Interactions of Paraoxonase-1 with Pharmacologically Relevant Carbamates
Abstract
1. Introduction
2. Results
2.1. rePON1 Arylesterase Activity
2.2. The Effect of Carbamates on rePON1 Activity
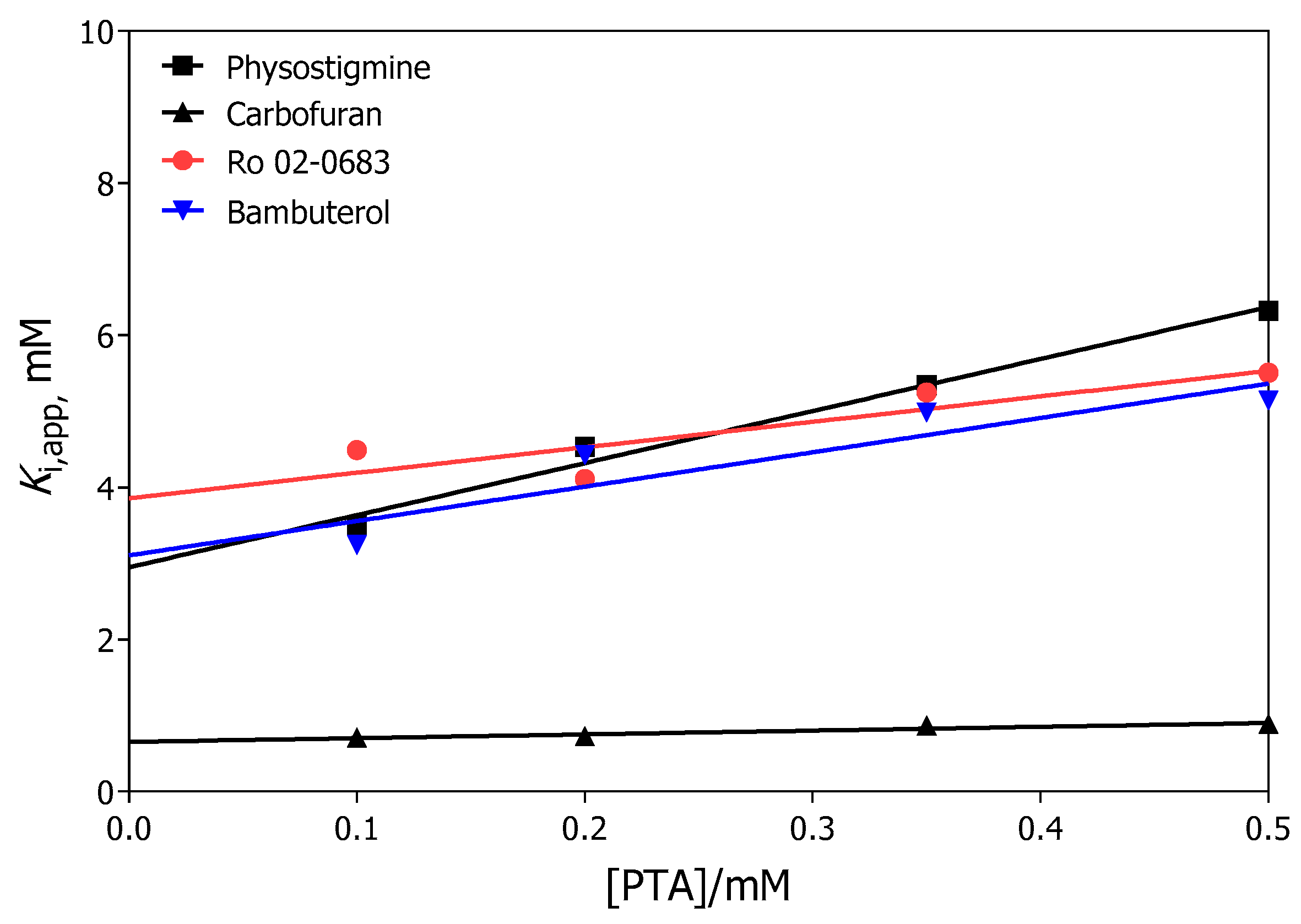
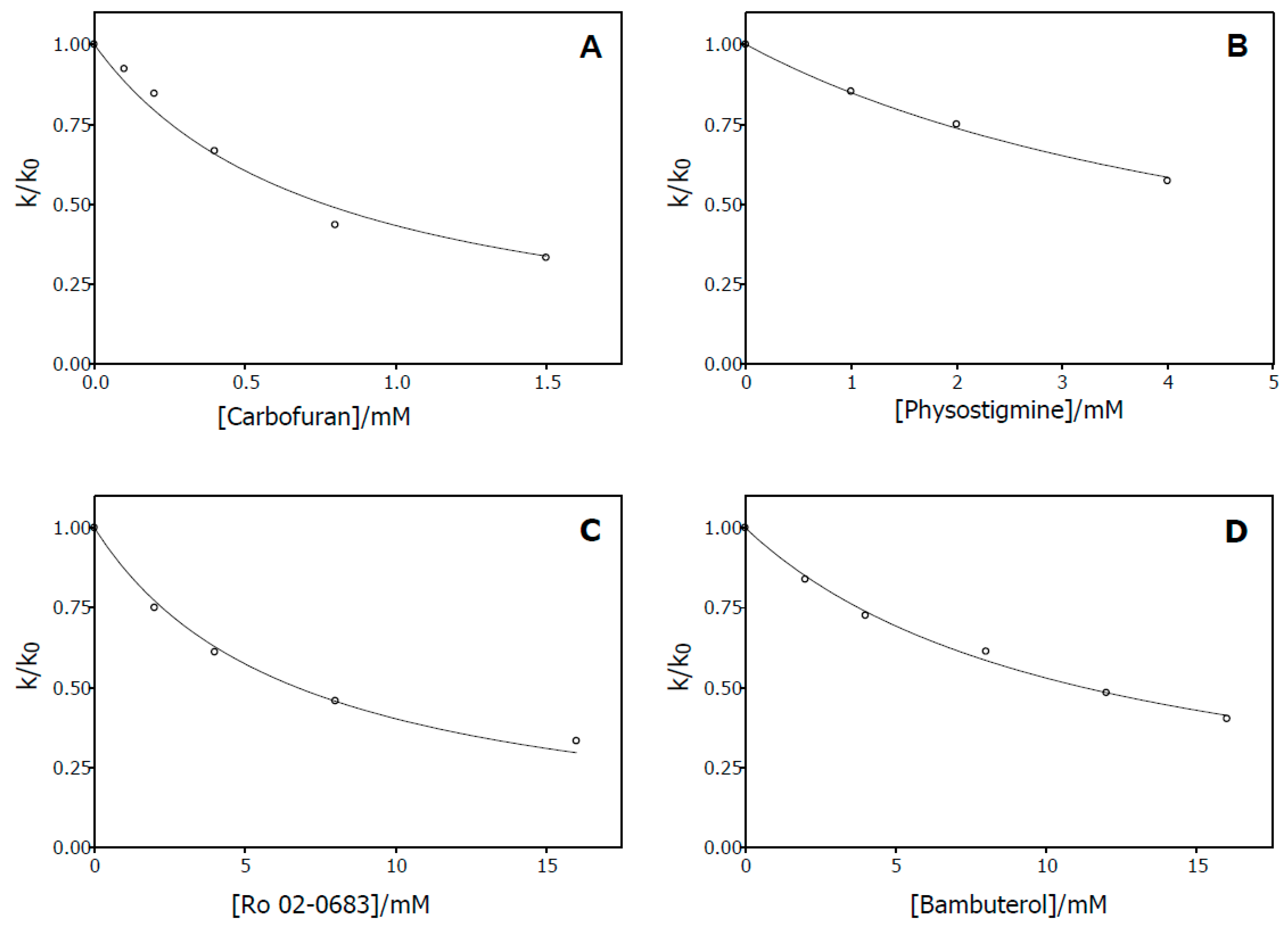
2.2.1. Dissociation Constants Determined from the Initial Rates
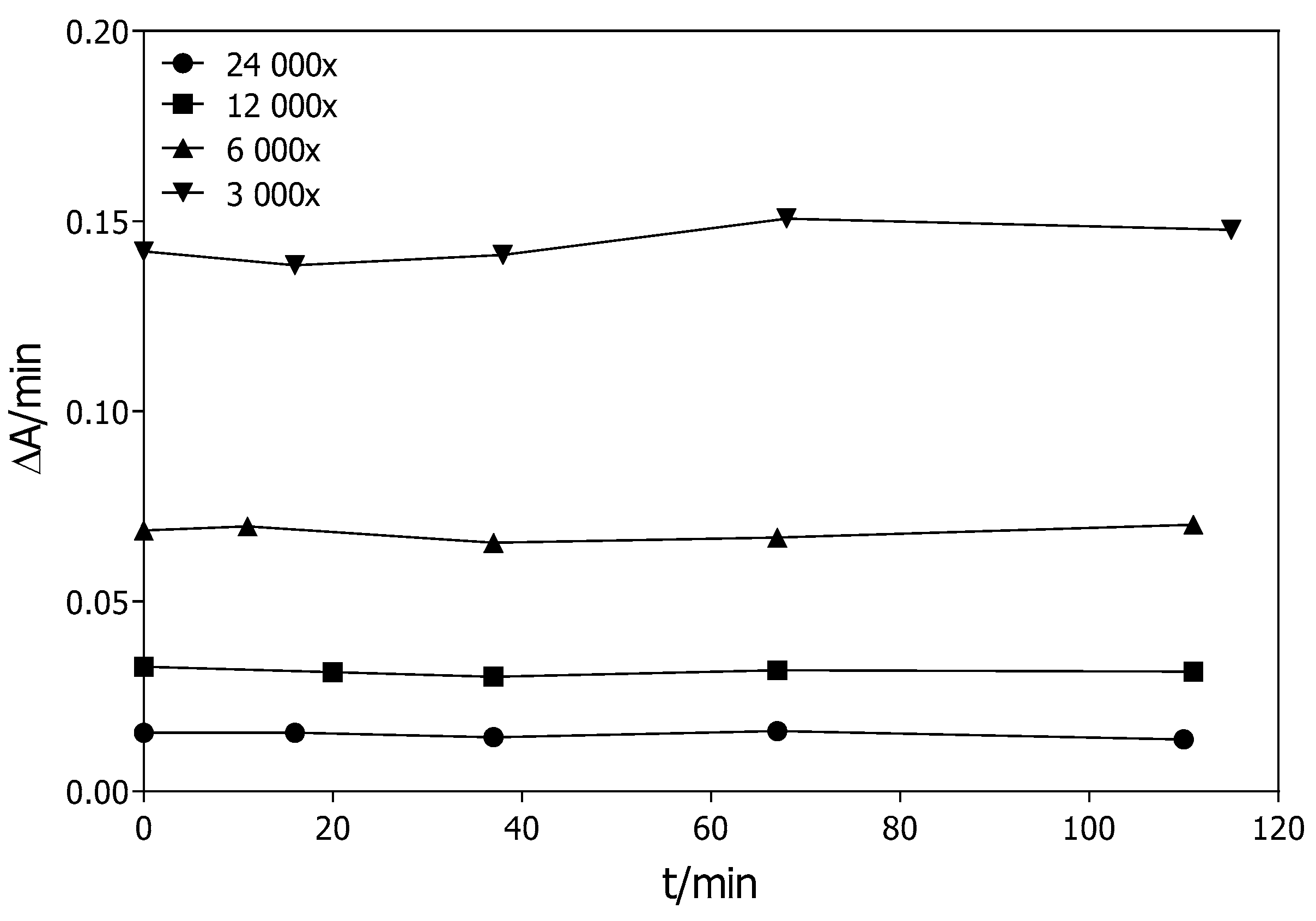
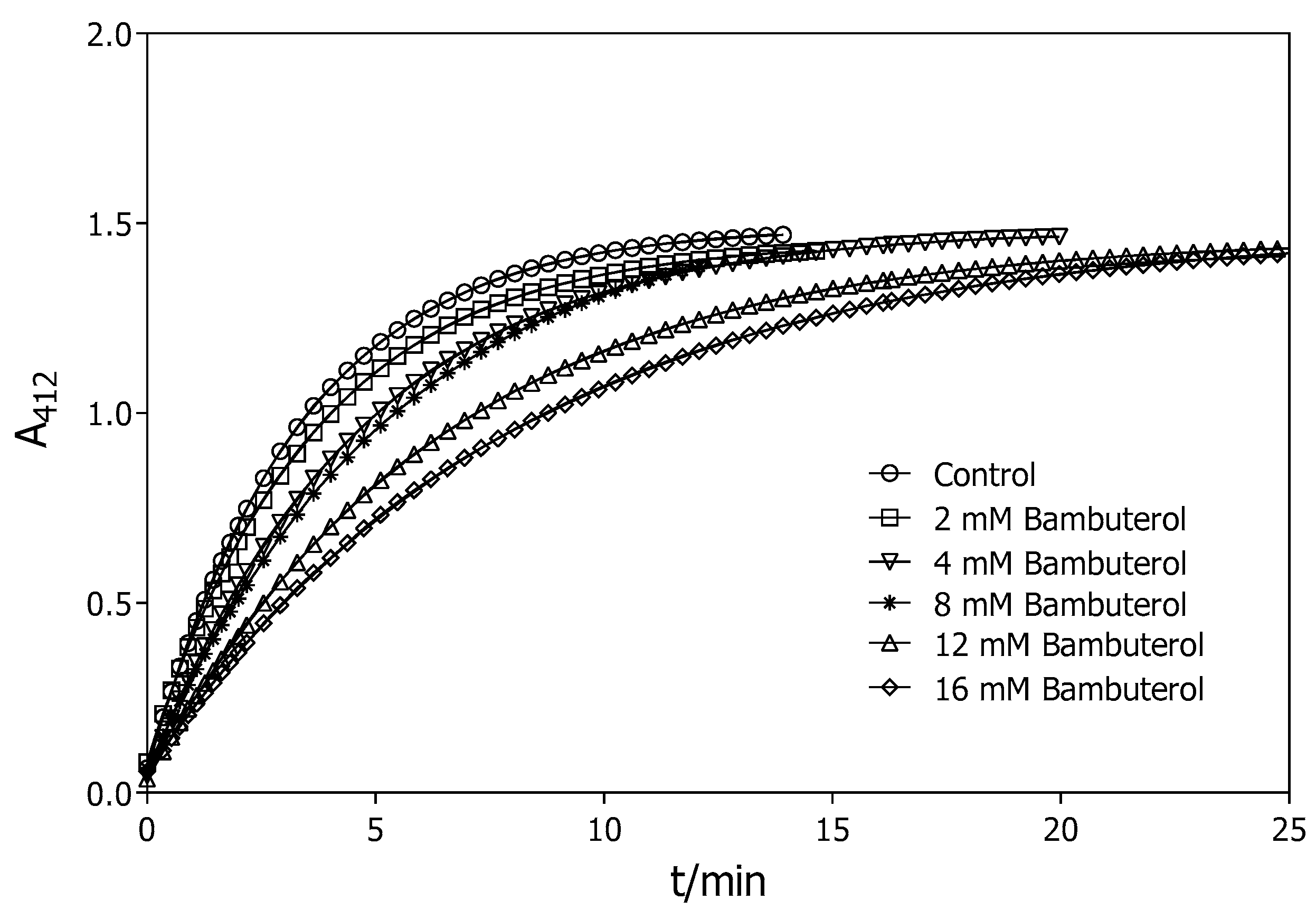
2.2.2. Stability of the G2E6 Variant of rePON1
2.2.3. Dissociation Constants Determined from the Progress Curves
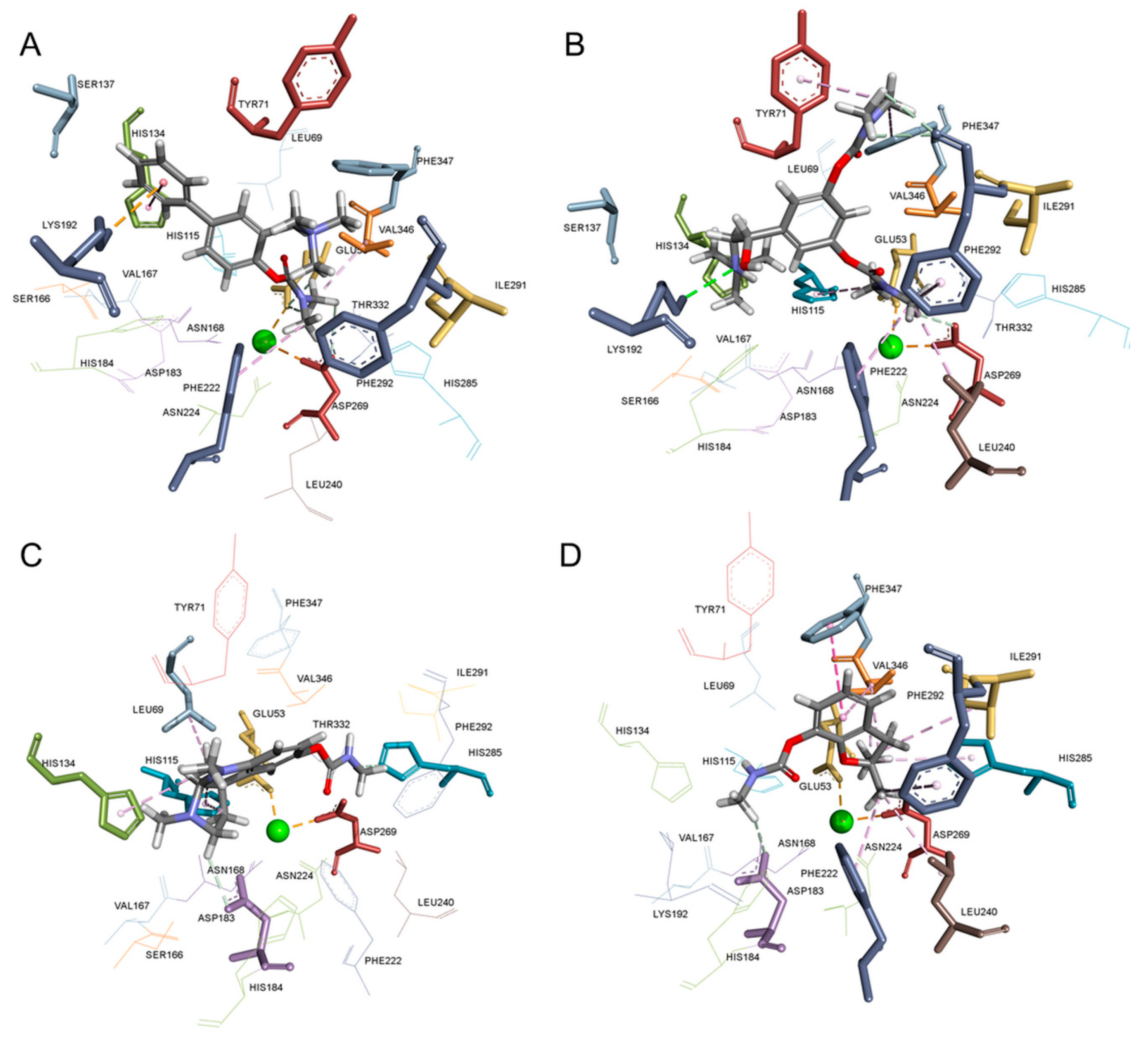
2.3. Molecular Modelling of the PON1-carbamate Complex
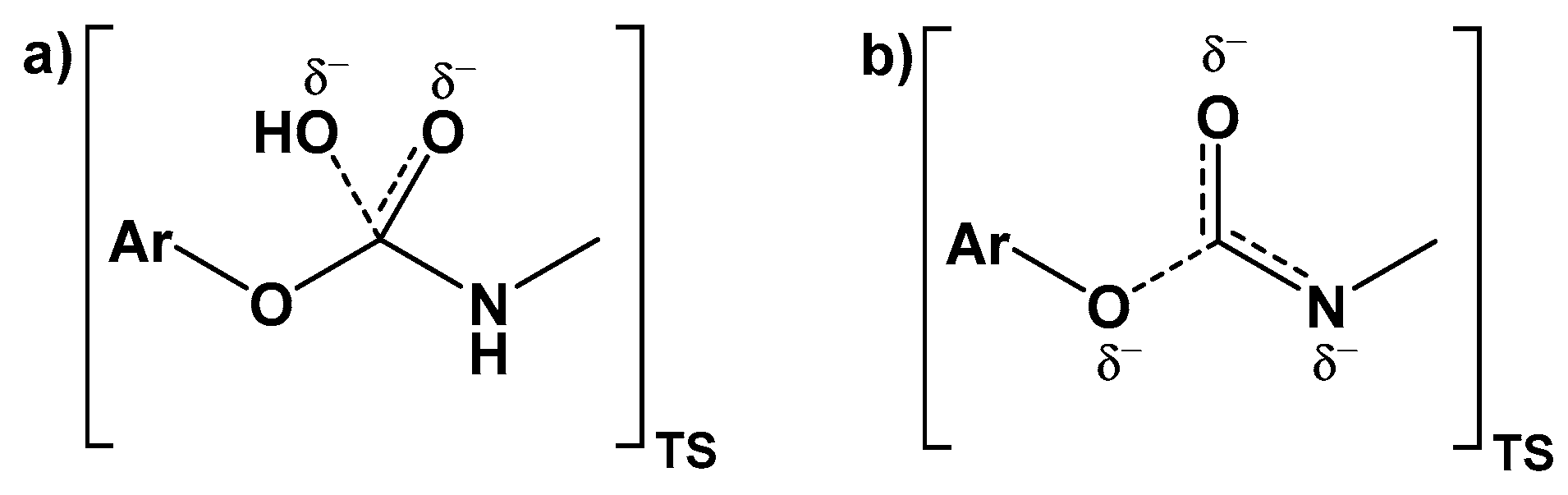
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Recombinant PON1 Expression and Purification
4.3. Determination of the Catalytic Constants of rePON1 from Initial Rate Measurements
4.4. Determination of the rePON1-carbamate Complex Dissociation Constants
4.5. Stability of the G2E6 Variant of rePON1
4.6. Determination of the Inhibition Constants of Carbamates from Progress Curve Measurements
4.7. Molecular Modelling of the rePON1-carbamate Complex
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mackness, M.; Mackness, B. Human paraoxonase-1 (PON1): Gene structure and expression, promiscuous activities and multiple physiological roles. Gene 2015, 567, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Luna, D.; Araceli Ortiz-Rodriguez, M.; Medina-Briseño, L.; Carreón-Torres, E.; Izquierdo-Vega, J.A.; Sharma, A.; Cancino-Díaz, J.C.; Pérez-Méndez, O.; Belefant-Miller, H.; Betanzos-Cabrera, G. Current therapies focused on high-density lipoproteins associated with cardiovascular disease. Molecules 2018, 23, 2730. [Google Scholar] [CrossRef] [PubMed]
