Identifying Reducing and Capping Sites of Protein-Encapsulated Gold Nanoclusters
Abstract
1. Introduction
2. Results and Discussion
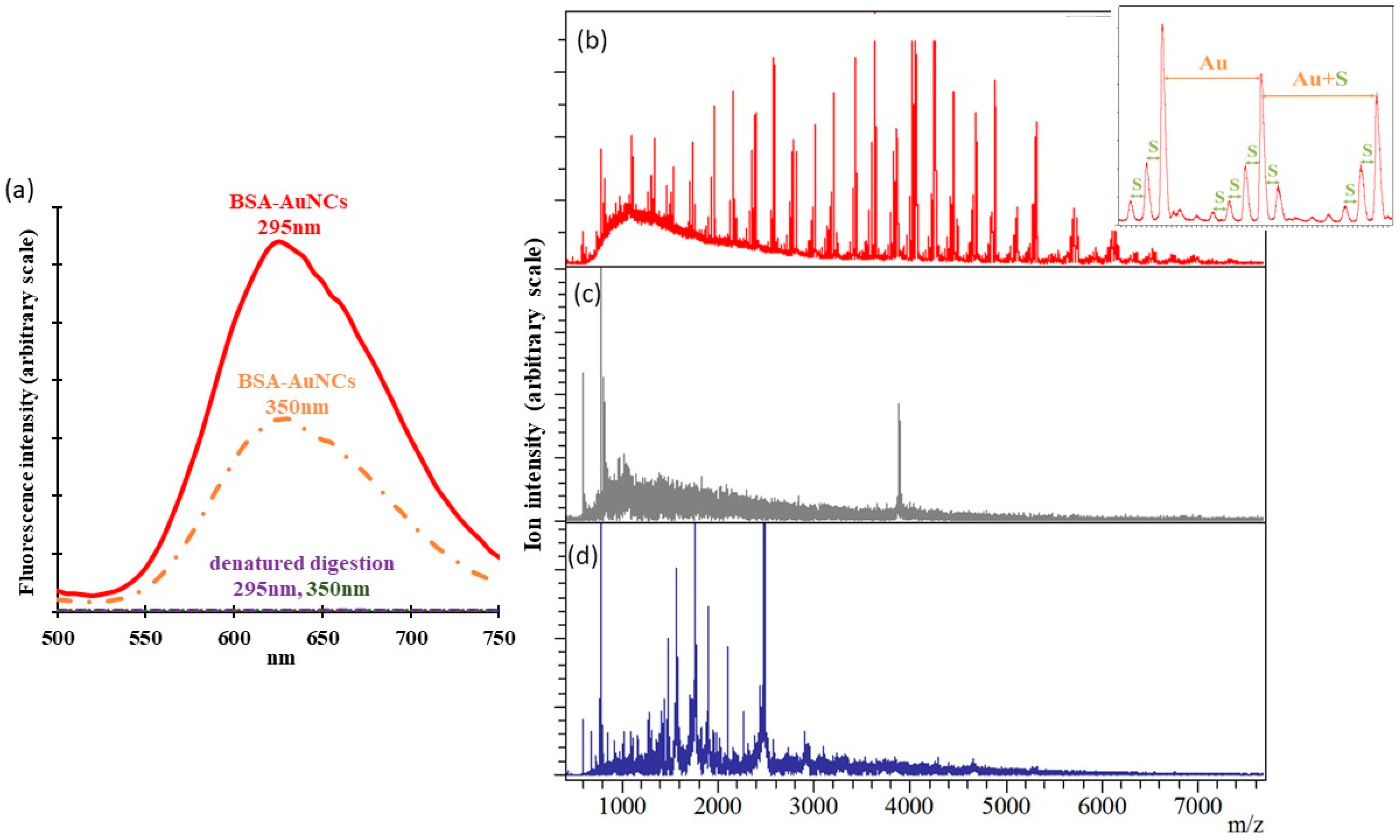
2.1. Characterization of As-synthesized BSA-AuNCs
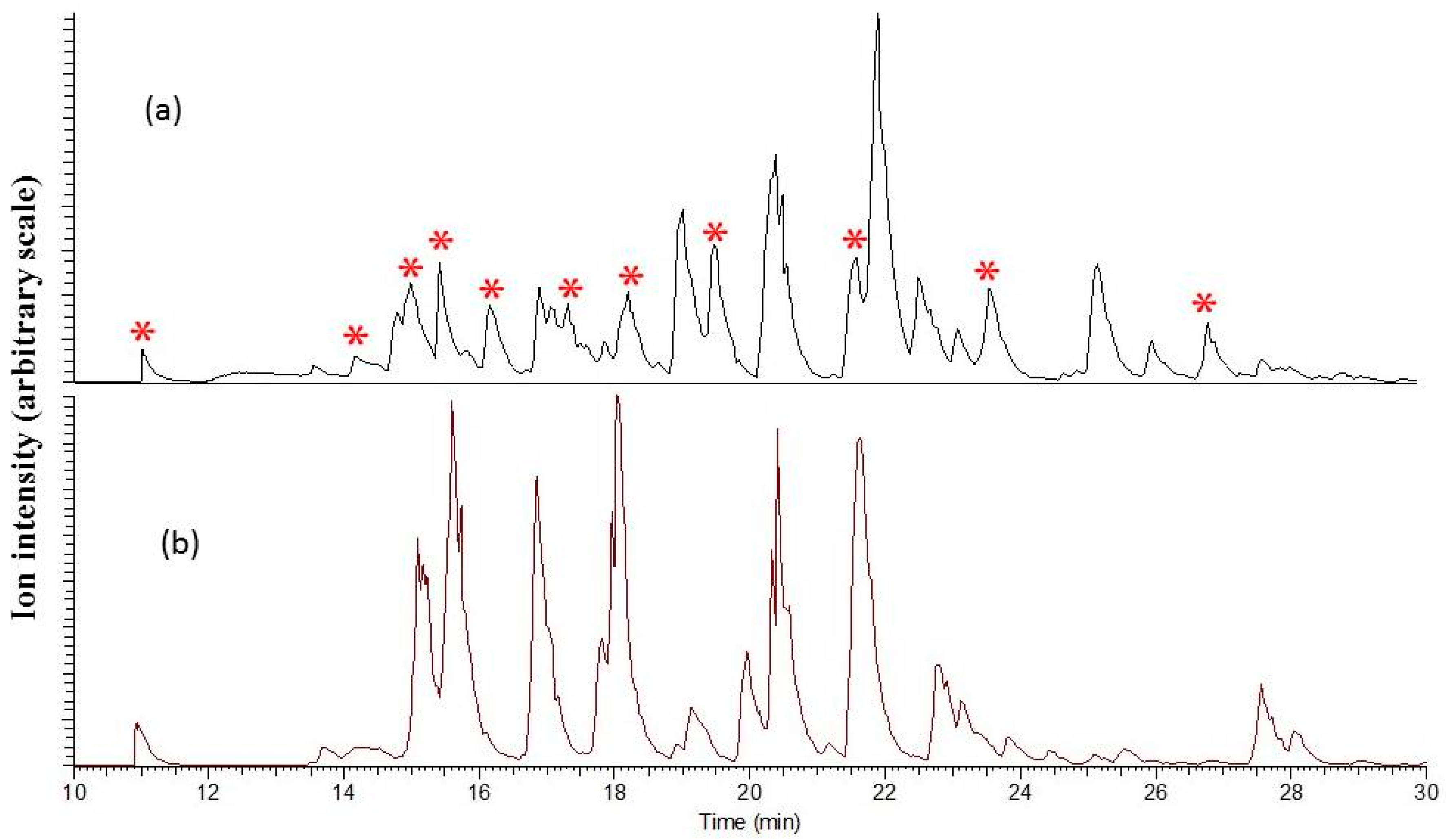
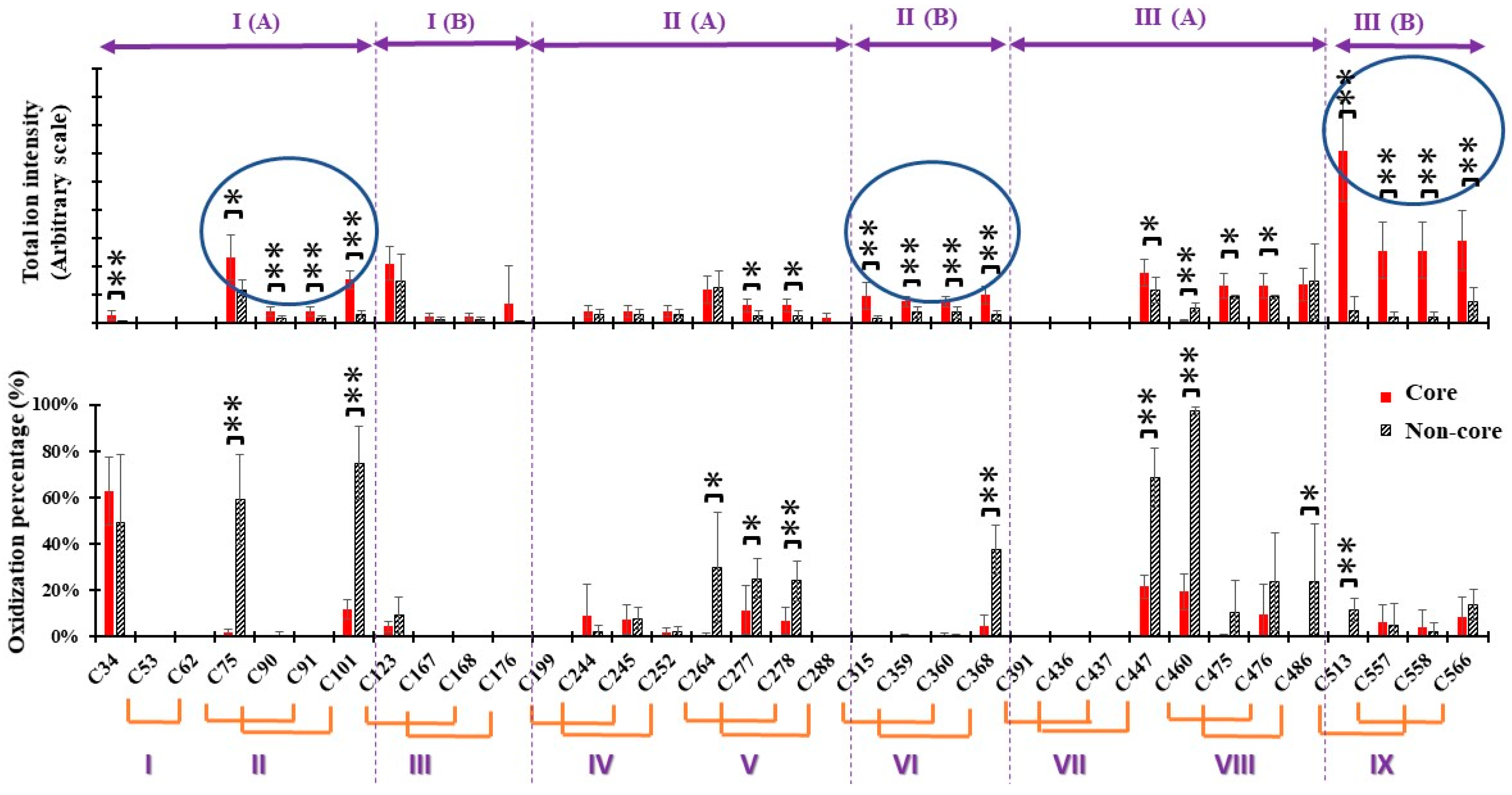
2.2. Identification of Global Reducing Sites
2.3. Disulfide Cleavages by Extensive Cysteine Oxidization
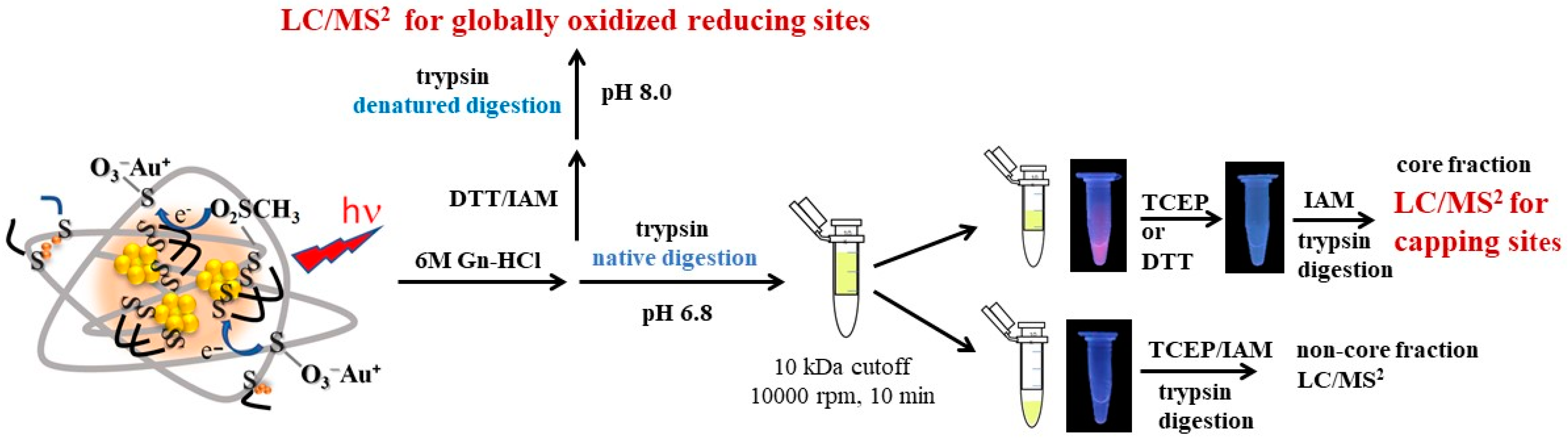
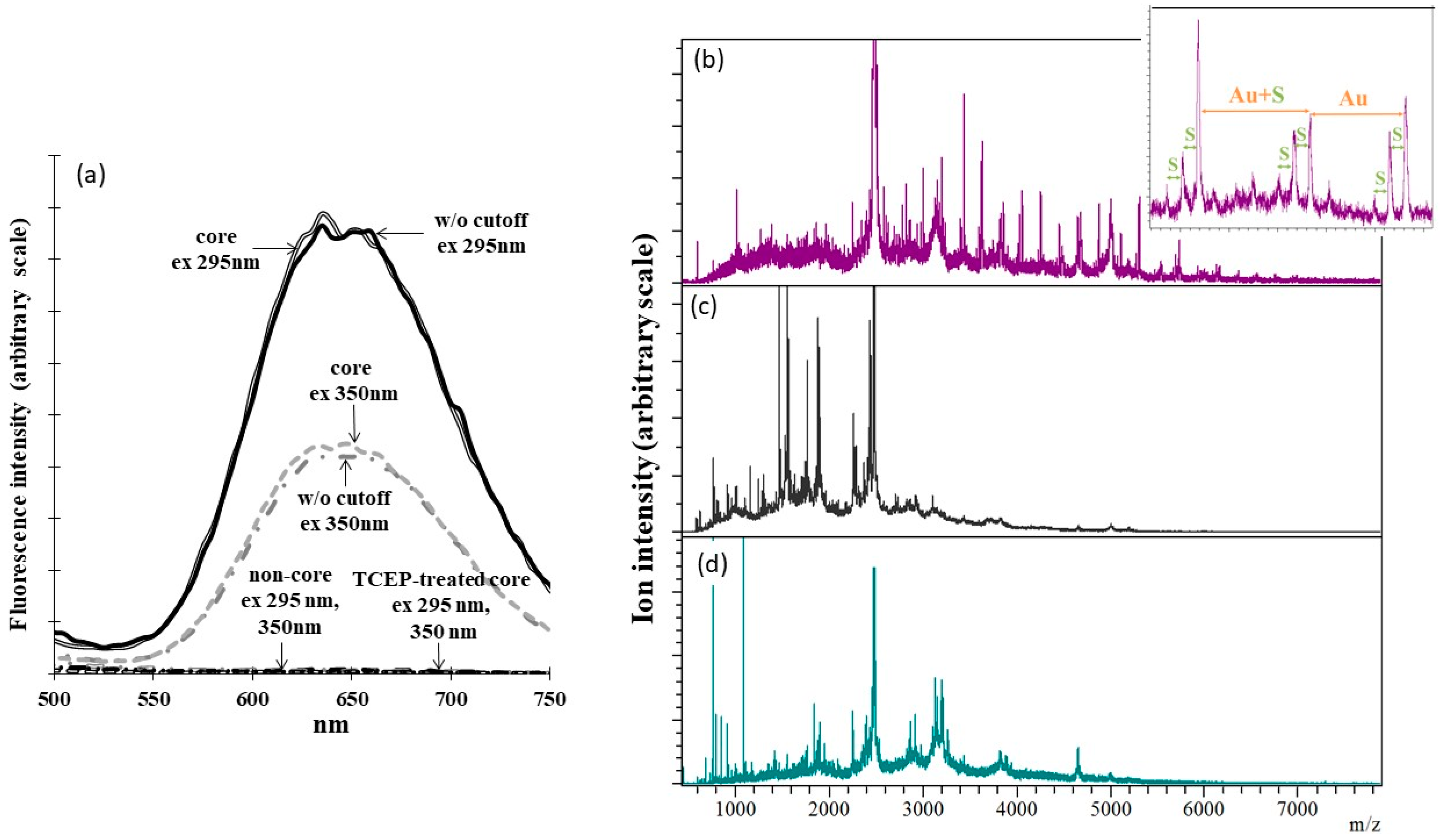
2.4. Identification of the Au-Core Capping Sites
2.5. Identification of C34 as Au+ Binding Sites Involved in Energy Transfer
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Synthesis of the Red BSA-AuNC
3.3. UV/Fluorescence Measurement
3.4. MALDI-MS Measurement
3.5. X -ray Photoelectron Spectroscopy (XPS)
3.6. Preparation of the Core and Non-Core Fraction of BSA-AuNC
3.7. Trypsin Digestion
3.8. LC-MS2 Analysis
3.9. Solvent Accessible Area
3.10. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lengke, M.F.; Southam, G. The effect of thiosulfate-oxidizing bacteria on the stability of the gold-thiosulfate complex. Geochim. Cosmochim. Acta 2005, 69, 3759–3772. [Google Scholar] [CrossRef]
