Comparison of Stir Bar Sorptive Extraction and Solid Phase Microextraction of Volatile and Semi-Volatile Metabolite Profile of Staphylococcus Aureus
Abstract
1. Introduction
2. Results
2.1. Methodology for Compound Identification
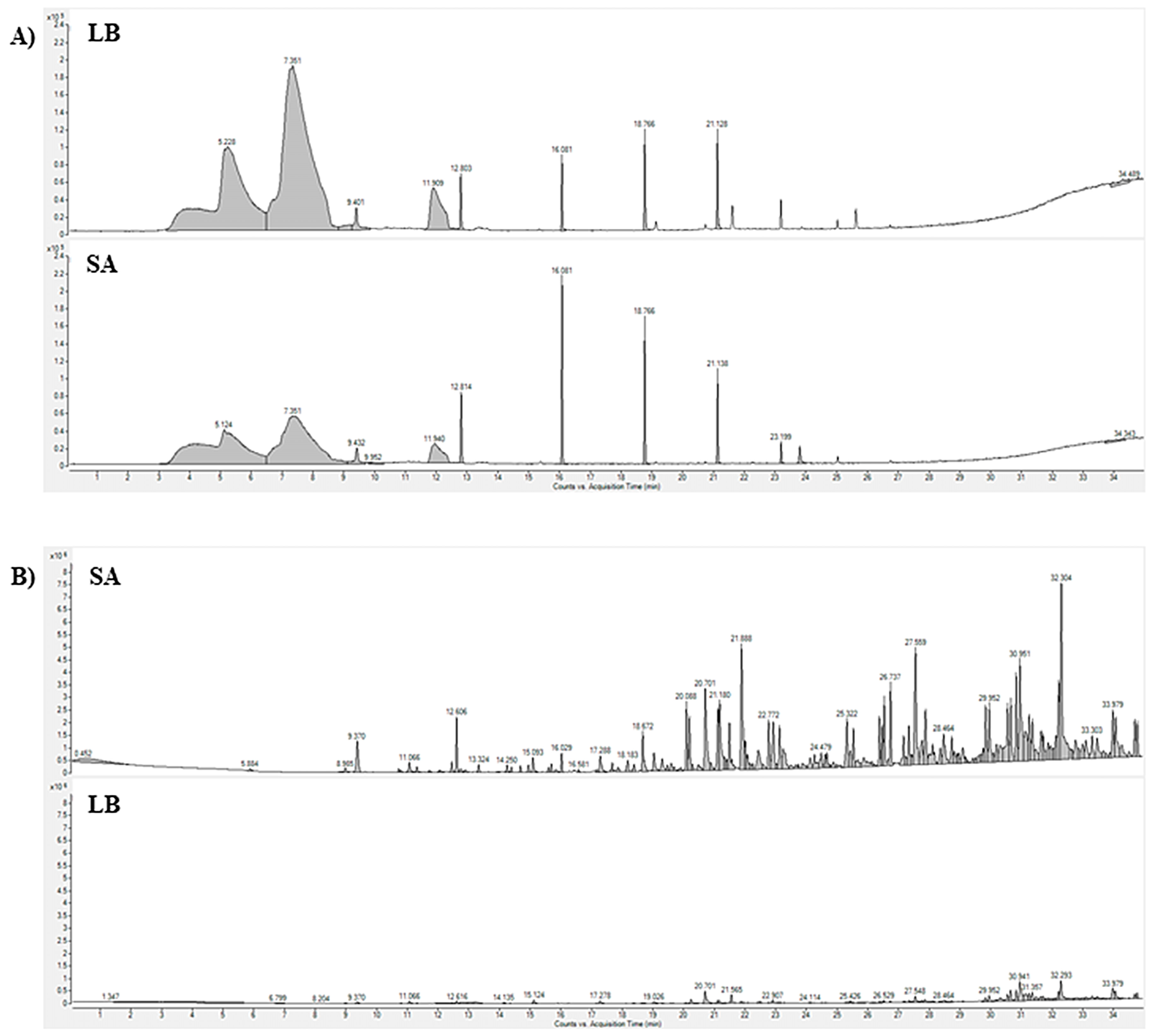
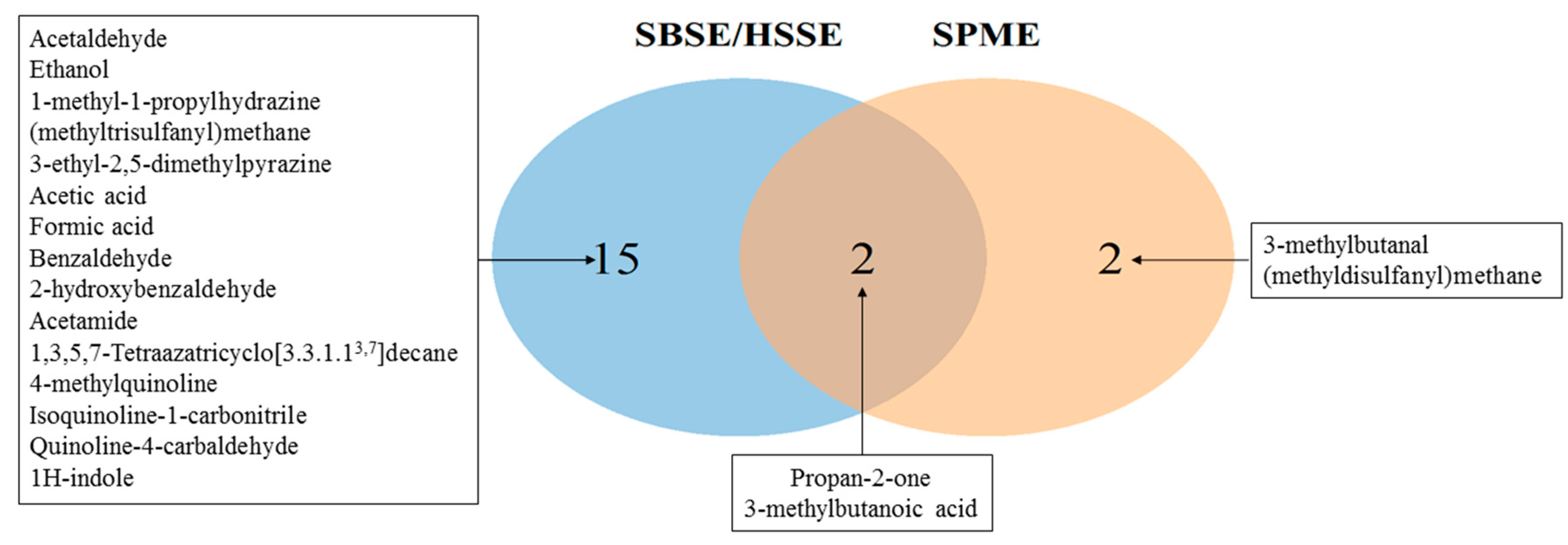
2.2. Comparison of Extraction Method (SPME and SBSE/HSSE)
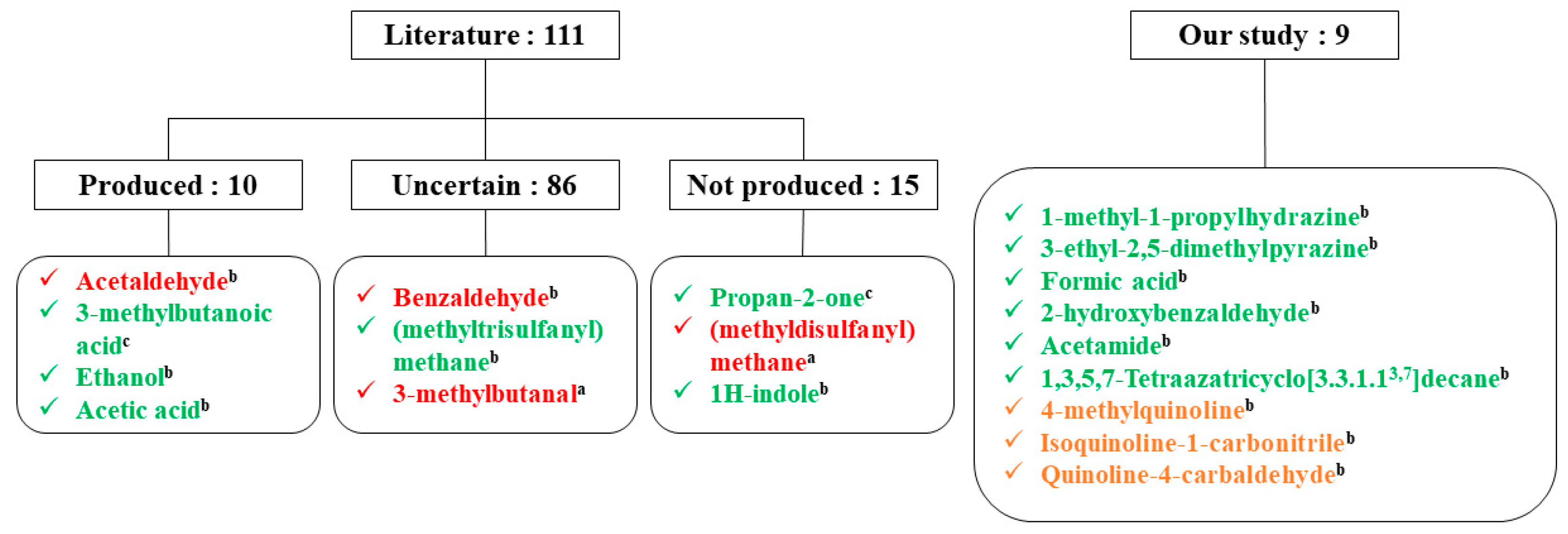
2.3. Validation of the Tentative Identification
2.4. Statistical Analysis to Determine Compounds Consumed and Produced by S. Aureus
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Culture Conditions
4.3. Headspace-SPME Extraction Procedure
4.4. SBSE/HSSE Extraction Procedure
4.5. GC–MS Analysis
4.6. Data Processing and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Snow, N.H. Solid-phase micro-extraction of drugs from biological matrices. J. Chromatogr. A 2000, 885, 445–455. [Google Scholar] [CrossRef]
- Ulrich, S. Solid-phase microextraction in biomedical analysis. J. Chromatogr. A 2000, 902, 167–194. [Google Scholar] [CrossRef]
- Filipiak, W.; Sponring, A.; Mikoviny, T.; Ager, C.; Schubert, J.; Miekisch, W.; Amann, A.; Troppmair, J. Release of volatile organic compounds (VOCs) from the lung cancer cell line CALU-1 in vitro. Cancer. Cell. Int. 2008, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Garner, C.E.; Smith, S.; de Lacy Costello, B.; White, P.; Spencer, R.; Probert, C.S.J.; Ratcliffe, N.M. Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. Faseb. J. 2007, 21, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Filipiak, W.; Sponring, A.; Filipiak, A.; Ager, C.; Schubert, J.; Miekisch, W.; Amann, A.; Troppmair, J. TD-GC-MS analysis of volatile metabolites of human lung cancer and normal cells in vitro. Cancer. Epidemiol. Biomark. Prev. 2010, 19, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Neerincx, A.H.; Geurts, B.P.; van Loon, J.; Tiemes, V.; Jansen, J.J.; Harren, F.J.M.; Kluijtmans, L.A.J.; Merkus, P.J.F.M.; Cristescu, S.M.; Buydens, L.M.C.; et al. Detection of Staphylococcus aureus in cystic fibrosis patients using breath VOC profiles. J. Breath. Res. 2016, 10, 046014. [Google Scholar] [CrossRef] [PubMed]