- Furlong, C.E.; Marsillach, J.; Jarvik, G.P.; Costa, L.G. Paraoxonases-1, -2 and -3: What are their functions? Chem. Biol. Interact. 2016, 259, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Koren-Gluzer, M.; Aviram, M.; Meilin, E.; Hayek, T. The antioxidant HDL-associated paraoxonase-1 (PON1) attenuates diabetes development and stimulates β-cell insulin release. Atherosclerosis 2011, 219, 510–518. [Google Scholar] [CrossRef]
- Ivanišević, J.; Kotur-Stevuljević, J.; Stefanović, A.; Miljković, M.; Jelić-Ivanović, Z.; Pejović, B.; Peco-Antić, A. Association of paraoxonase 1 and oxidative stress with acute kidney injury in premature asphyxiated neonates. Chem. Biol. Interact. 2017, 272, 47–52. [Google Scholar] [CrossRef]
- van den Berg, E.H.; Gruppen, E.G.; James, R.W.; Bakker, S.J.L.; Dullaart, R.P.F. Serum paraoxonase 1 activity is paradoxically maintained in nonalcoholic fatty liver disease despite low HDL cholesterol. J. Lipid Res. 2019, 60, 168–175. [Google Scholar] [CrossRef]
- Menini, T.; Gugliucci, A. Paraoxonase 1 in neurological disorders. Redox Rep. 2014, 19, 49–58. [Google Scholar] [CrossRef]
- Azizi, F.; Raiszadeh, F.; Solati, M.; Etemadi, A.; Rahmani, M.; Arabi, M. Serum paraoxonase 1 activity is decreased in thyroid dysfunction. J. Endocrinol. Invest. 2003, 26, 703–709. [Google Scholar] [CrossRef]
- Arenas, M.; Rodríguez, E.; Sahebkar, A.; Sabater, S.; Rizo, D.; Pallisé, O.; Hernández, M.; Riu, F.; Camps, J.; Joven, J. Paraoxonase-1 activity in patients with cancer: A systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 2018, 127, 6–14. [Google Scholar] [CrossRef]
- Billecke, S.; Draganov, D.; Counsell, R.; Stetson, P.; Watson, C.; Hsu, C.; La Du, B.N. Human serum paraoxonase (PON1) isozymes Q and R hydrolyze lactones and cyclic carbonate esters. Drug Metab. Dispos. 2000, 28, 1335–1342. [Google Scholar]
- Khersonsky, O.; Tawfik, D.S. Structure-reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry 2005, 44, 6371–6382. [Google Scholar] [CrossRef] [PubMed]