- Reith, F.; Etschmann, B.; Grosse, C.; Moors, H.; Benotmane, M.A.; Monsieurs, P.; Grass, G.; Doonan, C.; Vogt, S.; Lai, B.; et al. Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans. Proc. Natl. Acad. Sci. USA 2009, 106, 17757–17762. [Google Scholar] [CrossRef] [PubMed]
- Reith, F.; Rogers, S.L.; McPhail, D.C.; Webb, D. Biomineralization of gold: Biofilms on bacterioform gold. Science 2006, 313, 233–236. [Google Scholar] [CrossRef]
- Wei, H.; Wang, Z.; Zhang, J.; House, S.; Gao, Y.G.; Yang, L.; Robinson, H.; Tan, L.H.; Xing, H.; Hou, C.; et al. Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme. Nature Nanotech. 2011, 6, 93–97. [Google Scholar] [CrossRef]
- Maity, B.; Abe, S.; Ueno, T. Observation of gold sub-nanocluster nucleation within a crystalline protein cage. Nature Commun. 2017, 8, 14820. [Google Scholar] [CrossRef]
- Xie, J.; Zheng, Y.; Ying, J.Y. Protein-directed synthesis of highly fluorescent gold nanoclusters. J. Am. Chem. Soc. 2009, 131, 888–889. [Google Scholar] [CrossRef] [PubMed]
