Quantitative Structure–Activity Relationships for the Flavonoid-Mediated Inhibition of P-Glycoprotein in KB/MDR1 Cells
Abstract
1. Introduction
2. Results
2.1. Cytotoxicity
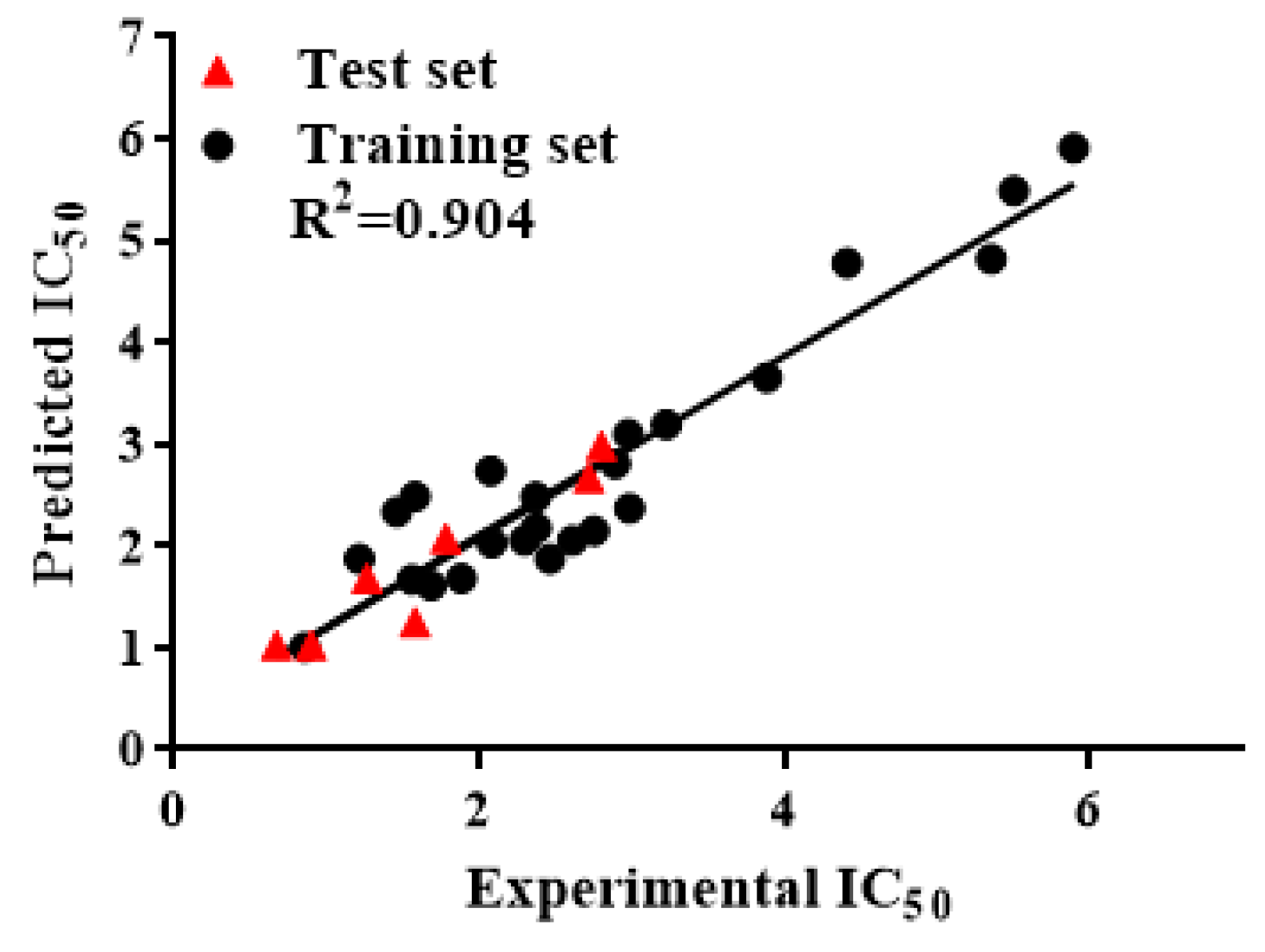
2.2. QSAR Study
- IC50 = 0.183vsurf_DW23 − 0.359E_sol − 3.181dipole + 10.627vsurf_G − 9 .859
- R2 = 0.892, Radj2 = 0.869, Q2 = 0.829, F = 39.073, p < 0.01, RMSE = 0.492, Rpred2 = 0.905
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture
4.3. Cytotoxicity Assay
4.4. 2D-QSAR Study
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| P-gp | P-glycoprotein |
| MDR | multidrug resistance |
| QSAR | quantitative structure–activity relationships |
| MRP | multidrug resistance protein |
| BCRP | breast cancer resistance protein |
| DMEM | minimum essential medium |
| MTT | 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide |
| FBS | Fetal bovine serum |
| PLS | partial least squares |
| LSD | least significant difference |
References
- Gadhe, C.G.; Cho, S.J. Flavonoids: An Emerging Lead in the P-glycoprotein Inhibition. J. Chosun Nat. Sci. 2016, 12, 2458–2470. [Google Scholar] [CrossRef][Green Version]
- Tsuruo, T.; Iida, H.; Tsukagoshi, S. Overcoming of vincristine resistance in P388 leukemia in vivo and in vitro through enhanced cytotoxicity of vincristine and vinblastine by verapamil. Cancer Res. 1981, 41, 1967–1972. [Google Scholar] [PubMed]
- Twentyman, P.R.; Fox, N.E.; White, D.J. Cyclosporin A and its analogues as modifiers of adriamycin and vincristine resistance in a multi-drug resistant human lung cancer cell line. Brit. J. Cancer. 1987, 56, 55–57. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M.; Pastan, I. Clinical trials of agents that reverse multidrug-resistance. J. Clin. Oncol. 1989, 7, 409–411. [Google Scholar] [CrossRef]
- Tran, C.D.; Timmins, P.; Conway, B.R.; Irwin, W.J. Investigation of the Coordinated Functional Activities of Cytochrome P450 3A4 and P-Glycoprotein in Limiting the Absorption of Xenobiotics in Caco-2 Cells. J. Pharm. Sci. 2002, 91, 117–128. [Google Scholar] [CrossRef]
- Shukla, S.; Wu, C.; Ambudkar, S.V. Development of inhibitors of ATP-binding cassette drug transporters: Present status and challenges. Expert Opin Drug Met. 2008, 4, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Mohana, S.; Ganesan, M.; Agilan, B.; Karthikeyan, R.; Srithar, G.; Mary, R.B.; Ambudkar, S.V. Screening dietary flavonoids for the reversal of P-glycoprotein-mediated multidrug resistance in cancer. Mol BioSyst. 2016, 12, 2458–2470. [Google Scholar] [CrossRef] [PubMed]