- Draganov, D.I. Lactonases with organophosphatase activity: Structural and evolutionary perspectives. Chem. Biol. Interact. 2010, 187, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, W.N. Serum esterases. II. An enzyme hydrolysing diethyl p-nitrophenyl phosphate (E600) and its identity with the A-esterase of mammalian sera. Biochem J. 1953, 53, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Harel, M.; Aharoni, A.; Gaidukov, L.; Brumshtein, B.; Khersonsky, O.; Meged, R.; Dvir, H.; Ravelli, R.B.; McCarthy, A.; Toker, L.; et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 2004, 11, 412–419. [Google Scholar] [CrossRef]
- Khersonsky, O.; Tawfik, D.S. The histidine 115-histidine 134 dyad mediates the lactonase activity of mammalian serum paraoxonases. J. Biol. Chem. 2006, 281, 7649–7656. [Google Scholar] [CrossRef]
- Ben-David, M.; Elias, M.; Filippi, J.J.; Duñach, E.; Silman, I.; Sussman, J.L.; Tawfik, D.S. Catalytic versatility and backups in enzyme active sites: The case of serum paraoxonase-1. J. Mol. Biol. 2012, 418, 181–196. [Google Scholar] [CrossRef]
- Ben-David, M.; Wieczorek, G.; Elias, M.; Silman, I.; Sussman, J.L.; Tawfik, D.S. Catalytic metal ion rearrangements underline promiscuity and evolvability of a metalloenzyme. J. Mol. Biol. 2013, 425, 1028–1038. [Google Scholar] [CrossRef]
- Bavec, A.; Knez, D.; Makovec, T.; Stojan, J.; Gobec, S.; Goličnik, M. Exploring the aryl esterase catalysis of paraoxonase-1 through solvent kinetic isotope effects and phosphonate-based isosteric analogues of the tetrahedral reaction intermediate. Biochimie 2014, 106, 184–186. [Google Scholar] [CrossRef]
- Sogorb, M.A.; Vilanova, E. Enzymes involved in the detoxification of organophosphorus, carbamate and pyrethroid insecticides through hydrolysis. Toxicol. Lett. 2002, 128, 215–228. [Google Scholar] [CrossRef]