- Barker, M.; Hengst, M.; Schmid, J.; Buers, H.-J.; Mittermaier, B.; Klemp, D.; Koppmann, R. Volatile organic compounds in the exhaled breath of young patients with cystic fibrosis. Eur. Respir. J. 2006, 27, 929–936. [Google Scholar] [CrossRef]
- Stoppacher, N.; Kluger, B.; Zeilinger, S.; Krska, R.; Schuhmacher, R. Identification and profiling of volatile metabolites of the biocontrol fungus Trichoderma atroviride by HS-SPME-GC-MS. J. Microbiol. Methods 2010, 81, 187–193. [Google Scholar] [CrossRef]
- Lemfack, M.C.; Nickel, J.; Dunkel, M.; Preissner, R.; Piechulla, B. mVOC: A database of microbial volatiles. Nucleic. Acids. Res. 2014, 42, D744–D748. [Google Scholar] [CrossRef]
- Arthur, C.L.; Pawliszyn, J. Solid phase microextraction with thermal desorption using fused silica optical fibers. Anal. Chem. 1990, 62, 2145–2148. [Google Scholar] [CrossRef]
- Musteata, F.M.; de Lannoy, I.; Gien, B.; Pawliszyn, J. Blood sampling without blood draws for in vivo pharmacokinetic studies in rats. J. Pharm. Biomed. Anal. 2008, 47, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Bojko, B.; Gorynski, K.; Gomez-Rios, G.A.; Knaak, J.M.; Machuca, T.; Cudjoe, E.; Spetzler, V.N.; Hsin, M.; Cypel, M.; Selzner, M.; et al. Low invasive in vivo tissue sampling for monitoring biomarkers and drugs during surgery. Lab. Invest. 2014, 94, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Julák, J.; Procházková-Francisci, E.; Stránská, E.; Rosová, V. Evaluation of exudates by solid phase microextraction-gas chromatography. J. Microbiol. Methods 2003, 52, 115–122. [Google Scholar] [CrossRef]
- Preti, G.; Thaler, E.; Hanson, C.W.; Troy, M.; Eades, J.; Gelperin, A. Volatile compounds characteristic of sinus-related bacteria and infected sinus mucus: Analysis by solid-phase microextraction and gas chromatography-mass spectrometry. J. Chromatogr. B 2009, 877, 2011–2018. [Google Scholar] [CrossRef]
- Tait, E.; Perry, J.D.; Stanforth, S.P.; Dean, J.R. Identification of volatile organic compounds produced by bacteria using HS-SPME-GC-MS. J. Chromatogr. Sci. 2014, 52, 363–373. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Shi, H.; Tang, C.; Zhang, R. Characteristics of volatile organic compounds produced from five pathogenic bacteria by headspace-solid phase micro-extraction/gas chromatography-mass spectrometry. J. Basic. Microbiol. 2017, 57, 228–237. [Google Scholar] [CrossRef]