- Govindaraju, S.; Ankireddy, S.R.; Viswanath, B.; Kim, J.; Yun, K. Fluorescent Gold Nanoclusters for Selective Detection of Dopamine in Cerebrospinal fluid. Sci. Rep. 2017, 7, 40298. [Google Scholar] [CrossRef]
- Saha, K.; Agasti, S.S.; Kim, C.; Li, X.; Rotello, V.M. Gold nanoparticles in chemical and biological sensing. Chem. Rev. 2012, 112, 2739–2779. [Google Scholar] [CrossRef]
- Chen, L.Y.; Wang, C.W.; Yuan, Z.; Chang, H.T. Fluorescent gold nanoclusters: Recent advances in sensing and imaging. Anal. Chem. 2015, 87, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Khandelia, R.; Bhandari, S.; Pan, U.N.; Ghosh, S.S.; Chattopadhyay, A. Gold Nanocluster Embedded Albumin Nanoparticles for Two-Photon Imaging of Cancer Cells Accompanying Drug Delivery. Small 2015, 11, 4075–4081. [Google Scholar] [CrossRef]
- Nune, S.K.; Gunda, P.; Thallapally, P.K.; Lin, Y.Y.; Forrest, M.L.; Berkland, C.J. Nanoparticles for biomedical imaging. Expert Opin. Drug Deliv. 2009, 6, 1175–1194. [Google Scholar] [CrossRef]
- Dorsey, J.F.; Sun, L.; Joh, D.Y.; Witztum, A.; Kao, G.D.; Alonso-Basanta, M.; Avery, S.; Hahn, S.M.; Al Zaki, A.; Tsourkas, A. Gold nanoparticles in radiation research: Potential applications for imaging and radiosensitization. Transl. Cancer Res. 2013, 2, 280–291. [Google Scholar]
- Egusa, S.; Ebrahem, Q.; Mahfouz, R.Z.; Saunthararajah, Y. Ligand exchange on gold nanoparticles for drug delivery and enhanced therapeutic index evaluated in acute myeloid leukemia models. Exp. Biol. Med. 2014, 239, 853–861. [Google Scholar] [CrossRef]
- Arvizo, R.; Bhattacharya, R.; Mukherjee, P. Gold nanoparticles: Opportunities and challenges in nanomedicine. Expert Opin. Drug Deliv. 2010, 7, 753–763. [Google Scholar] [CrossRef]
- Doane, T.L.; Burda, C. The unique role of nanoparticles in nanomedicine: Imaging, drug delivery and therapy. Chem. Soc. Rev. 2012, 41, 2885–2911. [Google Scholar] [CrossRef]
- Huang, X.; Jain, P.K.; El-Sayed, I.H.; El-Sayed, M.A. Gold nanoparticles: Interesting optical properties and recent applications in cancer diagnostics and therapy. Nanomedicine 2007, 2, 681–693. [Google Scholar] [CrossRef]
- Ferrari, M. Cancer nanotechnology: Opportunities and challenges. Nature Rev. Cancer 2005, 5, 161–171. [Google Scholar] [CrossRef]
- Daniel, M.C.; Astruc, D. Gold nanoparticles: Assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem. Rev. 2004, 104, 293–346. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Luo, Z.; Teo, C.S.; Tan, Y.N.; Xie, J. Tailoring the protein conformation to synthesize different-sized gold nanoclusters. Chem. Commun. 2013, 49, 9740–9742. [Google Scholar] [CrossRef]
- Dixon, J.M.; Egusa, S. Conformational Change-Induced Fluorescence of Bovine Serum Albumin-Gold Complexes. J. Am. Chem. Soc. 2018, 140, 2265–2271. [Google Scholar] [CrossRef]
- Zheng, J.; Nicovich, P.R.; Dickson, R.M. Highly fluorescent noble-metal quantum dots. Annu. Rev. Phys. Chem. 2007, 58, 409–431. [Google Scholar] [CrossRef]
- Tanaka, A.; Takeda, Y.; Imamura, M.; Sato, S. Dynamic final-state effect on the Au4fcore-level photoemission of dodecanethiolate-passivated Au nanoparticles on graphite substrates. Phys. Rev. B 2003, 68. [Google Scholar] [CrossRef]
- Le Guével, X.; Hötzer, B.; Jung, G.; Hollemeyer, K.; Trouillet, V.; Schneider, M. Formation of Fluorescent Metal (Au, Ag) Nanoclusters Capped in Bovine Serum Albumin Followed by Fluorescence and Spectroscopy. J. Phys. Chem. C 2011, 115, 10955–10963. [Google Scholar] [CrossRef]