- Moser, V.C.; Padilla, S. Esterase detoxication of acetylcholinesterase inhibitors using human liver samples in vitro. Toxicology. 2016, 353, 11–20. [Google Scholar] [CrossRef]
- Costa, L.G.; Giordano, G.; Cole, T.B.; Marsillach, J.; Furlong, C.E. Paraoxonase 1 (PON1) as a genetic determinant of susceptibility to organophosphate toxicity. Toxicology 2013, 307, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, C.E.; Shields, J.B.; Augustinsson, K.-B. Arylesterase from various mammalian sera in relation to cholinesterases, carboxylesterases and their activity towards some pesticides. Comp. Biochem. Physicol. 1976, 55C, 23–26. [Google Scholar] [CrossRef]
- Reiner, E.; Pavković, E.; Radić, Z.; Simeon, V. Diferentiation of esterases reacting with organophosphorous compounds. Chem. Biol. Interact. 1993, 87, 77–83. [Google Scholar] [CrossRef]
- Bahar, F.G.; Ohura, K.; Ogihara, T.; Imai, T. Species difference of esterase expression and hydrolase activity in plasma. J. Pharm. Sci. 2012, 101, 3979–3988. [Google Scholar] [CrossRef] [PubMed]
- Moser, V.C.; Padilla, S. Esterase metabolism of cholinesterase inhibitors using rat liver in vitro. Toxicology 2011, 281, 56–62. [Google Scholar] [CrossRef]
- Aldridge, W.N.; Reiner, E. Enzyme Inhibitors as Substrates. Interaction of Esterases with Esters of Organophosphorus and Carbamic Acids, 1st ed.; North Holland Publishing Co.: Amsterdam, The Netherlands, 1972. [Google Scholar]
- Reiner, E.; Radić, Z. Mechanism of action of cholinesterase inhibitor. In Cholinesterases and Cholinesterase Inhibitors; Giacobini, E., Ed.; Martin Dunitz Ltd.: London, UK, 2000; pp. 103–121. [Google Scholar]
- Avendaño, C.; Menéndez, J.C. Anticancer Drugs Trageting Tubulin and Microtubules. In Medicinal Chemistry of Anticancer Drugs; Avendaño, C., Menéndez, J.C., Eds.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 229–247. [Google Scholar]