- Baltussen, E.; Sandra, P.; David, F.; Cramers, C. Stir bar sorptive extraction (SBSE), a novel extraction technique for aqueous samples: Theory and principles. J. Microcolumn. Sep. 1999, 11, 737–747. [Google Scholar] [CrossRef]
- Berrou, K.; Dunyach-Remy, C.; Lavigne, J.-P.; Roig, B.; Cadiere, A. Multiple stir bar sorptive extraction combined with gas chromatography-mass spectrometry analysis for a tentative identification of bacterial volatile and/or semi-volatile metabolites. Talanta 2019, 195, 245–250. [Google Scholar] [CrossRef]
- Dong, Y.; Glaser, K.; Speer, C.P. New Threats from an Old Foe: Methicillin-Resistant Staphylococcus aureus Infections in Neonates. Neonatology 2018, 114, 127–134. [Google Scholar] [CrossRef]
- Fisher, E.L.; Otto, M.; Cheung, G.Y.C. Basis of Virulence in Enterotoxin-Mediated Staphylococcal Food Poisoning. Front. Microbiol. 2018, 9, 436. [Google Scholar] [CrossRef]
- Dayan, G.H.; Mohamed, N.; Scully, I.L.; Cooper, D.; Begier, E.; Eiden, J.; Jansen, K.U.; Gurtman, A.; Anderson, A.S. Staphylococcus aureus: The current state of disease, pathophysiology and strategies for prevention. Expert. Rev. Vaccines. 2016, 15, 1373–1392. [Google Scholar] [CrossRef] [PubMed]
- Krismer, B.; Weidenmaier, C.; Zipperer, A.; Peschel, A. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol. 2017, 15, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Thomer, L.; Schneewind, O.; Missiakas, D. Pathogenesis of Staphylococcus aureus Bloodstream Infections. Annu. Rev. Pathol. 2016, 11, 343–364. [Google Scholar] [CrossRef] [PubMed]
- Bos, L.D.J.; Sterk, P.J.; Schultz, M.J. Volatile metabolites of pathogens: A systematic review. Plos. Pathog. 2013, 9, e1003311. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Dickschat, J.S. Bacterial volatiles: The smell of small organisms. Nat. Prod. Rep. 2007, 24, 814–842. [Google Scholar] [CrossRef]
- Sethi, S.; Nanda, R.; Chakraborty, T. Clinical application of volatile organic compound analysis for detecting infectious diseases. Clin. Microbiol. Rev. 2013, 26, 462–475. [Google Scholar] [CrossRef]