- Middleton, E.; Kandaswami, C.; Theoharides, T.C. The Effects of Plant Flavonoids on Mammalian Cells: Implications for Inflammation, Heart Disease, and Cancer. Pharmacol Rev. 2000, 52, 673–751. [Google Scholar]
- Pietro, A.D.; Conseil, G.; Perezvictoria, J.M.; Dayan, G.; Baubichoncortay, H.; Trompier, D.; Barron, D. Modulation by flavonoids of cell multidrug resistance mediated by P-glycoprotein and related ABC transporters. Cell Mol Life Sci. 2002, 59, 307–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Morris, M.E. Effects of the Flavonoids Biochanin A, Morin, Phloretin, and Silymarin on P-Glycoprotein-Mediated Transport. J. Pharmacol Exp. Ther. 2002, 304, 1258–1267. [Google Scholar] [CrossRef]
- Ferreira, A.; Rodrigues, M.; Fortuna, A.; Falcao, A.; Alves, G. Flavonoid compounds as reversing agents of the P-glycoprotein-mediated multidrug resistance: An in vitro evaluation with focus on antiepileptic drugs. Food Res. Int. 2018, 103, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Limtrakul, P.; Khantamat, O.; Pintha, K. Inhibition of P-Glycoprotein Function and Expression by Kaempferol and Quercetin. J. Chemotherapy. 2005, 17, 86–95. [Google Scholar] [CrossRef]
- Ofer, M.; Wolffram, S.; Koggel, A.; Spahn-Langguth, H.; Langguth, P. Modulation of drug transport by selected flavonoids: Involvement of P-gp and OCT? Eur. J. Pharm. Sci. 2005, 25, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Lies, B.; Martens, S.; Schmidt, S.; Boll, M.; Wenzel, U. Flavone potently stimulates an apical transporter for flavonoids in human intestinal Caco-2 cells. Mol. Nutr. Food Res. 2012, 56, 1627–1635. [Google Scholar] [CrossRef]
- Feng, S.; Yuan, Z.; Yao, X.; Ma, W.; Liu, L.; Liu, Z.; Xie, Y. Tangeretin, a citrus pentamethoxyflavone, antagonizes ABCB1-mediated multidrug resistance by inhibiting its transport function. Pharm. Res. 2016, 110, 193–204. [Google Scholar] [CrossRef]
- Kitagawa, S.; Nabekura, T.; Takahashi, T.; Nakamura, Y.; Sakamoto, H.; Tano, H.; Tsukahara, G. Structure–Activity Relationships of the Inhibitory Effects of Flavonoids on P-Glycoprotein-Mediated Transport in KB-C2 Cells. Biol. Pharm. Bull. 2005, 28, 2274–2278. [Google Scholar] [CrossRef]