- Shih, J.C.; Tatum, O.W.; Rudzinski, L.A. New drug classes for the treatment of partial onset epilepsy: Focus on perampanel. Ther. Clin. Risk Manag. 2013, 9, 285–293. [Google Scholar] [CrossRef]
- Rautio, J.; Kumpulainen, H.; Heimbach, T.; Oliyal, R. Produgs: Design and clinical applications. Nat. Rev. Drug Discov. 2008, 7, 255–270. [Google Scholar] [CrossRef]
- Aviram, M.; Hardak, E.; Vaya, J.; Mahmood, S.; Milo, S.; Hoffman, A.; Billicke, S.; Draganov, D.; Rosenblat, M. Human serum paraoxonases (PON1) Q and R selectively decrease lipid peroxides in human coronary and carotid atherosclerotic lesions-PON1 esterase and peroxidase-like activities. Circulation 2000, 101, 2510–2517. [Google Scholar] [CrossRef]
- Eyer, P.; Worek, F.; Kiderlen, D.; Šinko, G.; Stuglin, A.; Simeon-Rudolf, V.; Reiner, E. Molar absorption coefficients for the reduced Ellman reagent: Reassessment. Anal. Biochem. 2003, 312, 224–227. [Google Scholar] [CrossRef]
- Kademi, A.; Aıt-Abdelkader, N.; Fakhreddine, L.; Baratti, J.C. Characterization of a new thermostable esterase from the moderate thermophilic bacterium Bacillus circulans. J. Mol. Catal. B-enzym. 2000, 10, 395–401. [Google Scholar] [CrossRef]
- Khersonsky, O.; Tawfik, D.S. Chromogenic and fluorogenic assays for the lactonase activity of serum paraoxonases. Chembiochem. 2006, 7, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Dahli, L.; Atrahimovich, D.; Vaya, J.; Khatib, S. Lyso-DGTS lipid isolated from microalgae enhances PON1 activities in vitro and in vivo, increases PON1 penetration into macrophages and decreases cellular lipid accumulation. Biofactors. 2018, 44, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Perla-Kaján, J.; Jakubowski, H. Paraoxonase 1 protects against protein N-homocysteinylation in humans. FASEB J. 2010, 24, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.B.; Hatch, M.A.; Ginsburg, S. Carbamylation of acetylcholinesterase. J. Biol. Chem. 1960, 235, 2312–2315. [Google Scholar] [PubMed]