- Martinez-Lozano Sinues, P.; Landoni, E.; Miceli, R.; Dibari, V.F.; Dugo, M.; Agresti, R.; Tagliabue, E.; Cristoni, S.; Orlandi, R. Secondary electrospray ionization-mass spectrometry and a novel statistical bioinformatic approach identifies a cancer-related profile in exhaled breath of breast cancer patients: A pilot study. J. Breath. Res. 2015, 9, 031001. [Google Scholar] [CrossRef]
- Ochiai, N.; Sasamoto, K.; Ieda, T.; David, F.; Sandra, P. Multi-stir bar sorptive extraction for analysis of odor compounds in aqueous samples. J. Chromatogr. A 2013, 1315, 70–79. [Google Scholar] [CrossRef]
- Ye, Q.; Zheng, D.; Liu, L.; Hong, L. Rapid analysis of aldehydes by simultaneous microextraction and derivatization followed by GC-MS. J. Sep. Sci. 2011, 34, 1607–1612. [Google Scholar] [CrossRef]
- Scotter, J.M.; Allardyce, R.A.; Langford, V.S.; Hill, A.; Murdoch, D.R. The rapid evaluation of bacterial growth in blood cultures by selected ion flow tube-mass spectrometry (SIFT-MS) and comparison with the BacT/ALERT automated blood culture system. J. Microbiol. Methods 2006, 65, 628–631. [Google Scholar] [CrossRef]
- Zhu, J.; Bean, H.D.; Kuo, Y.-M.; Hill, J.E. Fast detection of volatile organic compounds from bacterial cultures by secondary electrospray ionization-mass spectrometry. J. Clin. Microbiol. 2010, 48, 4426–4431. [Google Scholar] [CrossRef] [PubMed]
- Jünger, M.; Vautz, W.; Kuhns, M.; Hofmann, L.; Ulbricht, S.; Baumbach, J.I.; Quintel, M.; Perl, T. Ion mobility spectrometry for microbial volatile organic compounds: A new identification tool for human pathogenic bacteria. Appl. Microbiol. Biotechnol. 2012, 93, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Thorn, R.M.S.; Reynolds, D.M.; Greenman, J. Multivariate analysis of bacterial volatile compound profiles for discrimination between selected species and strains in vitro. J. Microbiol. Methods 2011, 84, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Dolch, M.E.; Hornuss, C.; Klocke, C.; Praun, S.; Villinger, J.; Denzer, W.; Schelling, G.; Schubert, S. Volatile organic compound analysis by ion molecule reaction mass spectrometry for Gram-positive bacteria differentiation. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3007–3013. [Google Scholar] [CrossRef] [PubMed]