- Santos, R.D.; Kuhnen, C.A.; Yunes, R.A. Molecular Structure and QSAR Study on Antispasmodic Activity of some Xanthoxyline Derivatives. Arch. Pharm. 2006, 339, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Wongrattanakamon, P.; Lee, V.S.; Nimmanpipug, P.; Jiranusornkul, S. 3D-QSAR modelling dataset of bioflavonoids for predicting the potential modulatory effect on P-glycoprotein activity. Data in Brief. 2016, 9, 35–42. [Google Scholar] [CrossRef]
- Kupsakova, I.; Rybar, A.; Docolomanský, P.; Drobna, Z.; Stein, U.; Walther, W.; Breier, A. Reversal of P-glycoprotein mediated vincristine resistance of L1210/VCR cells by analogues of pentoxifylline: A QSAR study. Eur. J. Pharm. Sci. 2004, 21, 283–293. [Google Scholar] [CrossRef]
- Srivastava, S.; Choudhary, B.S.; Sharma, M.; Malik, R. Pharmacophore modeling and 3D-QSAR studies of galloyl benzamides as potent P-gp inhibitors. Med. Chem. Res. 2016, 25, 1140–1147. [Google Scholar] [CrossRef]
- Myint, K.Z.; Xie, X. Recent Advances in Fragment-Based QSAR and Multi-Dimensional QSAR Methods. Int. J. Mol. Sci. 2010, 11, 3846–3866. [Google Scholar] [CrossRef]
- Borska, S.; Sopel, M.; Chmielewska, M.; Zabel, M.; Dziegiel, P. Quercetin as a potential modulator of P-glycoprotein expression and function in cells of human pancreatic carcinoma line resistant to daunorubicin. Molecules 2010, 15, 857–870. [Google Scholar] [CrossRef]
- Cruciani, G.; Crivori, P.; Carrupt, P.; Testa, B. Molecular fields in quantitative structure-permeation relationships: The VolSurf approach. J. Mol. Struc-theochem. 2000, 503, 17–30. [Google Scholar] [CrossRef]
- Yang, R.; Yu, L.; Zeng, H.; Liang, R.; Chen, X.; Qu, L. The Interaction of Flavonoid-Lysozyme and the Relationship between Molecular Structure of Flavonoids and Their Binding Activity to Lysozyme. J. Fluoresc. 2012, 22, 1449–1459. [Google Scholar] [CrossRef] [PubMed]
- Gadhe, C.G. Comparative Modeling of Human P-gp NBD2 and Docking and Binding Mode Analysis of 8-Geranyl Chrysin as a P-gp Modulator. J. Chosun. Natural Sci. 2012, 1, 18–21. [Google Scholar] [CrossRef]
- Fang, Y.; Xia, M.; Liang, F.; Cao, W.; Pan, S.; Xu, X. Establishment and use of human mouth epidermal carcinoma (KB) cells overexpressing P-glycoprotein to characterize structure requirements for flavonoids transported by the efflux transporter. J. Agr. Food Chem. 2019, 67, 2350–2360. [Google Scholar] [CrossRef]