- Goličnik, M.; Masson, P. Time-course of enzyme-catalyzed competing substrate degradation for michaelian behavior and for enzymes showing activation/inhibition by excess substrate. Chem. Biol. Interact. 2019, 309, 108704. [Google Scholar] [CrossRef] [PubMed]
- Mukhametgalieva, A.R.; Aglyamova, A.R.; Lushchekina, S.V.; Goličnik, M.; Masson, P. Time-course of human cholinesterases-catalyzed competing substrate kinetics. Chem. Biol. Interact. 2019, 310, 108702. [Google Scholar] [CrossRef]
- Lorentz, K.; Wirtz, W.; Wei, T. Continuous monitoring of arylesterase in human serum. Clin. Chim. Acta 2001, 308, 69–78. [Google Scholar] [CrossRef]
- Browne, R.W.; Koury, S.T.; Marion, S.; Wilding, G.; Muti, P.; Trevisan, M. Accuracy and biological variation of human serum paraoxonase 1 activity and polymorphism (Q192R) by kinetic assay. Clin. Chem. 2007, 532, 310–317. [Google Scholar] [CrossRef]
- Ceron, J.J.; Tecles, F.; Tvarijonaviciute, A. Serum paraoxonase 1 (PON1) measurement: Un update. BMC Vet. Res. 2014, 10, 74. [Google Scholar] [CrossRef]
- Debord, J.; Bollinger, J.C.; Harel, M.; Dantoine, T. Temperature dependence of binding and catalysis for human serum arylesterase/paraoxonase. Biochimie. 2014, 97, 72–77. [Google Scholar] [CrossRef]
- Stojan, J. The significance of low substrate concentration measurements for mechanistic interpretation in cholinesterases. Chem. Biol. Interact. 2013, 203, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Bosak, A.; Barlović-Tušek, B.; Reiner, E. Differentiation of EDTA-sensitive from EDTA-insensitive human serum esterases hydrolysing phenylacetate. J. Enzym. Inhib. Med. Chem. 2008, 23, 521–525. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tomić, S.; Trešec, A.; Tomašić, J.; Petrović, B.; Simeon Rudolf, V.; Škrinjarić-Špoljar, M.; Reiner, E. Catalytic properties of rabbit serum esterases hydrolyzing esterified monosaccharides. Biochim. Biophys. Acta 1995, 1251, 11–16. [Google Scholar]