- Allardyce, R.A.; Hill, A.L.; Murdoch, D.R. The rapid evaluation of bacterial growth and antibiotic susceptibility in blood cultures by selected ion flow tube mass spectrometry. Diagn. Microbiol. Infect. Dis. 2006, 55, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Storer, M.K.; Hibbard-Melles, K.; Davis, B.; Scotter, J. Detection of volatile compounds produced by microbial growth in urine by selected ion flow tube mass spectrometry (SIFT-MS). J. Microbiol. Methods 2011, 87, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Mesureur, J.; Ranaldi, S.; Monnin, V.; Girard, V.; Arend, S.; Welker, M.; O’Callaghan, D.; Lavigne, J.-P.; Keriel, A. A simple and safe protocol for preparing Brucella samples for Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry analysis. J. Clin. Microbiol. 2016, 54, 449–452. [Google Scholar] [CrossRef]
- Van Den Dool, H.; Kratz, P.D. A generalization of the retention index system including linear temperature programmed gas—liquid partition chromatography. J. Chromatogr. A 1963, 11, 463–471. [Google Scholar] [CrossRef]
- Eom, I.-Y.; Risticevic, S.; Pawliszyn, J. Simultaneous sampling and analysis of indoor air infested with Cimex lectularius L. (Hemiptera: Cimicidae) by solid phase microextraction, thin film microextraction and needle trap device. Anal. Chim. Acta. 2012, 716, 2–10. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors because there are destroyed by the analysis. Strains are available and cultures and analysis can be reproduced. |
| Volatile/Semi-Volatile Metabolite | Identification | Characterisation | ||||
|---|---|---|---|---|---|---|
| LRI a | Percentage ΔLRI | Area Variation | p-Value | Produced/Consumed by S. aureus | ||
| Measured | Literature | |||||
| Acetaldehyde c | 803 | 714 | 12.5% | ↘ | 3.12 × 10−3 | Consumed |
| Propan-2-one c | 843 | 818 | 3.1% | ↗ | 4.80 × 10−5 | Produced |
| 3-methylbutanal | 866 | 876 | 1.1% | ↘ | 6.38 × 10−5 | Consumed |
| Ethanol c | 938 | 940 | 0.2% | ↗ | 5.32 × 10−6 | Produced |
| (methyldisulfanyl)methane | 1059 | 1039 | 1.9% | ↘ | 2.92 × 10−4 | Consumed |
| 1-methyl-1-propylhydrazine | 1066 | - b | Produced | |||
| (methyltrisulfanyl)methane | 1376 | 1376 | 0.0% | ↗ | 3.38 × 10−2 | Produced |
| 3-ethyl-2,5-dimethylpyrazine | 1454 | 1450 | 0.3% | Produced | ||
| Acetic acid c | 1464 | 1479 | 1.0% | ↗ | 7.54 × 10−3 | Produced |
| Formic acid | 1531 | 1510 | 1.4% | ↗ | 4.89 × 10−6 | Produced |
| Benzaldehyde c | 1542 | 1534 | 0.5% | ↘ | 1.97 × 10−4 | Consumed |
| 3-methylbutanoic acid c | 1645 | 1655 | 0.6% | Produced | ||
| 2-hydroxybenzaldehyde | 1702 | 1680 | 1.3% | ↗ | 2.41 × 10−7 | Produced |
| Acetamide | 1767 | 1773 | 0.3% | Produced | ||
| 1,3,5,7-Tetraazatricyclo[3.3.1.13,7]decane | 1946 | - b | ↗ | 1.88 × 10−3 | Produced | |
| 4-methylquinoline | 2152 | 2108 | 2.1% | = | 6.90 × 10−1 | No variation |
| Isoquinoline-1-carbonitrile | 2391 | - b | = | 6.12 × 10−1 | No variation | |
| Quinoline-4-carbaldehyde | 2500 | - b | = | 5.37 × 10−1 | No variation | |
| 1H-indole | 2502 | 2451 | 2.1% | ↗ | 2.76 × 10−2 | Produced |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berrou, K.; Dunyach-Remy, C.; Lavigne, J.-P.; Roig, B.; Cadiere, A. Comparison of Stir Bar Sorptive Extraction and Solid Phase Microextraction of Volatile and Semi-Volatile Metabolite Profile of Staphylococcus Aureus. Molecules 2020, 25, 55. https://doi.org/10.3390/molecules25010055
Berrou K, Dunyach-Remy C, Lavigne J-P, Roig B, Cadiere A. Comparison of Stir Bar Sorptive Extraction and Solid Phase Microextraction of Volatile and Semi-Volatile Metabolite Profile of Staphylococcus Aureus. Molecules. 2020; 25(1):55. https://doi.org/10.3390/molecules25010055
Chicago/Turabian StyleBerrou, Kevin, Catherine Dunyach-Remy, Jean-Philippe Lavigne, Benoit Roig, and Axelle Cadiere. 2020. "Comparison of Stir Bar Sorptive Extraction and Solid Phase Microextraction of Volatile and Semi-Volatile Metabolite Profile of Staphylococcus Aureus" Molecules 25, no. 1: 55. https://doi.org/10.3390/molecules25010055
APA StyleBerrou, K., Dunyach-Remy, C., Lavigne, J.-P., Roig, B., & Cadiere, A. (2020). Comparison of Stir Bar Sorptive Extraction and Solid Phase Microextraction of Volatile and Semi-Volatile Metabolite Profile of Staphylococcus Aureus. Molecules, 25(1), 55. https://doi.org/10.3390/molecules25010055