- Castro, A.F.; Altenberg, G.A. Inhibition of drug transport by genistein in multidrug-resistant cells expressing P-glycoprotein. Biochem Pharmacol. 1997, 53, 89–93. [Google Scholar] [CrossRef]
- Choi, C.; Kim, J.; Kim, S. Reversal of P-glycoprotein-mediated MDR by 5,7,3′,4′,5′-pentamethoxyflavone and SAR. Biochem. Bioph. Res. Co. 2004, 320, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Liu, Y.; Zang, X.; Wu, T.; Qi, X.; Pan, S.; Xu, X. 3D-QSAR and docking studies of flavonoids as potent Escherichia coli inhibitors. Sci Rep.-UK 2016, 6, 23634. [Google Scholar] [CrossRef]
- Zhang, S.; We, L.; Bastow, K.; Zheng, W.; Brossi, A.; Lee, K.H.; Tropsha, A. Antitumor Agents 252. Application of validated QSAR models to database mining: discovery of novel tylophorine derivatives as potential anticancer agents. J. Comput aid Mol Des. 2007, 21, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Cao, W.; Xia, M.; Pan, S.; Xu, X. Study of structure and permeability relationship of flavonoids in Caco-2 cells. Nutrients 2017, 9, 1301. [Google Scholar] [CrossRef]
- Fang, Y.; Liang, F.; Liu, K.; Qaiser, S.; Pan, S.; Xu, X. Structure characteristics for intestinal uptake of flavonoids in Caco-2 cells. Food Res Int. 2018, 105, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.E.; Kadioglu, O.; Khalid, H.; Sugimoto, Y.; Efferth, T. Activity of the dietary flavonoid, apigenin, against multidrug-resistant tumor cells as determined by pharmacogenomics and molecular docking. J. Nutr. Biochem. 2015, 26, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.B.; Ling, V. Effect of quercetin on hoechst 33342 transport by purified and reconstituted p-glycoprotein. Biochem Pharmacol. 1997, 53, 587–596. [Google Scholar] [CrossRef]
- Szakacs, G.; Varadi, A.; Ozvegylaczka, C.; Sarkadi, B. The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME–Tox). Drug Discov Today. 2008, 13, 379–393. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
| No | Flavonoids | CAS | Core Structure | Substructure |
|---|---|---|---|---|
| 1 | 5-Methoxyflavone | 42079-78-7 | 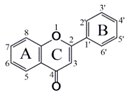 | R5=OMe |
| 2 | 5,7-Dimethoxyflavone | 21392-57-4 | R5, R7=OMe | |
| 3 | 5,3′-Dimethoxyflavone | R5, R3′=OMe | ||
| 4 | 5,7,3′-Trimethoxyflavone | R5, R7, R3′=OMe | ||
| 5 | 5,7,3′,4′-Tetramethoxyflavone | 855-97-0 | R5, R7, R3′, R4′=OMe | |
| 6 | Tangertin | 481-53-8 | R5, R6, R7, R3′, R4′=OMe | |
| 7 | Chrysin | 480-40-0 | R5, R7=OH | |
| 8 | Baicalein | 491-67-8 | R5, R6, R7 =OH | |
| 9 | Wogonin | 632-85-9 | R5, R7 =OH, R8=OMe | |
| 10 | Apigenin | 520-36-5 | R5, R7, R4′=OH | |
| 11 | Luteolin | 491-70-3 | R5, R7,, R3′, R4′=OH | |
| 12 | Vitexin | 3681-93-4 | R5, R7, R4′ =OH, R8=Cglc | |
| 13 | Schaftoside | 51938-32-0 | R5, R7, R4′ =OH, R6=Cglc, R8=Carb | |
| 14 | Galangin | 548-83-4 | R3, R5, R7=OH | |
| 15 | Kaempferide | 491-54-3 | R3, R5, R7 =OH, R4′=OMe | |
| 16 | Fisetin | 528-48-3 | R3, R7, R3′, R4′=OH | |
| 17 | Quercetin | 117-39-5 | R3, R5, R7, R3′, R4′=OH | |
| 18 | Morin | 480-16-0 | R3, R5, R7, R2′, R4′=OH | |
| 19 | Isorhamnetin | 480-19-3 | R3, R5, R7, R4′ =OH, R3′=OMe | |
| 20 | Myricetin | 529-44-2 | R3, R5, R7, R3′, R4′, R5′=OH | |
| 21 | Rutin | 153-18-4 | R3=ORG, R5, R7, R3′, R4′=OH | |
| 22 | Liquiritigenin | 578-86-9 |  | R7,, R4′=OH |
| 23 | Naringenin | 480-41-1 | R5, R7, R4′ =OH | |
| 24 | Hesperetin | 520-33-2 | R5, R7, R3′ =OH, R4′=OMe | |
| 25 | Taxifolin | 24198-97-8 | R3, R5, R7, R3′, R4′=OH | |
| 26 | Dihydromyricetin | 27200-12-0 | R3, R5, R7, R3′, R4′, R5′=OH | |
| 27 | Silibinin | 22888-70-6 | 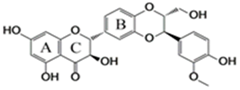 | |
| 28 | Daidzein | 40957-83-3 |  | R7, R4′=OH |
| 29 | Puerarin | 3681-99-0 | R7, R4′ =OH, R8=Cglc | |
| 30 | Genistein | 446-72-0 | R5, R7, R4′=OH | |
| 31 | Biochanin A | 491-80-5 | R5, R7 =OH, R4′=OMe | |
| No | KB/MDR1 Cells | KB Cells | RF | ||
|---|---|---|---|---|---|
| IC50(μM) | RFKB/MDR1 | IC50 (μM) | RFKB | ||
| C | 3.102 ± 0.441 | 1.000 | 0.715 ± 0.056 | 1.000 | 4.338 |
| E | 0.473 ± 0.005 | 6.818 | 0.455 ± 0.036 | 1.512 | 0.962 |
| 1 | 2.373 ± 0.970 | 1.307 | 0.993 ± 0.127 | 0.720 | 2.390 |
| 2 t | 1.579 ± 0.05 | 1.965 | 0.612 ± 0.107 | 1.169 | 2.582 |
| 3 | 1.580 ± 0.23 | 1.963 | 0.664 ± 0.051 | 1.077 | 2.380 |
| 4 t | 0.901 ± 0.042 | 3.443 | 0.955 ± 0.007 | 0.749 | 0.943 |
| 5 | 2.752 ± 0.211 | 1.127 | 0.905 ± 0.035 | 0.790 | 3.041 |
| 6 | 1.884 ± 0.243 | 1.646 | 2.610 ± 0.671 | 0.274 | 0.722 |
| 7 | 1.685 ± 0.623 | 1.841 | 0.916 ± 0.048 | 0.780 | 1.839 |
| 8 | 0.859 ± 0.137 | 3.613 | 0.413 ± 0.099 | 1.731 | 2.079 |
| 9 t | 0.676 ± 0.035 | 4.586 | 0.679 ± 0.023 | 1.053 | 0.996 |
| 10 | 3.226 ± 0.068 | 0.962 | 1.983 ± 0.078 | 0.361 | 1.627 |
| 11 | 5.894 ± 0.083 | 0.526 | 1.368 ± 0.077 | 0.523 | 4.310 |
| 12 | 5.501 ± 0.672 | 0.564 | 0.781 ± 0.133 | 0.916 | 7.046 |
| 13 | 1.459 ± 0.529 | 2.126 | 0.904 ± 0.158 | 0.791 | 1.613 |
| 14 | 1.560 ± 0.258 | 1.989 | 0.768 ± 0.113 | 0.931 | 2.030 |
| 15 | 2.891 ± 0.100 | 1.073 | 1.319 ± 0.041 | 0.542 | 2.192 |
| 16 | 2.463 ± 0.320 | 1.260 | 1.409 ± 0.225 | 0.508 | 1.748 |
| 17 | 5.353 ± 0.001 | 0.580 | 0.514 ± 0.006 | 1.391 | 10.414 |
| 18 | 1.214 ± 0.219 | 2.556 | 0.916 ± 0.064 | 0.781 | 1.325 |
| 19 | 2.976 ± 0.035 | 1.042 | 1.953 ± 0.197 | 0.366 | 1.524 |
| 20 t | 2.721 ± 0.067 | 1.140 | 0.567 ± 0.069 | 1.261 | 4.799 |
| 21 | 2.295 ± 0.054 | 1.352 | 1.312 ± 0.114 | 0.545 | 1.749 |
| 22 t | 2.803 ± 0.203 | 1.107 | 2.439 ± 0.189 | 0.293 | 1.149 |
| 23 | 2.076 ± 0.041 | 1.494 | 0.335 ± 0.015 | 2.138 | 6.205 |
| 24 | 2.084 ± 0.146 | 1.489 | 1.513 ± 0.057 | 0.473 | 1.377 |
| 25 | 2.371 ± 0.106 | 1.308 | 2.703 ± 0.182 | 0.264 | 0.877 |
| 26 | 2.611 ± 0.116 | 1.188 | 1.127 ± 0.092 | 0.635 | 2.317 |
| 27 t | 1.783 ± 0.055 | 1.740 | 0.962 ± 0.019 | 0.744 | 1.854 |
| 28 | 4.409 ± 0.540 | 0.704 | 0.203 ± 0.093 | 3.531 | 21.773 |
| 29 | 3.882 ± 0.172 | 0.799 | 2.393 ± 0.313 | 0.299 | 1.623 |
| 30 t | 1.267 ± 0.108 | 2.448 | 0.458 ± 0.072 | 1.561 | 2.767 |
| 31 | 2.986 ± 0.298 | 1.039 | 2.432 ± 0.227 | 0.294 | 1.228 |
| IC50 | vsurf_DW23 | E_sol | dipole | vsurf_G | |
|---|---|---|---|---|---|
| IC50 | 1.000 | 0.674 ** | −0.432 * | −0.297 | −0.041 |
| vsurf_DW23 | 1.000 | 0.006 | −0.268 | −0.212 | |
| E_sol | 1.000 | −0.464 * | 0.230 | ||
| dipole | 1.000 | 0.164 | |||
| vsurf_G | 1.000 |
| No. | vsurf_DW23 | E_sol | dipole | vsurf_G | IC50 (Experimental) | IC50 (Predicted) | Residuals |
|---|---|---|---|---|---|---|---|
| 1 | 0.707 | −3.559 | 0.841 | 1.252 | 2.373 | 2.180 | 0.193 |
| 2 | 1.000 | −3.054 | 1.237 | 1.295 | 1.579 | 1.252 | 0.327 |
| 3 | 1.000 | −3.711 | 0.896 | 1.288 | 1.580 | 2.490 | −0.910 |
| 4 | 1.581 | −2.211 | 1.348 | 1.326 | 0.901 | 1.026 | −0.125 |
| 5 | 1.000 | −1.574 | 0.972 | 1.351 | 2.752 | 2.149 | 0.603 |
| 6 | 3.391 | −0.953 | 1.133 | 1.335 | 1.884 | 1.684 | 0.200 |
| 7 | 1.000 | −4.867 | 1.158 | 1.244 | 1.685 | 1.612 | 0.073 |
| 8 | 0.707 | −2.935 | 1.161 | 1.259 | 0.859 | 1.009 | −0.150 |
| 9 | 1.000 | −4.269 | 1.262 | 1.240 | 0.676 | 1.025 | −0.349 |
| 10 | 1.000 | −5.855 | 0.829 | 1.262 | 3.226 | 3.196 | 0.030 |
| 11 | 14.221 | −5.762 | 0.769 | 1.275 | 5.894 | 5.914 | −0.020 |
