Prediction of Protein Tertiary Structure via Regularized Template Classification Techniques
Abstract
1. Introduction
2. Methods
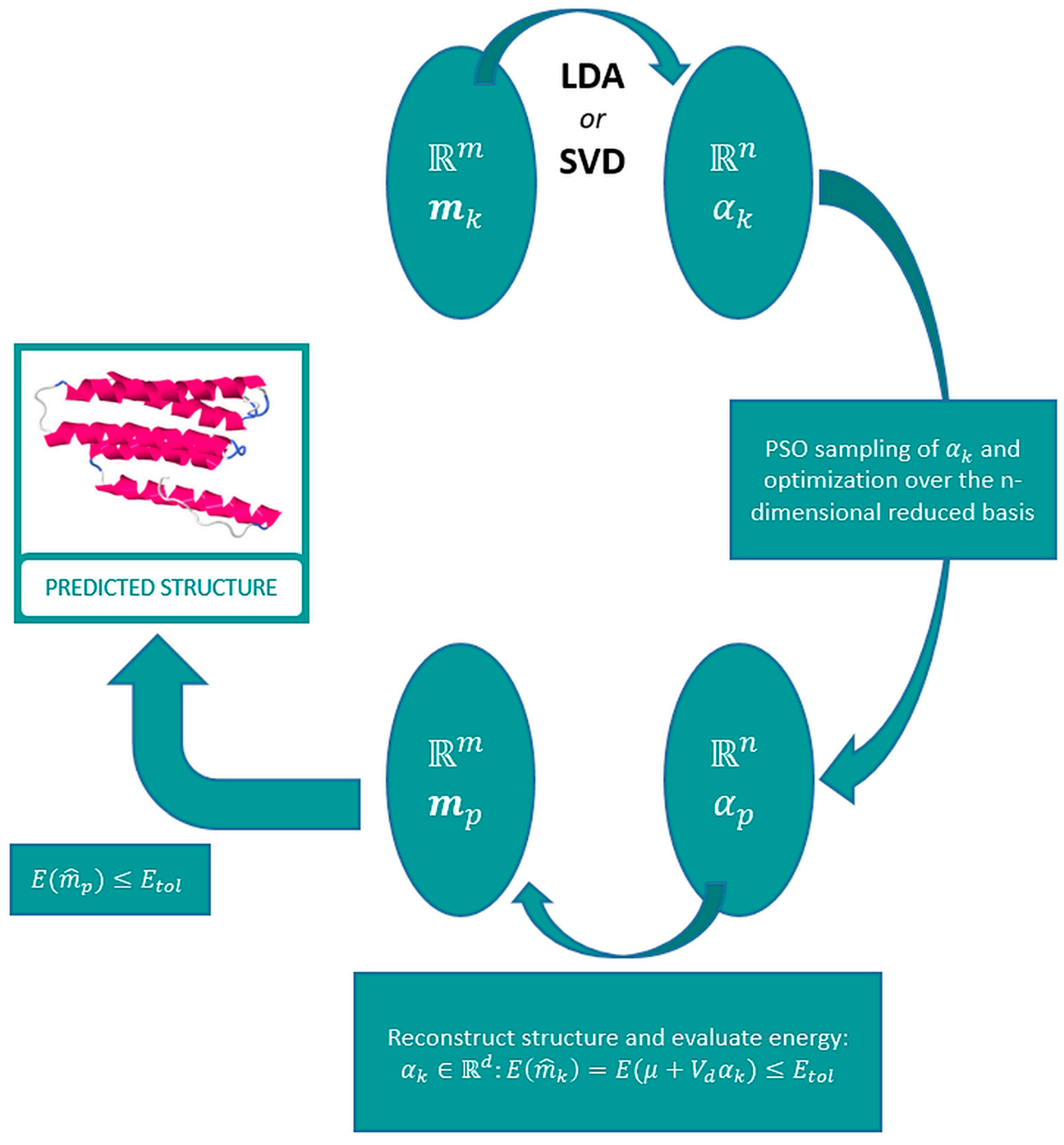
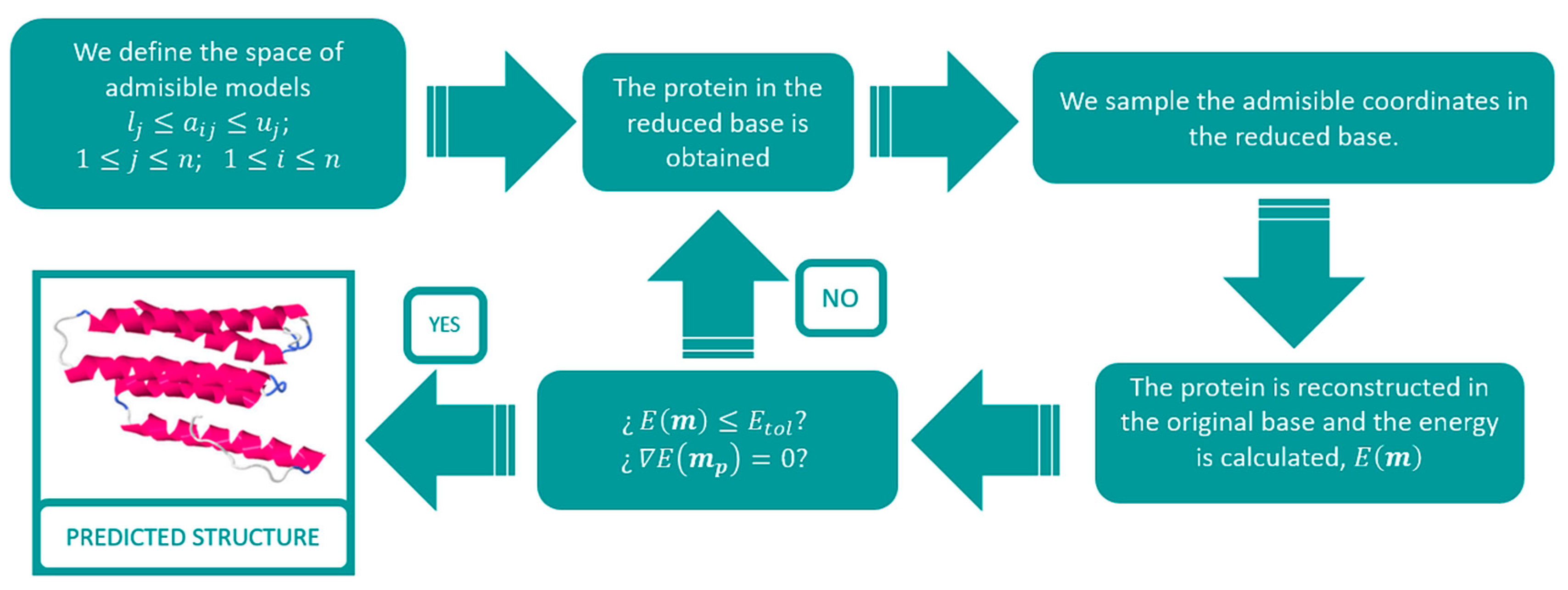
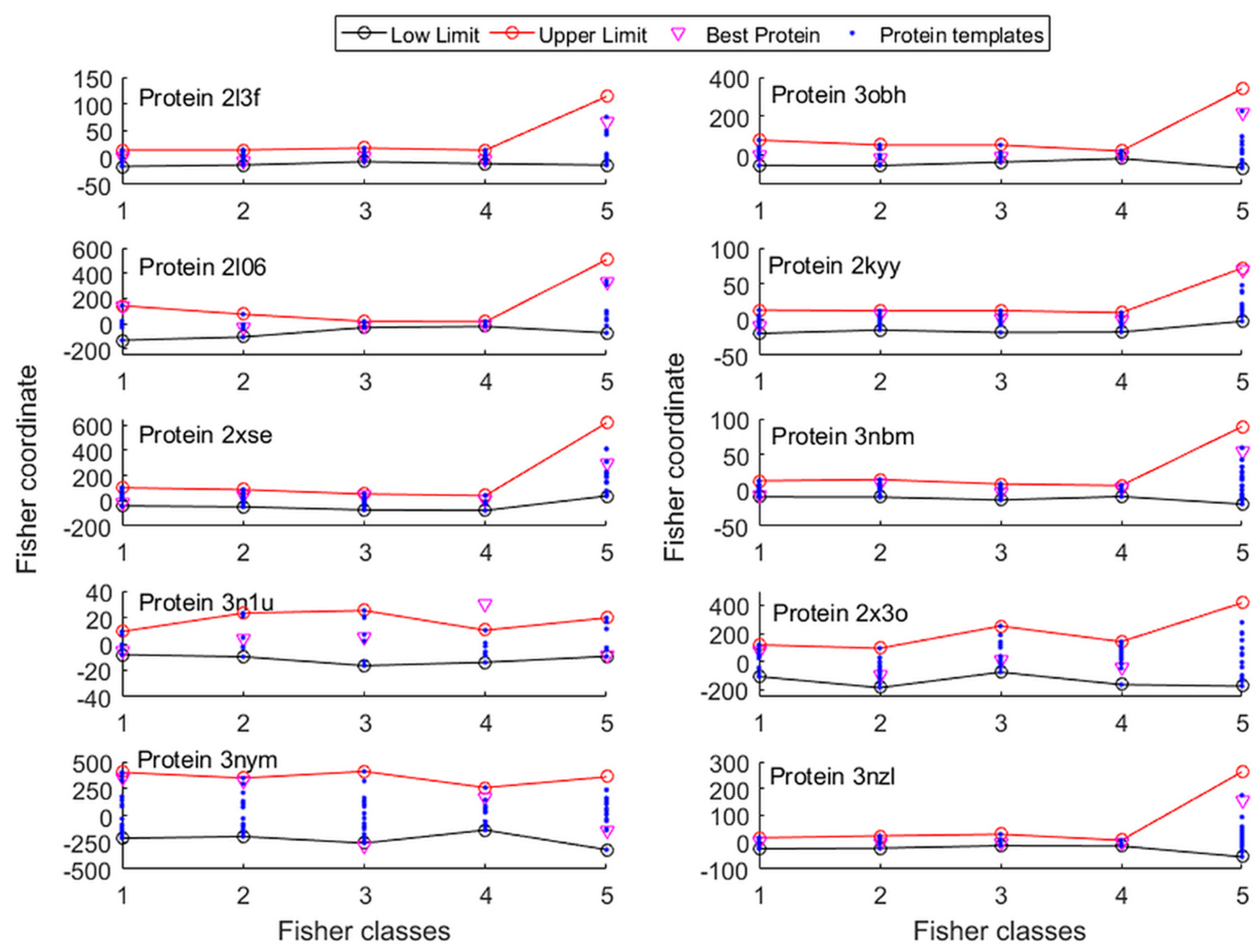
2.1. Template Selection and Model Reduction via an L2-Regularized LDA Discriminant Classifier
2.2. Protein Modelling
2.3. Optimization of the Protein Energy Function
2.4. Protein Refinement via Singular Value Decomposition
3. Results
3.1. Overview of Computational Experiments
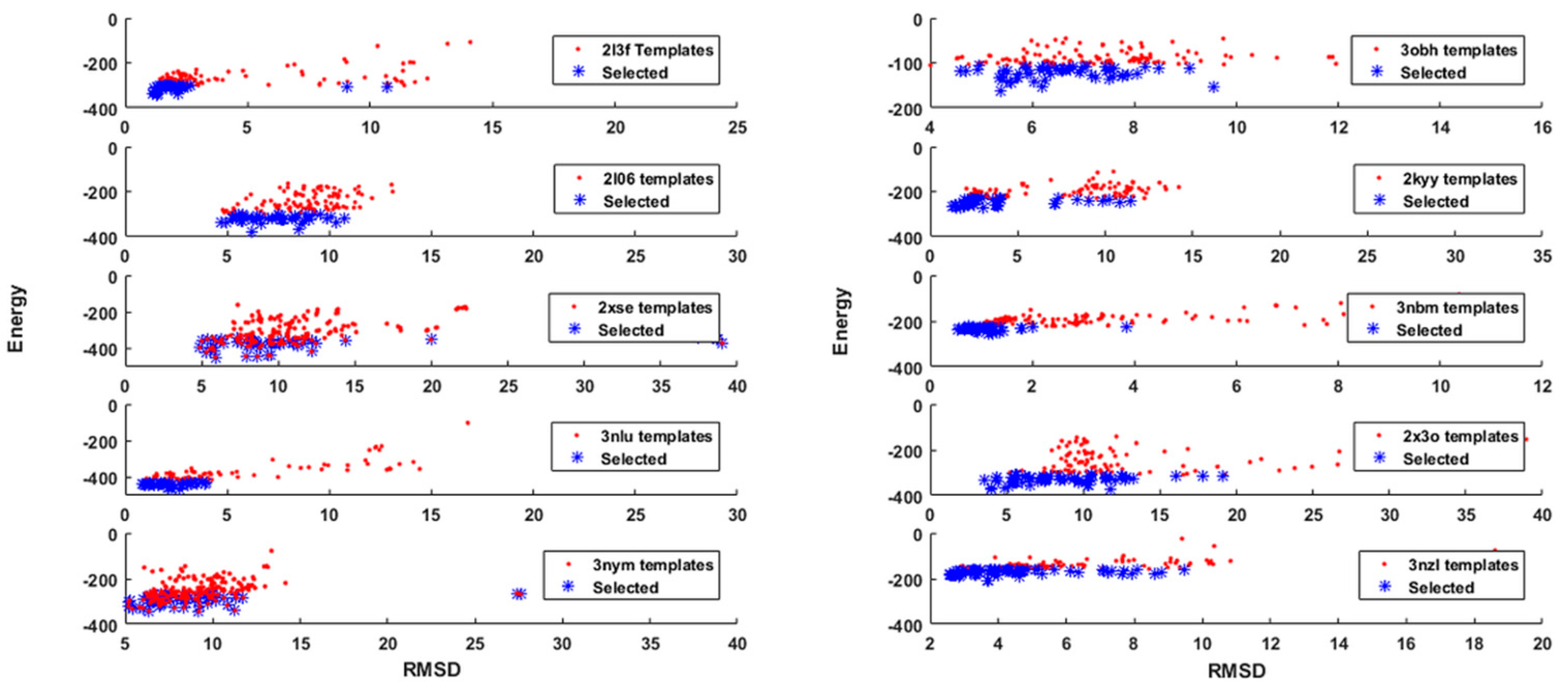
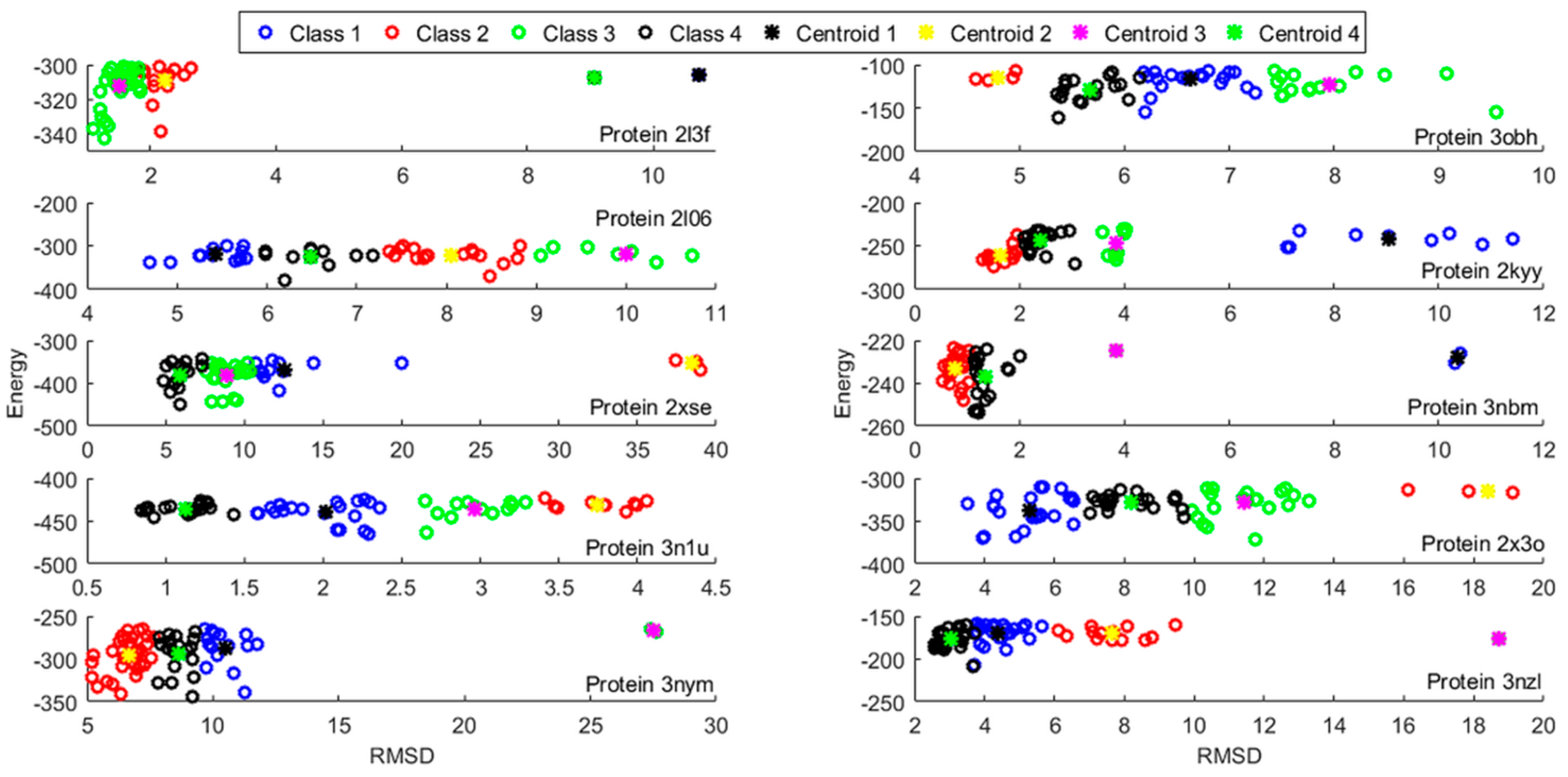
3.2. Template Selection and Protein Model Reduction
3.3. Protein Model Optimization and Refinement
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rose, P.W.; Prlić, A.; Altunkaya, A.; Bi, C.; Bradley, A.R.; Christie, C.H.; Costanzo, L.D.; Duarte, J.M.; Dutta, S.; Feng, Z.; et al. The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017, 45, D271–D281. [Google Scholar] [CrossRef]
- Zhang, Y. Progress and challenges in protein structure prediction. Curr. Opin. Struct. Boil. 2008, 18, 342–348. [Google Scholar] [CrossRef]
- Tyka, M.D.; Keedy, D.; André, I.; DiMaio, F.; Song, Y.; Richardson, D.C.; Richardson, J.S.; Baker, D. Alternate states of proteins revealed by detailed energy landscape mapping. J. Mol. Boil. 2010, 405, 607–618. [Google Scholar] [CrossRef]
- Fiser, A. Protein structure modeling in the proteomics era. Expert Rev. Proteom. 2004, 1, 97–110. [Google Scholar] [CrossRef]
- Marti-Renom, M.A.; Stuart, A.C.; Sali, A.; Sánchez, R.; Melo, F.; Sali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. [Google Scholar] [CrossRef]
- Chothia, C.; Lesk, A. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986, 5, 823–826. [Google Scholar] [CrossRef]
- Lesk, A.; Chothia, C. How different amino acid sequences determine similar protein structures: The structure and evolutionary dynamics of the globins. J. Mol. Boil. 1980, 136, 225–270. [Google Scholar] [CrossRef]
- Pieper, U. MODBASE: A database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006, 34, D291–D295. [Google Scholar] [CrossRef]
- Saraswathi, S.; Fernández-Martínez, J.L.; Kolinski, A.; Jernigan, R.L.; Kloczkowski, A. Fast learning optimized prediction methodology (FLOPRED) for protein secondary structure prediction. J. Mol. Model. 2012, 18, 4275–4289. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins Struct. Funct. Bioinform. 2007, 69, 108–117. [Google Scholar] [CrossRef]
- Das, R.; Qian, B.; Raman, S.; Vernon, R.; Thompson, J.; Bradley, P.; Khare, S.; Tyka, M.D.; Bhat, D.; Chivian, D.; et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins Struct. Funct. Bioinform. 2007, 69, 118–128. [Google Scholar] [CrossRef]
- Andreeva, A.; Howorth, D.; Chandonia, J.-M.; Brenner, S.E.; Hubbard, T.; Chothia, C.; Murzin, A.G. Data growth and its impact on the SCOP database: New developments. Nucleic Acids Res. 2007, 36, D419–D425. [Google Scholar] [CrossRef]
- Chothia, C.; Gough, J.; Vogel, C.; Teichmann, S. Evolution of the protein repertoire. Science 2003, 300, 1701–1703. [Google Scholar] [CrossRef]
- Greene, L.H.; Lewis, T.E.; Addou, S.; Cuff, A.L.; Dallman, T.; Dibley, M.; Redfern, O.; Pearl, F.M.; Nambudiry, R.; Reid, A.J.; et al. The CATH domain structure database: New protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2006, 35, D291–D297. [Google Scholar] [CrossRef]
- Battey, J.N.D.; Kopp, J.; Bordoli, L.; Read, R.; Clarke, N.D.; Schwede, T. Automated server predictions in CASP7. Proteins Struct. Funct. Bioinform. 2007, 69, 68–82. [Google Scholar] [CrossRef]
- Fernandez-Fuentes, N.; Madrid-Aliste, C.J.; Rai, B.K.; Fajardo, J.E.; Fiser, A. M4T: A comparative protein structure modeling server. Nucleic Acids Res. 2007, 35, W363–W368. [Google Scholar] [CrossRef]
- Rai, B.K.; Madrid-Aliste, C.J.; Fajardo, J.E.; Fiser, A. MMM: A sequence-to-structure alignment protocol. Bioinformatics 2006, 22, 2691–2692. [Google Scholar] [CrossRef][Green Version]
- Kopp, J.; Bordoli, L.; Battey, J.N.; Kiefer, F.; Schwede, T. Assessment of CASP7 predictions for template-based modeling targets. Proteins Struct. Funct. Bioinform. 2007, 69, 38–56. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol. 2003, 374, 461–491. [Google Scholar] [CrossRef]
- Contreras-Moreira, B.; Fitzjohn, P.W.; Offman, M.; Smith, G.R.; Bates, P.A. Novel use of a genetic algorithm for protein structure prediction: Searching template and sequence alignment space. Proteins Struct. Funct. Bioinform. 2003, 53, 424–429. [Google Scholar] [CrossRef]
- Schaffer, A.A.; Aravind, L.; Madden, T.L.; Shavirin, S.; Spouge, J.L.; Wolf, Y.I.; Koonin, E.V.; Altschul, S.F. Improving the accuracy of PSI-BLAST protein databasae searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001, 29, 2994–3005. [Google Scholar] [CrossRef]
- Brenner, S.E.; Chothia, C.; Hubbard, T. Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Nat. Acad. Sci. USA 1998, 95, 6073–6078. [Google Scholar] [CrossRef] [PubMed]
- Sauder, J.M.; Arthur, J.W.; Dunbrack, R.L., Jr. Large-scale comparison of protein sequence alignment algorithms with structure alignments. Proteins Struct. Funct. Bioinform. 2000, 40, 6–22. [Google Scholar] [CrossRef]
- Venclovas, C.; Margelevičius, M. Comparative modeling in CASP6 using consensus approach to template selection, sequence-structure alignment, and structure assessment. Proteins Struct. Funct. Bioinform. 2005, 61, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, R.; Sali, A. Evaluation of comparative protein structure modeling by MODELLER-3. Proteins Struct. Funct. Bioinform. 1997, 29, 50–58. [Google Scholar] [CrossRef]
- Eisenberg, D.; Lüthy, R.; Bowie, J.U. [20] VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997, 277, 396–404. [Google Scholar] [CrossRef]
- Petrey, D.; Xiang, Z.; Tang, C.L.; Xie, L.; Gimpelev, M.; Mitros, T.; Soto, C.S.; Goldsmith-Fischman, S.; Kernytsky, A.; Schlessinger, A.; et al. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins Struct. Funct. Bioinform. 2003, 53, 430–435. [Google Scholar] [CrossRef]
- Al-Lazikani, B.; Sheinerman, F.B.; Honig, B. Combining multiple structure and sequence alignments to improve sequence detection and alignment: Application to the SH2 domains of Janus kinases. PNAS 2001, 98, 14796–14801. [Google Scholar] [CrossRef]
- Reddy, B.V.; Li, W.W.; Shindyalov, I.N.; Bourne, P.E. Conserved key amino acid positions (CKAAPs) derived from the analysis of common substructures in proteins. Proteins Struct. Funct. Bioinform. 2001, 42, 148–163. [Google Scholar] [CrossRef]
- Rai, B.K.; Fiser, A. Multiple mapping method: A novel approach to the sequence-to-structure alignment problem in comparative protein structure modeling. Proteins Struct. Funct. Bioinform. 2006, 63, 644–661. [Google Scholar] [CrossRef]
- Morales-Cordovilla, J.A.; Sanchez, V.; Ratajczak, M. Protein alignment based on higher order conditional random fields for template-based modeling. PLoS ONE 2018, 13, e0197912. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, M.; Haneef, I.; Carney, D.; Blundell, T. Knowledge based modelling of homologous proteins, part I: Three-dimensional frameworks derived from the simultaneous superposition of multiple structures. Protein Eng. Des. Sel. 1987, 1, 377–384. [Google Scholar] [CrossRef] [PubMed]
- John, B. Comparative protein structure modeling by iterative alignment, model building and model assessment. Nucleic Acids Res. 2003, 31, 3982–3992. [Google Scholar] [CrossRef] [PubMed]
- Chivian, D.; Baker, D. Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic Acids Res. 2006, 34, e112. [Google Scholar] [CrossRef]
- Bruccoleri, R.E.; Karplus, M. Prediction of the folding of short polypeptide segments by uniform conformational sampling. Biopolym. Orig. Res. Biomol. 1987, 26, 137–168. [Google Scholar] [CrossRef]
- Collura, V.; Higo, J.; Garnier, J. Modeling of protein loops by simulated annealing. Protein Sci. 1993, 2, 1502–1510. [Google Scholar] [CrossRef]
- Studer, G.; Tauriello, G.; Bienert, S.; Waterhouse, A.M.; Bertoni, M.; Bordoli, L.; Schwede, T.; Lepore, R. Modeling of protein tertiary and quaternary structures based on evolutionary information. Adv. Struct. Saf. Stud. 2018, 301–316. [Google Scholar] [CrossRef]
- Ciemny, M.P.; Badaczewska-Dawid, A.E.; Pikuzinska, M.; Kolinski, A.; Kmiecik, S. Modeling of Ddisordered protein structures using monte carlo simulations and knowledge-based statistical force fields. Int. J. Mol. Sci. 2019, 20, 606. [Google Scholar] [CrossRef]
- Hou, J.; Wu, T.; Cao, R.; Cheng, J. Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13. Proteins Struct. Funct. Bioinform. 2019, 87, 1165–1178. [Google Scholar] [CrossRef]
- Fine, R.M.; Wang, H.; Shenkin, P.S.; Yarmush, D.L.; Levinthal, C. Predicting antibody hypervariable loop conformations II: Minimization and molecular dynamics studies of MCPC603 from many randomly generated loop conformations. Proteins Struct. Funct. Bioinform. 1986, 1, 342–362. [Google Scholar] [CrossRef]
- Zheng, Q.; Rosenfeld, R.; Vajda, S.; DeLisi, C. Determining protein loop conformation using scaling-relaxation techniques. Protein Sci. 1993, 2, 1242–1248. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Machancoses, Ó.; Fernández-Martínez, J.; Fernández-Brillet, C.; Cernea, A.; Fernández-Muñiz, Z.; Kloczkowski, A. Principal component analysis in protein tertiary structure prediction. J. Bioinform. Comput. Boil. 2018, 16, 1850005. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Machancoses, Ó.; Fernández-Martínez, J.L.; Corbeanu, A.C.; Fernández-Muñiz, Z.; Kloczkowski, A. Predicting protein tertiary structure and its uncertainty analysis via particle swarm sampling. J. Mol. Model. 2019, 25, 79. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Dudoit, S.; Fridlyand, J.; Speed, T.P. Comparison of discrimination methods for the classification of tumors using gene expression data. J. Am. Stat. Assoc. 2002, 97, 77–87. [Google Scholar] [CrossRef]
- Ye, J.; Li, T.; Xiong, T.; Janardan, R. Using uncorrelated discriminant analysis for tisue classification with gene expression data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2004, 1, 181–190. [Google Scholar] [CrossRef]
- Sharma, A.; Paliwal, K.K. Cancer classification by gradient LDA technique using microarray gene expression data. Data Knowl. Eng. 2008, 66, 338–347. [Google Scholar] [CrossRef]
- Kalina, J.; Matonoha, C. A sparse pair-preserving centroid-based supervised learning method for high-dimensional biomedical data or images. Biocybern. Biomed. Eng. 2020, 40, 774–786. [Google Scholar] [CrossRef]
- Cernea, A.; Fernández-Martínez, J.; De Andrés-Galiana, E.J.; Fernández-Ovies, F.J.; Fernández-Muñiz, Z.; Álvarez-Machancoses, Ó.; Saligan, L.; Sonis, S.T. Sampling defective pathways in phenotype prediction problems via the fisher’s ratio sampler. In Computer Vision; Springer Science and Business Media: Berlin, Germany, 2018; Volume 10814, pp. 15–23. [Google Scholar]
- Yang, Y.; Zhou, Y. Specific interactions for ab initio folding of protein terminal regions with secondary structures. Proteins Struct. Funct. Bioinform. 2008, 72, 793–803. [Google Scholar] [CrossRef]
- Qiu, D.; Shenkin, P.S.; Hollinger, F.P.; Still, W.C. The GB/SA continuum model for solvation. A fast analytical method for the calculation of approximate born radii. J. Phys. Chem. A 1997, 101, 3005–3014. [Google Scholar] [CrossRef]
- Kalina, J.; Tebbens, E.J.D. Algorithms for regularized linear discriminant analysis. BIOINFORMATICS 2015, 1, 128–133. [Google Scholar]
- Schäfer, J.; Strimmer, K. A shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat. Appl. Genet. Mol. Boil. 2005, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Tarantola, A. Inverse Problem Theory and Methods for Model Parameter Estimation; Society for Industrial & Applied Mathematics (SIAM): Philadelphia, PA, USA, 2005. [Google Scholar]
- Fernández-Martínez, J. Model reduction and uncertainty analysis in inverse problems. Lead. Edge 2015, 34, 1006–1016. [Google Scholar] [CrossRef]
- Gniewek, P.; Kolinski, A.; Kloczkowski, A.; Gront, D. BioShell-threading: Versatile monte carlo package for protein 3D threading. BMC Bioinform. 2014, 15, 22. [Google Scholar] [CrossRef] [PubMed]
- Gniewek, P.; Koliński, A.; Jernigan, R.L.; Kloczkowski, A. How noise in force fields can affect the structural refinement of protein models? Proteins Struct. Funct. Bioinform. 2011, 80, 335–341. [Google Scholar] [CrossRef]
- Gront, D.; Kolinski, A. BioShell–A package of tools for structural biology prediction. Bioinformatics 2006, 22, 621–622. [Google Scholar] [CrossRef]
- Gront, D.; Kolinski, A. Utility library for structural bioinformatics. Bioinformatics 2008, 24, 584–585. [Google Scholar] [CrossRef]
- Price, S.L. From crystal structure prediction to polymorph prediction: Interpreting the crystal energy landscape. Phys. Chem. Chem. Phys. 2008, 10, 1996. [Google Scholar] [CrossRef]
- Fernández-Martínez, J.L.; Pallero, J.L.G.; Fernández-Muñiz, Z. Pedruelo-González, L.M. The effect of the noise and Tikhonov’s regularization in inverse problems. Part I: The linear case. J. Appl. Geophys. 2014, 108, 176–185. [Google Scholar] [CrossRef]
- Fernández-Martínez, J.L.; Pallero, J.L.G.; Fernández-Muñiz, Z. Pedruelo-González, L.M. The effect of the noise and Tikhonov’s regularization in inverse problems. Part II: The nonlinear case. J. Appl. Geophys. 2014, 108, 186–193. [Google Scholar] [CrossRef]
- García-Gonzalo, E.; Fernández-Martínez, J. A brief historical review of particle sSwarm optimization (PSO). J. Bioinform. Intell. Control. 2012, 1, 3–16. [Google Scholar] [CrossRef]
- Fernández-Martínez, J.; Álvarez, J.P.F.; García-Gonzalo, M.E.; Pérez, C.O.M.; Kuzma, H.A.; Stark, T.P.C.T.J. Particle Swarm Optimization (PSO): A simple and powerful algorithm family for geophysical inversion. 2008 SEG Annu. Meet. 2008, 3568–3571. [Google Scholar] [CrossRef]
- Fernández-Martínez, J.; García-Gonzalo, E. Stochastic stability and numerical analysis of two novel algorithms of the PSO family: PP-GPSO and RR-GPSO. Int. J. Artif. Intell. Tools 2012, 21, 1240011. [Google Scholar] [CrossRef]
- Kennedy, J.; Eberhart, R. A new optimizer using particle swarm theory. In Proceedings of the MHS’95. Proceedings of the Sixth International Symposium on Micro Machine and Human Science, Nagoya, Japan, 4–6 October 1995; pp. 39–43. [Google Scholar] [CrossRef]
- Álvarez-Machancoses, Ó.; Fernández-Martínez, J.; Cernea, A.; Kloczkowski, A. Protein tertiary structure prediction via SVD and PSO sampling. In Bioinformatics and Biomedical Engineering. IWBBIO 2018. Lecture Notes in Computer Science; Springer Science and Business Media: Berlin, Germany, 2018; Volume 10813, pp. 211–220. [Google Scholar]
- Qian, B.; Ortiz, A.R.; Baker, D. Improvement of comparative model accuracy by free-energy optimization along principal components of natural structural variation. Proc. Natl. Acad. Sci. USA 2004, 101, 15346–15351. [Google Scholar] [CrossRef]
Sample Availability: Templates utilized for the predictions are available at https://predictioncenter.org/. |

| Protein (CASP Code) | Number of Residues | Number of Templates | Number of Classes |
|---|---|---|---|
| 2l3f (T0545) | 166 | 185 | 4 |
| 3obh (T0551) | 82 | 199 | 4 |
| 2l06 (T0555) | 155 | 182 | 4 |
| 2kyy (T0557) | 153 | 183 | 4 |
| 2xse (T0561) | 170 | 180 | 4 |
| 3nbm (T0580) | 108 | 195 | 4 |
| 3n1u (T0635) | 191 | 181 | 4 |
| 2x3o (T0637) | 240 | 194 | 4 |
| 3nym (T0639) | 128 | 206 | 4 |
| 3nzl (T0643) | 82 | 178 | 4 |
| 4pqx (T0760) | 217 | 94 | 4 |
| 4q69 (T0770) | 462 | 100 | 4 |
| 4qdy (T0780) | 227 | 103 | 4 |
| 4l4w (T0790) | 295 | 107 | 4 |
| 4qrk (T0800) | 220 | 277 | 4 |
| Q6MI90_BDEBA (T0810) | 383 | 164 | 4 |
| VCID6010 (T0820) | 140 | 333 | 4 |
| 5f15 (T0830) | 575 | 225 | 4 |
| 4gt8 (T0840) | 669 | 96 | 4 |
| U1 Protein (T0850) | 190 | 268 | 4 |
| 5d9g (T0864) | 246 | 264 | 4 |
| 5j5v (T0870) | 323 | 268 | 4 |
| 1ctf (T0880) | 787 | 321 | 4 |
| 5t87 (T0885) | 116 | 122 | 4 |
| 3k1e (T0890) | 125 | 321 | 4 |
| 5aot (T0900) | 106 | 255 | 4 |
| 6c0t (T0910) | 347 | 105 | 4 |
| 5ere (T0920) | 568 | 91 | 4 |
| 5sy1 (T0930) | 149 | 187 | 4 |
| 1o6d (T0940) | 163 | 259 | 4 |
| Protein (CASP Code) | Number of Residues | Number of Classes | Reduced Basis Terms | Percentile of Decoys | Number of Iterations | Swarm Size | Energy Obtained |
|---|---|---|---|---|---|---|---|
| 2l3f (T0545) | 166 | 4 | 5 | 30 | 50 | 40 | −343.86 |
| 3obh (T0551) | 82 | 4 | 5 | 30 | 50 | 40 | −163.42 |
| 2l06 (T0555) | 155 | 4 | 5 | 30 | 50 | 40 | −381.96 |
| 2kyy (T0557) | 153 | 4 | 5 | 30 | 50 | 40 | −152.77 |
| 2xse (T0561) | 170 | 4 | 5 | 30 | 50 | 40 | −449.50 |
| 3nbm (T0580) | 108 | 4 | 5 | 30 | 50 | 40 | −255.42 |
| 3n1u (T0635) | 191 | 4 | 5 | 30 | 50 | 40 | −369.47 |
| 2x3o (T0637) | 240 | 4 | 5 | 30 | 50 | 40 | −372.10 |
| 3nym (T0639) | 128 | 4 | 5 | 30 | 50 | 40 | −343.22 |
| 3nzl (T0643) | 82 | 4 | 5 | 30 | 50 | 40 | −210.34 |
| 4pqx (T0760) | 217 | 4 | 5 | 30 | 50 | 40 | −496.11 |
| 4q69 (T0770) | 462 | 4 | 5 | 30 | 50 | 40 | −992.46 |
| 4qdy (T0780) | 227 | 4 | 5 | 30 | 50 | 40 | −425.77 |
| 4l4w (T0790) | 295 | 4 | 5 | 30 | 50 | 40 | −598.56 |
| 4qrk (T0800) | 220 | 4 | 5 | 30 | 50 | 40 | −502.35 |
| Q6MI90_BDEBA (T0810) | 383 | 4 | 5 | 30 | 50 | 40 | −902.65 |
| VCID6010 (T0820) | 140 | 4 | 5 | 30 | 50 | 40 | −356.56 |
| 5f15 (T0830) | 575 | 4 | 5 | 30 | 50 | 40 | −1214.65 |
| 4gt8 (T0840) | 669 | 4 | 5 | 30 | 50 | 40 | −1115.98 |
| U1 Protein (T0850) | 190 | 4 | 5 | 30 | 50 | 40 | −448.13 |
| 5d9g (T0864) | 246 | 4 | 5 | 30 | 50 | 40 | −545.61 |
| 5j5v (T0870) | 323 | 4 | 5 | 30 | 50 | 40 | −408.32 |
| 1ctf (T0880) | 787 | 4 | 5 | 30 | 50 | 40 | −398.39 |
| 5t87 (T0885) | 116 | 4 | 5 | 30 | 50 | 40 | −298.43 |
| 3k1e (T0890) | 125 | 4 | 5 | 30 | 50 | 40 | −561.94 |
| 5aot (T0900) | 106 | 4 | 5 | 30 | 50 | 40 | −208.01 |
| 6c0t (T0910) | 347 | 4 | 5 | 30 | 50 | 40 | −838.89 |
| 5ere (T0920) | 568 | 4 | 5 | 30 | 50 | 40 | −1229.68 |
| 5sy1 (T0930) | 149 | 4 | 5 | 30 | 50 | 40 | −1812.17 |
| 1o6d (T0940) | 163 | 4 | 5 | 30 | 50 | 40 | −627.02 |
| Protein (CASP Code) | RMSD LDA–SVD | RMSD Zhang Server | RMSD Rosetta Server |
|---|---|---|---|
| 2l3f (T0545) | 1.27 | 2.17 | 2.38 |
| 3obh (T0551) | 5.30 | 2.75 | 2.65 |
| 2l06 (T0555) | 6.16 | 2.99 | 3.20 |
| 2kyy (T0557) | 1.16 | 2.54 | 2.08 |
| 2xse (T0561) | 5.88 | 3.01 | 3.09 |
| 3nbm (T0580) | 1.16 | 1.80 | 1.37 |
| 3n1u (T0635) | 1.31 | 0.74 | 1.08 |
| 2x3o (T0637) | 5.18 | 2.44 | 2.61 |
| 3nym (T0639) | 6.70 | 2.74 | 2.11 |
| 3nzl (T0643) | 3.67 | 2.72 | 2.75 |
| 4pqx (T0760) | 2.73 | 2.93 | 3.21 |
| 4q69 (T0770) | 5.01 | 4.53 | 4.47 |
| 4qdy (T0780) | 3.12 | 2.97 | 2.93 |
| 4l4w (T0790) | 3.81 | 4.96 | 4.48 |
| 4qrk (T0800) | 12.25 | 7.25 | 9.80 |
| Q6MI90_BDEBA (T0810) | 13.85 | 8.30 | 14.76 |
| VCID6010 (T0820) | 12.97 | 9.32 | 14.75 |
| 5f15 (T0830) | 26.57 | 20.77 | 11.15 |
| 4gt8 (T0840) | 3.03 | 2.71 | 4.99 |
| U1 Protein (T0850) | 3.65 | 3.48 | 4.03 |
| 5d9g (T0864) | 2.81 | 2.58 | 2.10 |
| 5j5v (T0870) | 17.12 | 12.67 | 11.84 |
| 1ctf (T0880) | 6.66 | 8.69 | 7.59 |
| 5t87 (T0885) | 2.87 | 3.92 | 2.93 |
| 3k1e (T0890) | 8.93 | 3.99 | 8.02 |
| 5aot (T0900) | 3.32 | 7.63 | 4.67 |
| 6c0t (T0910) | 1.74 | 1.58 | 1.83 |
| 5ere (T0920) | 2.38 | 2.40 | 2.32 |
| 5sy1 (T0930) | 4.56 | 3.27 | 4.25 |
| 1o6d (T0940) | 4.13 | 3.71 | 3.07 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez-Machancoses, Ó.; Fernández-Martínez, J.L.; Kloczkowski, A. Prediction of Protein Tertiary Structure via Regularized Template Classification Techniques. Molecules 2020, 25, 2467. https://doi.org/10.3390/molecules25112467
Álvarez-Machancoses Ó, Fernández-Martínez JL, Kloczkowski A. Prediction of Protein Tertiary Structure via Regularized Template Classification Techniques. Molecules. 2020; 25(11):2467. https://doi.org/10.3390/molecules25112467
Chicago/Turabian StyleÁlvarez-Machancoses, Óscar, Juan Luis Fernández-Martínez, and Andrzej Kloczkowski. 2020. "Prediction of Protein Tertiary Structure via Regularized Template Classification Techniques" Molecules 25, no. 11: 2467. https://doi.org/10.3390/molecules25112467
APA StyleÁlvarez-Machancoses, Ó., Fernández-Martínez, J. L., & Kloczkowski, A. (2020). Prediction of Protein Tertiary Structure via Regularized Template Classification Techniques. Molecules, 25(11), 2467. https://doi.org/10.3390/molecules25112467







