https://www.bioinformatics.babraham.ac.uk). Sequencing reads were aligned to the human reference genome assembly (hg19) using TopHat [
66]. Further analysis was performed according to workflow for Cufflinks version 2.2.0 and higher [
67]. Transcript assembly and estimation of the relative abundances were carried out with Cufflinks [
68] using the following settings: Quartile normalization method for the libraries, blind dispersion method for replicates, and biased correction using canonical hg19 as reference. Cuffmerge and Cuffquant were then performed, followed by final comparison analysis in Cuffdiff. Transcripts that revealed significant and consistent differences between conditions in our dataset (
p-value < 0.05) were subjected to further interpretation by ingenuity pathway analysis. Differentially expressed genes (DGE) were grouped in functional pathways using ingenuity pathway analysis (IPA Spring Release 2017, QIAGEN, Hilde, Germany) on a gene level, using RefSeq annotation [
26,
56].
4.18. Animals
Experiments were performed on 8–12 week-old female BALB/c (24 mice in total) and C57BL/6 mice (36 mice in total). All mice originated from the Centre for Experimental Medicine of the Medical University of Białystok, Poland, and were kept in the animal facility at the Nencki Institute of Experimental Biology throughout the course of the study. After delivery to the Nencki Institute animal facility, mice rested for at least two weeks for acclimatization before starting the experiment. Experiments were approved by the First Local Ethics Committee in Warsaw (permit No. 491/2013 and 542/2018). Animal experimentation was performed in accordance with EU directive 2010/63/EU, and all experiments were undertaken in such a way as to minimize discomfort and the number of animals used in the study.
4.19. Ear Pinna Injury and IM Peptide Treatment
Mice were placed individually in separate cages, weighed, and randomly divided into two groups within each mouse strain: The treatment group and the control group. Before the study all mice were randomly given numerical equivalents, and all experimenters performing further analysis were not aware of the experimental group and referred solely to those numbers. Similarly, vials containing IM (treatment) or saline (control) were encrypted and assigned to particular mice, so the person performing injections and ear photographs was not aware of mice background. Lastly, the person quantifying each ear hole closure was not aware of the group. Numerical equivalents of each mice were only decrypted at the end of the study. Before study each animal was anesthetized with Isoflurane (Iso-Vet; Piramal, UK), ears were quickly disinfected with 70% isopropanol and circular punches 2 mm in diameter were made in the center of both ear pinnae using disposable biopsy punches (Integra Miltex; Integra Life Sciences, Princeton, NJ, USA). Due to some occasional self-inflicted injuries (tearing off one or both ear lobes) during the course of the study, the measurement data from destroyed lobes were excluded from comparisons of the next time points.
Immediately after pinna lesion, mice in the treatment group received subcutaneous injections of a sterile solution of IM dissolved in saline (0.5 mg/mL), at a dose of 5.0 µg/g body weight, whereas mice in control groups received equivalent volumes of sterile saline. After recovery, all mice were returned to their home cages. Subcutaneous injections of IM solution or saline, respectively, were continued in the mice on days 1, 2, 3, 4, 7, 14, 21, 28, and 35 after ear pinna injury. Thus, each mouse received 10 injections in total. All subcutaneous injections of IM or saline were made in nape of the neck.
To monitor the progress of wound healing, ears were photographed on the day of the injury (day 0, before and right after pinna injury), and on days 7, 14, 21, 28, 35, and 42. To this end, the mice were anesthetized, their ears were gently flattened using glass cover slips on the flat surface of a sterile plastic block and each ear was photographed from the dorsal side parallel to the glass. A millimeter scale was present within each photographed area for length reference and to allow standardized comparison between each measuring point and between mice. Finally, the area of the wound hole was outlined by hand on digitized images as an overlay, and the hole areas in the ears were evaluated using computer-assisted image analysis (ImageJ Software [
69]). Experiments were conducted separately on BALB/c and C57BL/6 mice strain in an identical manner. In total, 12 mice (24 ears) per treatment group and 12 mice (24 ears) per control group entered the analysis in case of BALB/c strain. The final number of analyzed ears was 23 per group on day 42. In C57BL/6 strain, the total number of mice entering the analysis was 8 in the treatment group and 18 in the control group. The final number of analyzed ears on day 42 was 32 in the control group and 16 in the treatment group.
4.20. Tissue Isolation for Histological Analyses
On day 42 post-injury, the mice were anesthetized with isoflurane and euthanized by cervical dislocation. The ear pinna tissues were dissected, transferred to Petri dishes, a small microscopic cover glass was placed with small spacers above the pinna to prevent further tissue rolling, and all tissue samples were fixed overnight in 4% paraformaldehyde solution buffered with 0.1 mM PBS in preparation for histopathological analyses. Next, the skin tissue was embedded in paraffin, fixed and cut into 10 µm-thick sections. The morphology of the skin slices was investigated by standard hematoxylin and eosin staining, followed by microscopic analysis using an image analysis system (Olympus BX61VC, Tokyo, Japan).
4.21. Statistics
Statistical significance was determined with a non-parametric Mann–Whitney U test. Statistical significance was accepted to be p≤ 0.05. Analyses were performed with Statistica 13.3 software (Statsoft, Warsaw, Poland) and graphs were prepared with GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA).