Comparative Phytochemical Characterization, Genetic Profile, and Antiproliferative Activity of Polyphenol-Rich Extracts from Pigmented Tubers of Different Solanum tuberosum Varieties
Abstract
:1. Introduction
2. Results
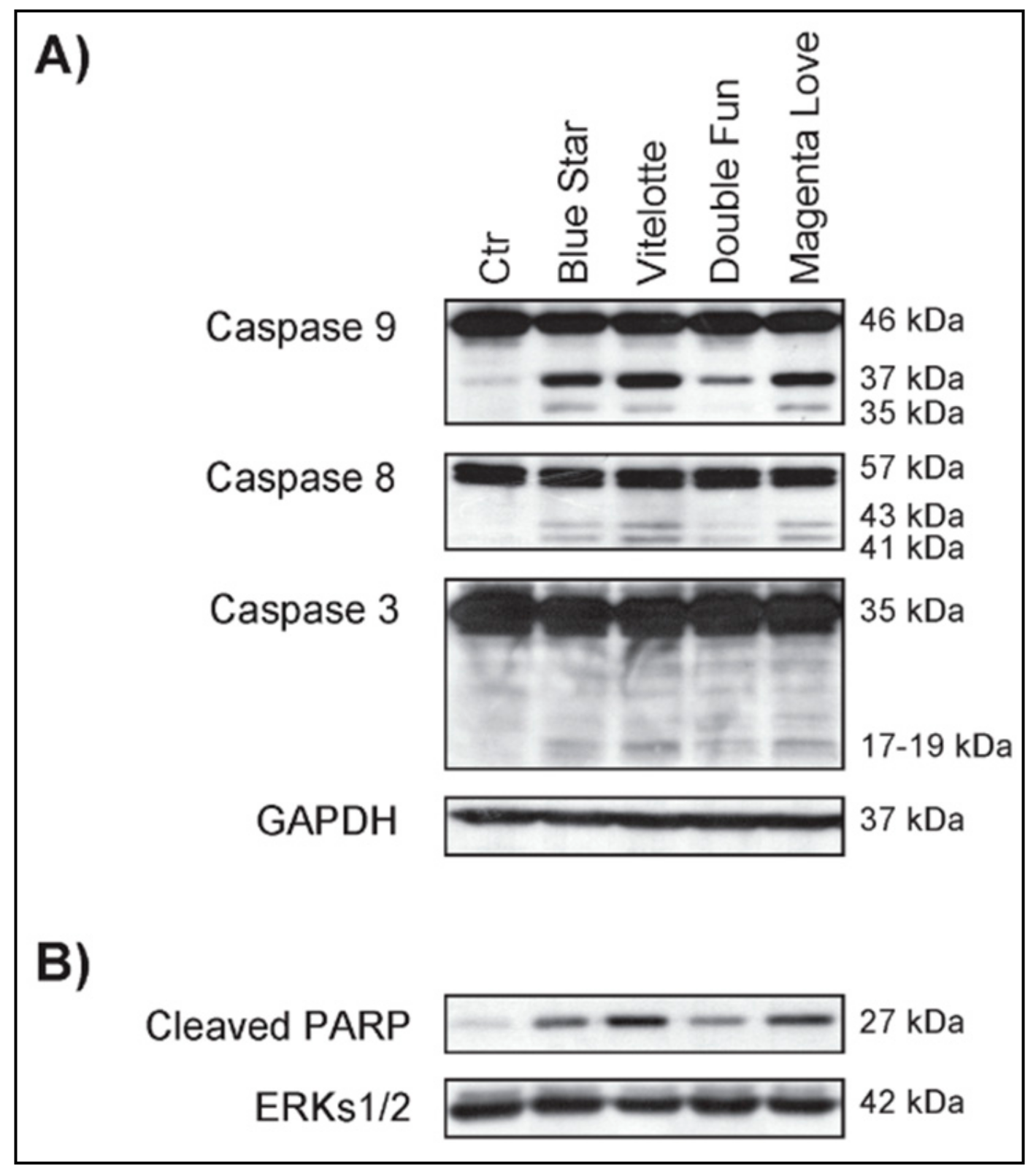
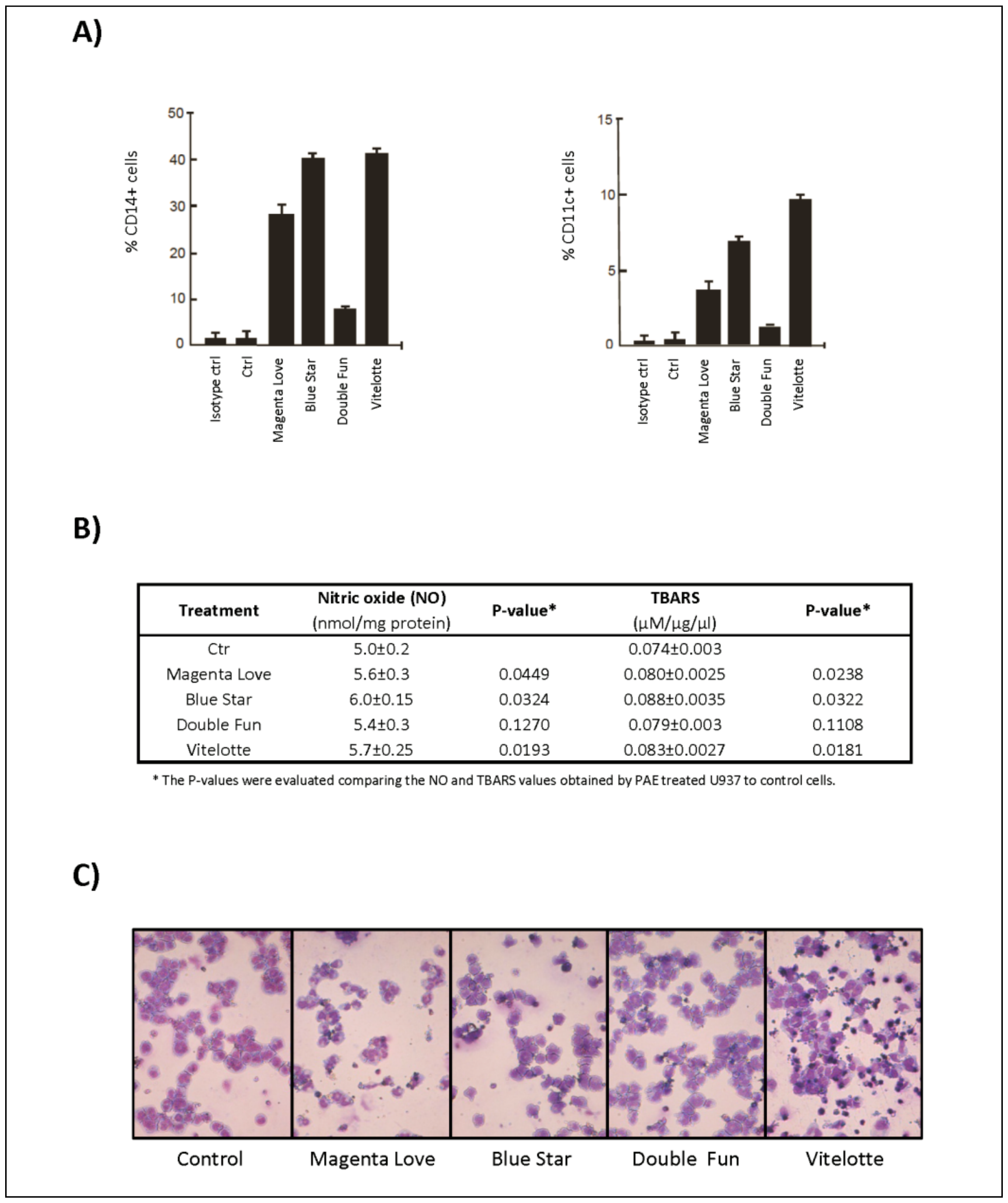
2.1. Polyphenol/Anthocyanin-Rich Extracts from Pigmented Tubers of Different Potato Varieties Induce Antiproliferative and Apoptotic Effects in Hematological Cancer Cell Lines
2.2. MS Analyses of Polyphenol/Anthocyanin-Rich Extracts from Pigmented Tubers
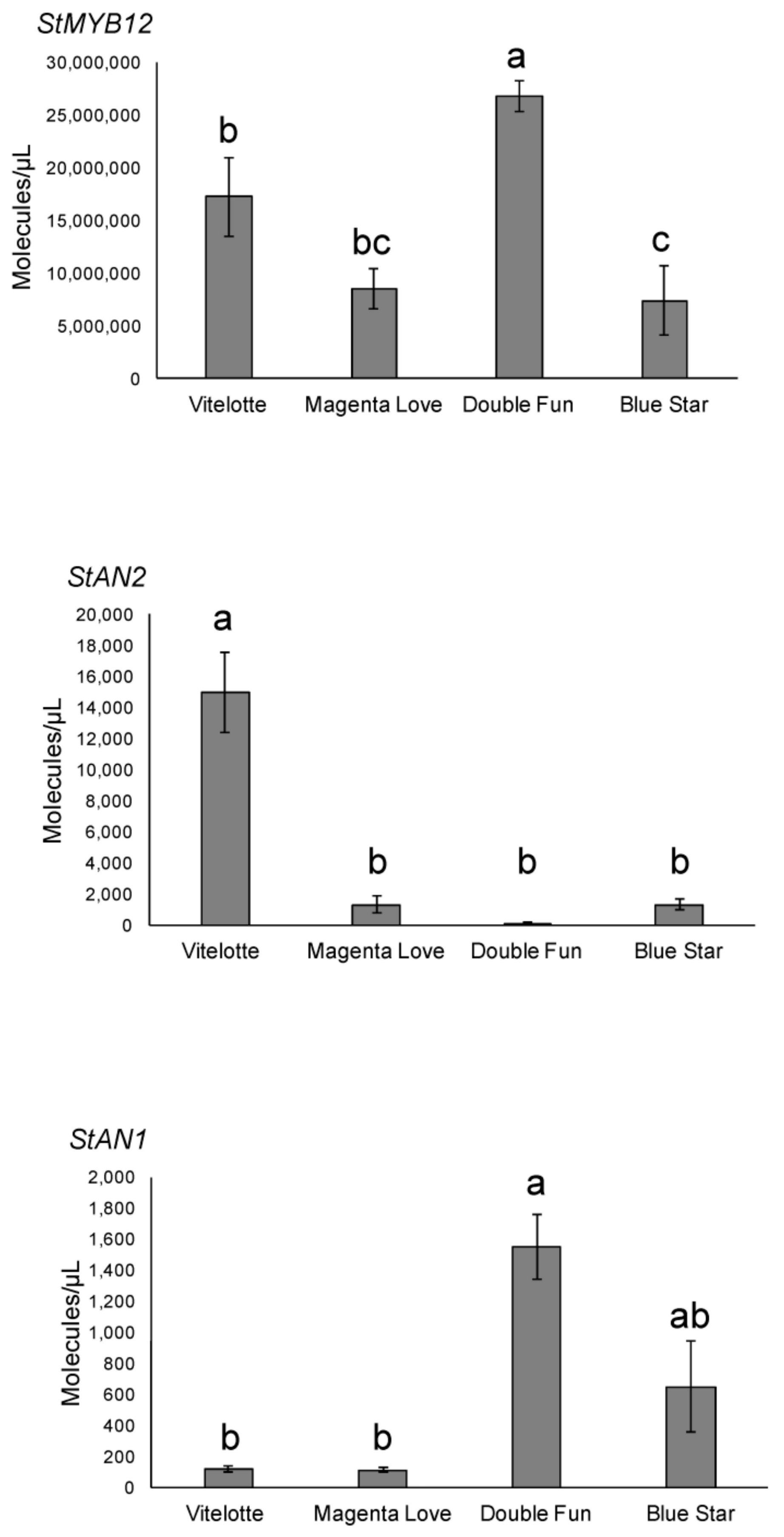
2.3. Molecular Genotyping of Pigmented Potato Varieties
3. Discussion
3.1. Comparative Bioactive Effects and Characterization of PAE from Pigmented Potato Tubers
3.2. Comparative Genetic Profiling of Pigmented Potato Varieties
4. Materials and Methods
4.1. Plant Material
4.2. Preparation of Crude Extracts from Pigmented Potato Tubers
4.3. Cell lines and Culture Conditions
4.4. Cell Morphology Analysis
4.5. Nitrite Levels
4.6. TBARS Assay
4.7. Western Blot Analysis
4.8. Differentiation Assay
4.9. Chemicals and Sample Preparation for LC-MS Analysis
4.10. LC-ESI-Orbitrap-MS Analysis, Qualitative and Quantitative Analyses
4.11. LC–ESI-QTrap-MS and LC–ESI-QTrap-MS/MS Analyses
4.12. Genotyping and Transcript Profiling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Furrer, A.; Cladis, D.P.; Kurilich, A.; Manoharan, R.; Ferruzzi, M.G. Changes in phenolic content of commercial potato varieties through industrial processing and fresh preparation. Food Chem. 2017, 218, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Aversano, R.; Contaldi, F.; Adelfi, M.G.; D’Amelia, V.; Diretto, G.; De Tommasi, N.; Vaccaro, C.; Vassallo, A.; Carputo, D. Comparative metabolite and genome analysis of tuber-bearing potato species. Phytochemistry 2017, 137, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Ezekiel, R.; Singh, N.; Sharma, S.; Kaur, A. Beneficial phytochemicals in potato—A review. Food Res. Int. 2013, 50, 487–496. [Google Scholar] [CrossRef]
- Lombardo, S.; Pandino, G.; Mauromicale, G. The influence of growing environment on the antioxidant and mineral content of “early” crop potato. J. Food Compos. Anal. 2013, 32, 28–35. [Google Scholar] [CrossRef]
- Ji, X.; Rivers, L.; Zielinski, Z.; Xu, M.; MacDougall, E.; Stephen, J.; Zhang, S.; Wang, Y.; Chapman, R.G.; Keddy, P.; et al. Quantitative analysis of phenolic components and glycoalkaloids from 20 potato clones and in vitro evaluation of antioxidant, cholesterol uptake, and neuroprotective activities. Food Chem. 2012, 133, 1177–1187. [Google Scholar] [CrossRef] [Green Version]
- Bontempo, P.; Carafa, V.; Grassi, R.; Basile, A.; Tenore, G.C.; Formisano, C.; Rigano, D.; Altucci, L. Antioxidant, antimicrobial and antiproliferative activities of Solanum tuberosum L. var. Vitelotte. Food Chem. Toxicol. 2013, 55, 304–312. [Google Scholar] [CrossRef]
- Bontempo, P.; De Masi, L.; Carafa, V.; Rigano, D.; Scisciola, L.; Iside, C.; Grassi, R.; Molinari, A.M.; Aversano, R.; Nebbioso, A.; et al. Anticancer activities of anthocyanin extract from genotyped Solanum tuberosum L. “Vitelotte”. J. Funct. Foods 2015, 19, 584–593. [Google Scholar] [CrossRef] [Green Version]
- Eichhorn, S.; Winterhalter, P. Anthocyanins from pigmented potato (Solanum tuberosum L.) varieties. Food Res. Int. 2005, 38, 943–948. [Google Scholar] [CrossRef]
- D’Amelia, V.; Aversano, R.; Chiaiese, P.; Carputo, D. The antioxidant properties of plant flavonoids: Their exploitation by molecular plant breeding. Phytochem. Rev. 2018, 17, 611–625. [Google Scholar] [CrossRef]
- Dai, J.; Mumper, R.J. Plant phenolics: Extraction, analysis and their antioxidant and anticancer properties. Molecules 2010, 15, 7313–7352. [Google Scholar] [CrossRef]
- Mueller, D.; Jung, K.; Winter, M.; Rogoll, D.; Melcher, R.; Kulozik, U.; Schwarz, K.; Richling, E. Encapsulation of anthocyanins from bilberries—Effects on bioavailability and intestinal accessibility in humans. Food Chem. 2018, 248, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Lachman, J.; Hamouz, K. Red and purple coloured potatoes as a significant antioxidant source in human nutrition—A review. Plant Soil Environ. 2005, 51, 477–482. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Liu, R.; Tsao, R. Anthocyanin-rich phenolic extracts of purple root vegetables inhibit pro-inflammatory cytokines induced by H2O2 and enhance antioxidant enzyme activities in Caco-2 cells. J. Funct. Foods 2016, 22, 363–375. [Google Scholar] [CrossRef]
- Ramdath, D.D.; Padhi, E.; Hawke, A.; Sivaramalingam, T.; Tsao, R. The glycemic index of pigmented potatoes is related to their polyphenol content. Food Funct. 2014, 5, 909–915. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef] [Green Version]
- Martin, C.; Li, J. Medicine is not health care, food is health care: Plant metabolic engineering, diet and human health. New Phytol. 2017, 216, 699–719. [Google Scholar] [CrossRef] [Green Version]
- Payyavula, R.S.; Singh, R.K.; Navarre, D.A. Transcription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J. Exp. Bot. 2013, 64, 5115–5131. [Google Scholar] [CrossRef] [Green Version]
- D’Amelia, V.; Aversano, R.; Ruggiero, A.; Batelli, G.; Appelhagen, I.; Dinacci, C.; Hill, L.; Martin, C.; Carputo, D. Subfunctionalization of duplicate MYB genes in Solanum commersonii generated the cold-induced ScAN2 and the anthocyanin regulator ScAN1. Plant Cell Environ. 2018, 41, 1038–1051. [Google Scholar] [CrossRef]
- Li, Y.; Tang, W.; Chen, J.; Jia, R.; Ma, L.; Wang, S.; Wang, J.; Shen, X.; Chu, Z.; Zhu, C.; et al. Development of marker-free transgenic potato tubers enriched in caffeoylquinic acids and flavonols. J. Agr. Food Chem. 2016, 64, 2932–2940. [Google Scholar] [CrossRef]
- Narvaez-Cuenca, C.-E.; Vincken, J.-P.; Zheng, C.; Gruppen, H. Diversity of (dihydro) hydroxycinnamic acid conjugates in Colombian potato tubers. Food Chem. 2013, 139, 1087–1097. [Google Scholar] [CrossRef]
- Matsuura, H.; Ohmori, F.; Kobayashi, M.; Sakurai, A.; Yoshihara, T. Qualitative and quantitative analysis of endogenous jasmonoids in potato plant (Solanum tuberosum L.). Biosci. Biotechnol. Biochem. 2000, 64, 2380–2387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Aoshima, K.; Oda, Y.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Villano, C.; Aversano, R.; Frusciante, L.; Garramone, R.; Iorizzo, M.; Carputo, D. Utilizzazione di marcatori molecolari SSR e AFLP per l’identificazione varietale in patata. Minerva Biotecnol. 2012, 24, 3–10. (In Italian) [Google Scholar]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Miceli, M.; Bontempo, P.; Nebbioso, A.; Altucci, L. Natural compounds in epigenetics: A current view. Food Chem. Toxicol. 2014, 73, 71–83. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, A.; Zappavigna, S.; Villa, M.R.; Improta, S.; Cesario, E.; Mastrullo, L.; Caraglia, M.; Stiuso, P. Prognostic role of translocator protein and oxidative stress markers in chronic lymphocytic leukemia patients treated with bendamustine plus rituximab. Oncol. Lett. 2015, 9, 1327–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magrinat, G.; Mason, S.N.; Shami, P.J.; Weinberg, J.B. Nitric oxide modulation of human leukemia cell differentiation and gene expression. Blood 1992, 80, 1880–1884. [Google Scholar] [CrossRef] [Green Version]
- Paixão, J.; Dinis, T.C.; Almeida, L.M. Malvidin-3-glucoside protects endothelial cells up-regulating endothelial NO synthase and inhibiting peroxynitrite-induced NF-κB activation. Chem. Biol. Interact. 2012, 199, 192–200. [Google Scholar] [CrossRef]
- Garcia-Alonso, M.; Rimbach, G.; Sasai, M.; Nakahara, M.; Matsugo, S.; Uchida, Y.; Rivas-Gonzalo, J.C.; De Pascual-Teresa, S. Electron spin resonance spectroscopy studies on the free radical scavenging activity of wine anthocyanins and pyranoanthocyanins. Mol. Nutr. Food Res. 2005, 49, 1112–1119. [Google Scholar] [CrossRef]
- Guo, H.; Liu, G.; Zhong, R.; Wang, Y.; Wang, D.; Xia, M. Cyanidin-3-O-β-glucoside regulates fatty acid metabolism via an AMP-activated protein kinase dependent signaling pathway in human HepG2 cells. Lipids Health Dis. 2012, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Rinaldi, M.; Barrera, G.; Aquino, A.; Spinsanti, P.; Pizzimenti, S.; Farace, M.G.; Dianzani, M.U.; Fazio, V.M. 4-Hydroxynonenal-induced MEL cell differentiation involves PKC activity translocation. Biochem. Biophys. Res. Commun. 2000, 272, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Niedzwiecki, A.; Roomi, M.W.; Kalinovsky, T.; Rath, M. Anticancer efficacy of polyphenols and their combinations. Nutrients 2016, 8, 552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalia, R.K.; Rai, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 2018, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Butelli, E.; Alseekh, S.; Tohge, T.; Rallapalli, G.; Luo, J.; Kawar, P.G.; Hill, L.; Santino, A.; Fernie, A.R.; et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015, 6, 8635. [Google Scholar] [CrossRef] [Green Version]
- Han, K.-H.; Matsumoto, A.; Shimada, K.-I.; Sekikawa, M.; Fukushima, M. Effects of anthocyanin-rich purple potato flakes on antioxidant status in F344 rats fed a cholesterol-rich diet. Br. J. Nutr. 2007, 98, 914–921. [Google Scholar] [CrossRef] [Green Version]
- Stiuso, P.; Bagarolo, M.L.; Ilisso, C.P.; Vanacore, D.; Martino, E.; Caraglia, M.; Porcelli, M.; Cacciapuoti, G. Protective effect of tyrosol and S-adenosylmethionine against ethanol-induced oxidative stress of HepG2 cells involves Sirtuin 1, P53 and ERK1/2 Signaling. Int. J. Mol. Sci. 2016, 17, 622. [Google Scholar] [CrossRef] [Green Version]
- Bontempo, P.; Doto, A.; Miceli, M.; Mita, L.; Benedetti, R.; Nebbioso, A.; Veglione, M.; Rigano, D.; Cioffi, M.; Sica, V.; et al. Psidium guajava L. anti-neoplastic effects: Induction of apoptosis and cell differentiation. Cell Proliferat. 2012, 45, 22–31. [Google Scholar] [CrossRef]
- Bontempo, P.; Rigano, D.; Doto, A.; Formisano, C.; Conte, M.; Nebbioso, A.; Carafa, V.; Caserta, G.; Sica, V.; Molinari, A.M.; et al. Genista sessilifolia DC. extracts induce apoptosis across a range of cancer cell lines. Cell Proliferat. 2013, 46, 183–192. [Google Scholar] [CrossRef]
- De Masi, L.; Siviero, P.; Castaldo, D.; Cautela, D.; Esposito, C.; Laratta, B. Agronomic, chemical and genetic profiles of hot peppers (Capsicum annuum ssp.). Mol. Nutr. Food Res. 2007, 51, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Masi, P.; Aversano, R.; Carucci, F.; Palomba, S.; Carputo, D. Microstructure and tuber properties of potato varieties with different genetic profiles. Food Chem. 2018, 239, 789–796. [Google Scholar] [CrossRef] [PubMed]
- D’Amelia, V.; Aversano, R.; Batelli, G.; Caruso, I.; Castellano Moreno, M.; Castro-Sanz, A.B.; Chiaiese, P.; Fasano, C.; Palomba, F.; Carputo, D. High AN1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014, 80, 527–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the four potato varieties are available from the authors. |
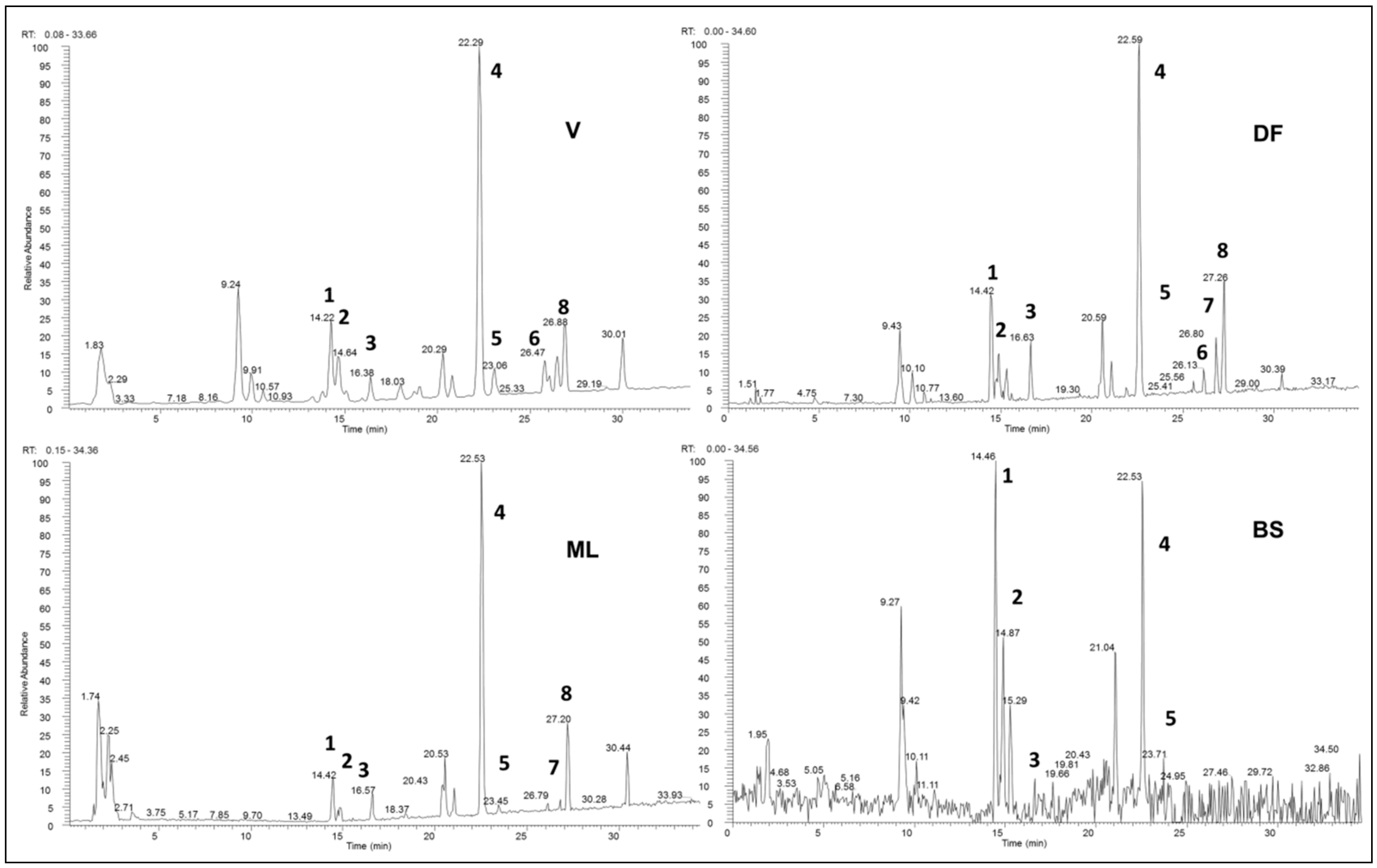
| Phenolics | Mass Parent ion (m/z) | MS/MS | Compound | Molecular Formula | V mg/100g DW | DF mg/100g DW | ML mg/100g DW | BS mg/100g DW |
|---|---|---|---|---|---|---|---|---|
| 1 | 353.0149 | 191.0235 | 3-O-caffeoylquinic acid | C16H18O9 | 32.2 ± 2.0 | 32.0 ± 2.0 | 12.0 ± 1.0 | 108.0 ± 30.0 |
| 2 | 353.0867 | 191.0151 | 5-O-caffeoylquinic acid | C16H18O9 | 31.0 ± 1.0 | 21.0 ± 6.0 | 11.0 ± 1.0 | 59.0 ± 3.0 |
| 3 | 387.1025 | 335.2375 299.0197 | tuberonic acid glc | C18H28O9 | 10.1 ± 0.06 | 10.7 ± 0.08 | 8.6 ± 0.08 | 6.7 ± 0.1 |
| 4 | 367.1026 | 191.0210 160.9260 134.8752 | feruloyl quinic acid | C17H20O9 | 105.0 ± 5.9 | 98.0 ± 9.7 | 97.0 ± 8.9 | 107.0 ± 25.0 |
| 5 | 771.2523 | 720.3513 711.2075 696.3084 662.3907 | quercetin-3-O-(2′-glucosyl)-rutinoside | C33H40O21 | 6.5 ± 0.2 | ND | 5.5 ± 0.1 | ND |
| 6 | 609.1450 | 301.1120 | rutin | C27H30O16 | 13.3 ± 0.6 | 8.5 ± 0.6 | 3.8 ± 0.2 | ND |
| 7 | 463.0901 | 301.9224 | quercetin-3-O-glucoside | C21H20O12 | 3.5 ± 0.2 | 7.8 ± 0.3 | 2.7 ± 0.2 | ND |
| 8 | 381.1183 | 301.1876 178.9705 134.8748 | quercetin-3-O-sulphate | C15H10O10S | 18.7 ± 0.2 | 29.0 ± 0.3 | 16.8 ± 0.2 | ND |
| An | Mass Parent Ion (m/z) | MS/MS | Tentative ID | Molecular Formula | V mg/100g DW | DF mg/100g DW | ML mg/100g DW | BS mg/100g DW |
|---|---|---|---|---|---|---|---|---|
| 1′ | 887.2609 | 271.0234 | pelargonidin 3-(4‴-p-coumaroylrutinoside)-5-glc | C42H47O21 | 16.32 ± 0.02 | 14.32 ± 0.09 | 18.12 ± 0.04 | 12.08 ± 0.05 |
| 2′ | 903.2558 | 287.0124 | cyanidin-3-(p-coumaroyl)-rutinoside-5-glc | C42H47O22 | 0.87 ± 0.03 | ND | ND | ND |
| 3′ | 917.2715 | 301.0051 | peonidin 3-O-[6-O-(4-O-(E)-p-coumaroyl-O-α-rha)-β-glc]-5-O-β-glc | C43H49O22 | 1.23 ± 0.08 | ND | ND | ND |
| 4′ | 917.2715 | 301.1581 | peonidin 3-caffeoylrutinoside-5-glc | C43H49O23 | 1.08 ± 0.06 | ND | 1.10 ± 0.07 | ND |
| 5′ | 933.2664 | 303.1961 | delphinidin 3-O-[6-O-(4-O-(E)-p-coumaroyl-O-α-rha)-β-glc]-5-O-β-glc | C42H47O23 | 0.85 ± 0.03 | ND | ND | ND |
| 6′ | 919.2508 | 317.1110 | petunidin 3-(4‴-p-coumarylrutinoside) | C37H39O18 | ND | 2.52 ± 0.03 | ND | 2.05 ± 0.04 |
| 7′ | 933.2664 | 317.0415 | petunidin 3-p-coumaroylrutinoside-5-glc | C43H49O23 | 3.78 ± 0.03 | 3.28 ± 0.03 | 3.05 ± 0.02 | 3.45 ± 0.03 |
| 8′ | 977.2926 | 331.1865 | malvidin 3-O-(6-O-(4-O-feruloyl-α-rha)-β-glc)-5-β-glc | C45H53O24 | 8.91 ± 0.05 | 6.57 ± 0.03 | ND | 1.02 ± 0.01 |
| 9′ | 917.2715 | 271.3245 | pelargonidin 3-(4‴-feruloylrutinoside)-5-glc | C43H49O22 | ND | 5.21 ± 0.04 | 4.07 ± 0.05 | 0.22 ± 0.01 |
| 10′ | 949.2614 | 317.1456 | petunidin 3-O-[6-O-(4-O-(E)-caffeoyl-O-α-rha)-β-glc]-5-O-β-glc | C43H49O24 | 0.32 ± 0.03 | 1.62 ± 0.04 | ND | 1.60 ± 0.03 |
| 11′ | 963.2773 | 317.1335 | petunidin 3-O-[6-O-(4-O-(4-O-(β-D-glc)-feruloyl)-α-L-rha)-β-D-glc]- 5-O- [β-D-glc] | C44H51O24 | ND | 0.42 ± 0.01 | ND | 0.65 ± 0.04 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Masi, L.; Bontempo, P.; Rigano, D.; Stiuso, P.; Carafa, V.; Nebbioso, A.; Piacente, S.; Montoro, P.; Aversano, R.; D’Amelia, V.; et al. Comparative Phytochemical Characterization, Genetic Profile, and Antiproliferative Activity of Polyphenol-Rich Extracts from Pigmented Tubers of Different Solanum tuberosum Varieties. Molecules 2020, 25, 233. https://doi.org/10.3390/molecules25010233
De Masi L, Bontempo P, Rigano D, Stiuso P, Carafa V, Nebbioso A, Piacente S, Montoro P, Aversano R, D’Amelia V, et al. Comparative Phytochemical Characterization, Genetic Profile, and Antiproliferative Activity of Polyphenol-Rich Extracts from Pigmented Tubers of Different Solanum tuberosum Varieties. Molecules. 2020; 25(1):233. https://doi.org/10.3390/molecules25010233
Chicago/Turabian StyleDe Masi, Luigi, Paola Bontempo, Daniela Rigano, Paola Stiuso, Vincenzo Carafa, Angela Nebbioso, Sonia Piacente, Paola Montoro, Riccardo Aversano, Vincenzo D’Amelia, and et al. 2020. "Comparative Phytochemical Characterization, Genetic Profile, and Antiproliferative Activity of Polyphenol-Rich Extracts from Pigmented Tubers of Different Solanum tuberosum Varieties" Molecules 25, no. 1: 233. https://doi.org/10.3390/molecules25010233
APA StyleDe Masi, L., Bontempo, P., Rigano, D., Stiuso, P., Carafa, V., Nebbioso, A., Piacente, S., Montoro, P., Aversano, R., D’Amelia, V., Carputo, D., & Altucci, L. (2020). Comparative Phytochemical Characterization, Genetic Profile, and Antiproliferative Activity of Polyphenol-Rich Extracts from Pigmented Tubers of Different Solanum tuberosum Varieties. Molecules, 25(1), 233. https://doi.org/10.3390/molecules25010233