- Dinaut, A.N.; Chen, M.J.; Marks, A.; Batey, R.A.; Taylor, S.D. Hydrolysis of an N-methylcarbamate by a catalytic antibody. Chem. Comm. 2000, 5, 385–386. [Google Scholar] [CrossRef]
- Wentworth, P.; Datta, A.; Smith, S.; Marshall, A.; Partridge, L.J.; Blackburn, G.M. Antibody catalysis of BAc2 aryl carbamate ester hydrolysis: A highly disfavored chemical process. J. Am. Chem. Soc. 1997, 119, 2315–2316. [Google Scholar] [CrossRef]
- Aharoni, A.; Gaidukov, L.; Yagur, S.; Toker, L.; Silman, I.; Tawfik, D.S. Directed evolution of mammalian paraoxonases PON1 and PON3 for bacterial expression and catalytic specialization. Proc. Natl. Acad. Sci. USA 2004, 101, 482–487. [Google Scholar] [CrossRef]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Feather-Stone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Cornish-Bowden, A. Fundamentals of Enzyme Kinetics, 3th ed.; Portland Press Ltd.: London, UK, 2004. [Google Scholar]
- Simeon-Rudolf, V.; Šinko, G.; Štuglin, A.; Reiner, E. Inhibition of human blood acetylcholinesterase and butyrylcholinesterase by ethopropazine. Croat. Chem. Acta 2001, 74, 173–182. [Google Scholar]
- Maraković, N.; Knežević, A.; Vinković, V.; Kovarik, Z.; Šinko, G. Design and synthesis of N-substituted-2-hydroxyiminoacetamides and interactions with cholinesterases. Chem. Biol. Interact. 2016, 259, 122–132. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
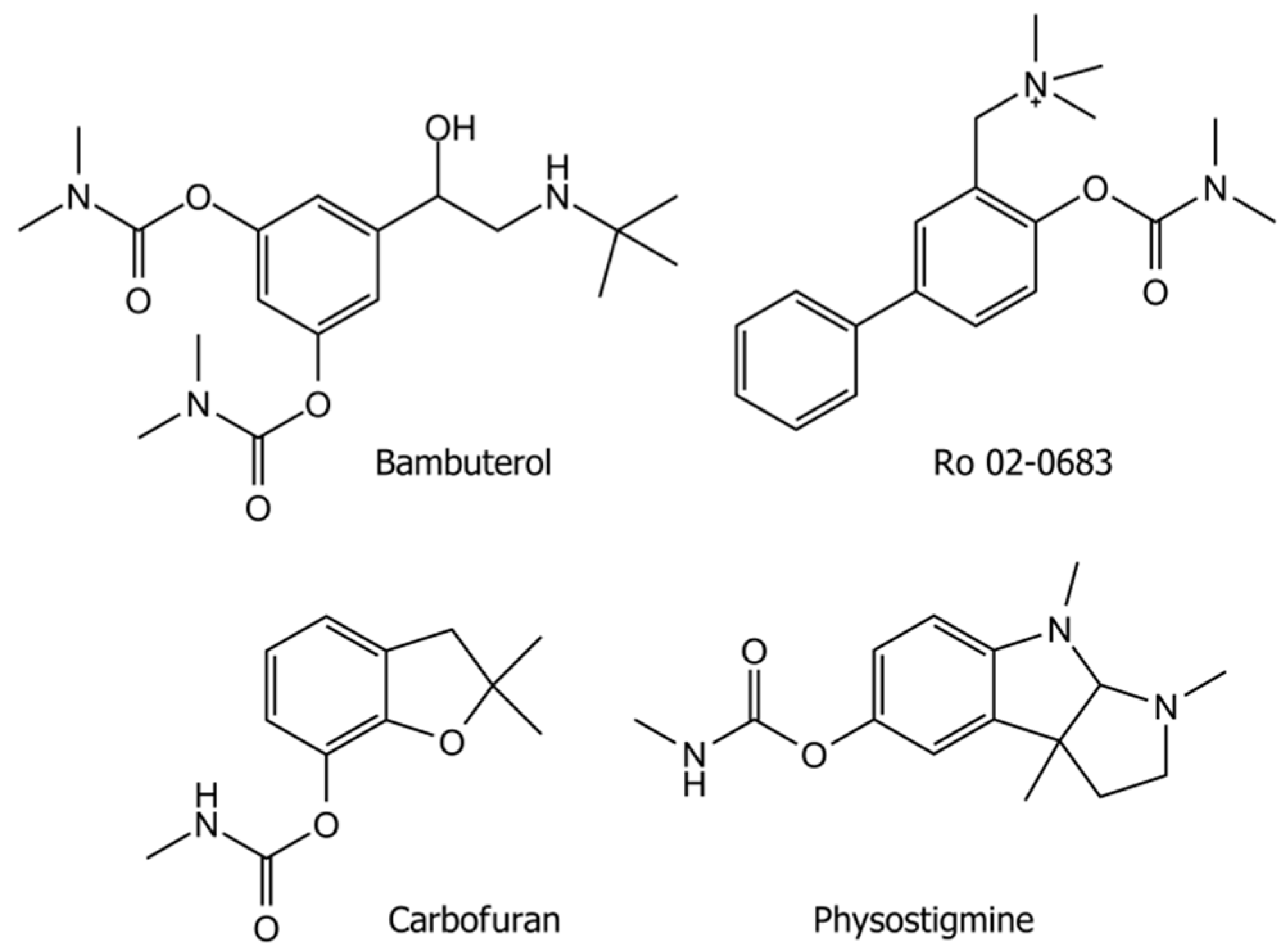
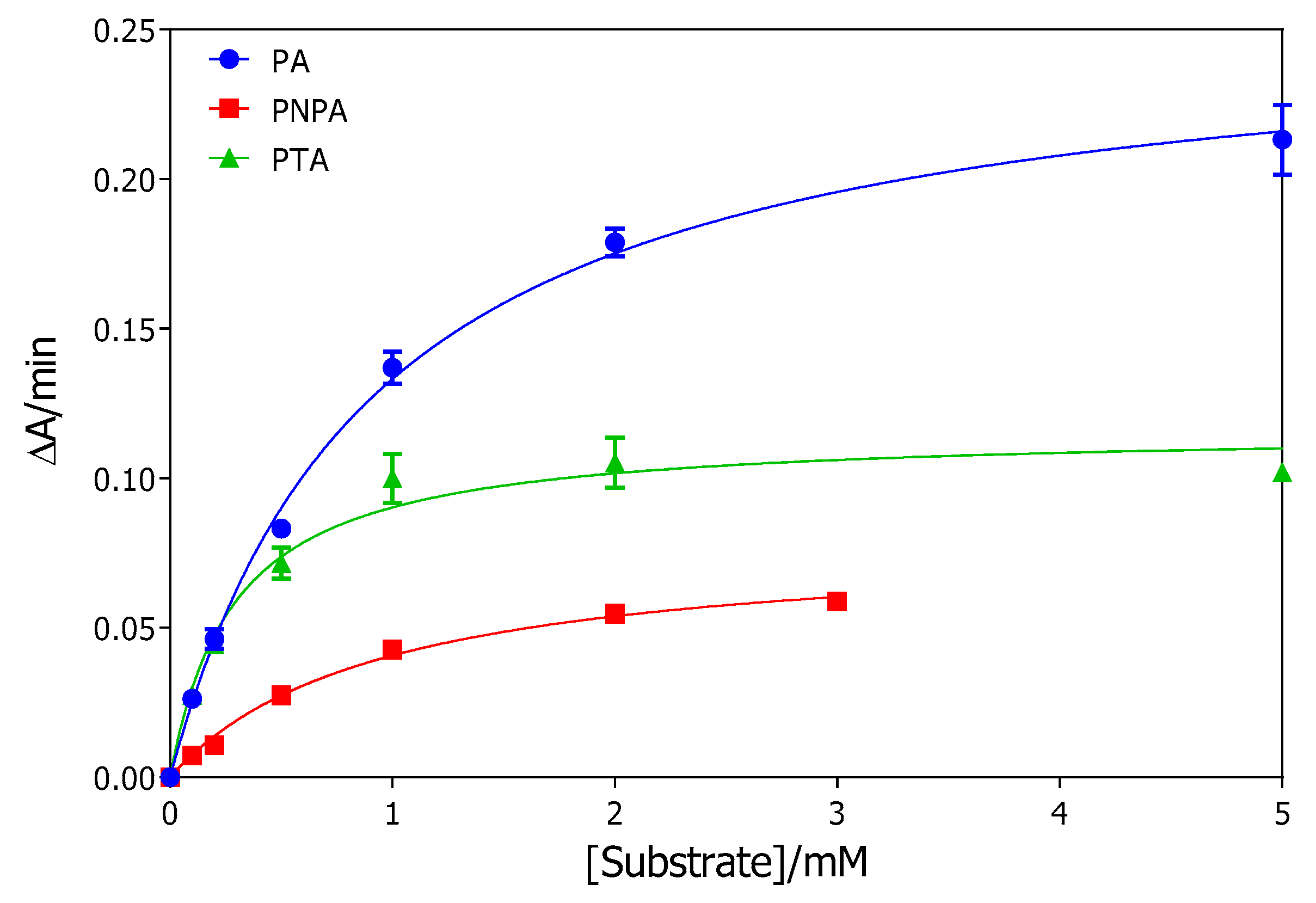

| Substrate | Km (mM) | ΔAmax/min | Vmax (µM∙min−1) * | SAmax (µmol∙min−1∙mL−1) |
|---|---|---|---|---|
| PA | 0.92 ± 0.06 | 0.255 ± 0.006 | 194 ± 5 | 582 ± 15 |
| PNPA | 0.95 ± 0.10 | 0.079 ± 0.003 | 6.2 ± 0.2 | 18.6 ± 0.6 |