| 12 | 1.118 | −11.881 | 0.997 | 1.322 | 5.501 | 5.494 | 0.007 |
| 13 | 0.707 | 1.216 | 0.776 | 1.409 | 1.459 | 2.336 | −0.877 |
| 14 | 0.707 | −0.596 | 0.679 | 1.256 | 1.560 | 1.670 | −0.110 |
| 15 | 0.707 | −0.369 | 0.420 | 1.293 | 2.891 | 2.808 | 0.083 |
| 16 | 0.500 | 1.410 | 0.440 | 1.275 | 2.463 | 1.874 | 0.589 |
| 17 | 11.597 | −0.871 | 0.438 | 1.283 | 5.353 | 4.819 | 0.534 |
| 18 | 0.866 | −3.328 | 1.024 | 1.284 | 1.214 | 1.878 | −0.664 |
| 19 | 3.606 | 0.379 | 0.434 | 1.300 | 2.976 | 3.101 | −0.125 |
| 20 | 0.500 | −0.215 | 0.446 | 1.297 | 2.721 | 2.671 | 0.050 |
| 21 | 1.581 | 0.317 | 1.021 | 1.410 | 2.295 | 2.055 | 0.240 |
| 22 | 1.000 | −2.655 | 0.523 | 1.258 | 2.803 | 2.982 | −0.179 |
| 23 | 1.000 | −2.064 | 0.551 | 1.264 | 2.076 | 2.740 | −0.664 |
| 24 | 0.500 | −0.711 | 0.690 | 1.293 | 2.084 | 2.032 | 0.052 |
| 25 | 1.118 | −3.043 | 0.796 | 1.278 | 2.371 | 2.483 | −0.112 |
| 26 | 1.000 | −1.461 | 0.793 | 1.292 | 2.611 | 2.054 | 0.557 |
| 27 | 1.000 | −3.440 | 1.426 | 1.416 | 1.783 | 2.071 | −0.288 |
| 28 | 16.523 | −1.351 | 0.657 | 1.244 | 4.409 | 4.782 | −0.373 |
| 29 | 1.000 | −4.664 | 0.811 | 1.339 | 3.882 | 3.652 | 0.230 |
| 30 | 0.500 | −5.127 | 1.167 | 1.253 | 1.267 | 1.681 | −0.414 |
| 31 | 1.000 | −5.328 | 1.082 | 1.277 | 2.986 | 2.369 | 0.617 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, M.; Fang, Y.; Cao, W.; Liang, F.; Pan, S.; Xu, X. Quantitative Structure–Activity Relationships for the Flavonoid-Mediated Inhibition of P-Glycoprotein in KB/MDR1 Cells. Molecules 2019, 24, 1661. https://doi.org/10.3390/molecules24091661
Xia M, Fang Y, Cao W, Liang F, Pan S, Xu X. Quantitative Structure–Activity Relationships for the Flavonoid-Mediated Inhibition of P-Glycoprotein in KB/MDR1 Cells. Molecules. 2019; 24(9):1661. https://doi.org/10.3390/molecules24091661
Chicago/Turabian StyleXia, Mengmeng, Yajing Fang, Weiwei Cao, Fuqiang Liang, Siyi Pan, and Xiaoyun Xu. 2019. "Quantitative Structure–Activity Relationships for the Flavonoid-Mediated Inhibition of P-Glycoprotein in KB/MDR1 Cells" Molecules 24, no. 9: 1661. https://doi.org/10.3390/molecules24091661
APA StyleXia, M., Fang, Y., Cao, W., Liang, F., Pan, S., & Xu, X. (2019). Quantitative Structure–Activity Relationships for the Flavonoid-Mediated Inhibition of P-Glycoprotein in KB/MDR1 Cells. Molecules, 24(9), 1661. https://doi.org/10.3390/molecules24091661