| PTA | 0.29 ± 0.04 | 0.116 ± 0.004 | 8.2 ± 0.3 | 123 ± 5 |
| PA | PNPA | PTA | |
|---|---|---|---|
| ksp (10−3 min−1) | 0.61 ± 0.07 | 12.4 ± 0.7 | 3.5 ± 0.3 |
| Carbamate | Ki (mM) | Ks (mM) |
|---|---|---|
| Ro 02-0683 | 3.9 ± 0.6 | 1.2 ± 0.7 |
| Bambuterol | 3.1 ± 0.4 | 0.6 ± 0.2 |
| Physostigmine | 3.0 ± 0.6 | 0.4 ± 0.1 |
| Carbofuran | 0.65 ± 0.05 | 1.3 ± 0.4 |
| Carbamate | Ki (mM) |
|---|---|
| Ro 02-0683 | 11.3 ± 0.4 |
| Bambuterol | 6.8 ± 0.4 |
| Physostigmine | 5.6 ± 0.2 |
| Carbofuran | 0.77 ± 0.07 |
| Carbamate | Type of Interaction | ||
|---|---|---|---|
| H-bond | π Interactions * | Aliphatic Non-polar Interactions | |
| Ro 02-0683 | Glu53, Asp269 | His134, Lys192, Phe222 | Val346 |
| Bambuterol | Lys192, Asp269, Phe292 | Tyr71, His115, Phe222, Phe292, Phe347 | Leu240 |
| Physostigmine | Asp183, His285 | His115, His134 | Leu69 |
| Carbofuran | Asp183 | Phe222, His285, Phe292, Phe347 | Leu240, Ile291, Val346 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bosak, A.; Bavec, A.; Konte, T.; Šinko, G.; Kovarik, Z.; Goličnik, M. Interactions of Paraoxonase-1 with Pharmacologically Relevant Carbamates. Molecules 2020, 25, 211. https://doi.org/10.3390/molecules25010211
Bosak A, Bavec A, Konte T, Šinko G, Kovarik Z, Goličnik M. Interactions of Paraoxonase-1 with Pharmacologically Relevant Carbamates. Molecules. 2020; 25(1):211. https://doi.org/10.3390/molecules25010211
Chicago/Turabian StyleBosak, Anita, Aljoša Bavec, Tilen Konte, Goran Šinko, Zrinka Kovarik, and Marko Goličnik. 2020. "Interactions of Paraoxonase-1 with Pharmacologically Relevant Carbamates" Molecules 25, no. 1: 211. https://doi.org/10.3390/molecules25010211
APA StyleBosak, A., Bavec, A., Konte, T., Šinko, G., Kovarik, Z., & Goličnik, M. (2020). Interactions of Paraoxonase-1 with Pharmacologically Relevant Carbamates. Molecules, 25(1), 211. https://doi.org/10.3390/molecules25010211