- Wu, Z.; Jin, R. On the ligand’s role in the fluorescence of gold nanoclusters. Nano letters 2010, 10, 2568–2573. [Google Scholar] [CrossRef]
- Raut, S.; Chib, R.; Butler, S.; Borejdo, J.; Gryczynski, Z.; Gryczynski, I. Evidence of energy transfer from tryptophan to BSA/HSA protected gold nanoclusters. Methods Appl. Fluoresc. 2014, 2, 035004. [Google Scholar] [CrossRef]
- Chaudhari, K.; Xavier, P.L.; Pradeep, T. Understanding the evolution of luminescent gold quantum clusters in protein templates. Acs Nano 2011, 5, 8816–8827. [Google Scholar] [CrossRef]
- Xu, Y.; Palchoudhury, S.; Qin, Y.; Macher, T.; Bao, Y. Make conjugation simple: A facile approach to integrated nanostructures. Langmuir 2012, 8, 8767–8772. [Google Scholar] [CrossRef]
- Simms, G.A.; Padmos, J.D.; Zhang, P. Structural and electronic properties of protein/thiolate-protected gold nanocluster with “staple” motif: A XAS, L-DOS, and XPS study. J. Chem. Phys. 2009, 131, 214703. [Google Scholar] [CrossRef]
- Chen, T.H.; Tseng, W.L. (Lysozyme type VI)-stabilized Au8 clusters: Synthesis mechanism and application for sensing of glutathione in a single drop of blood. Small 2012, 8, 1912–1919. [Google Scholar] [CrossRef]
- Kawasaki, H.; Hamaguchi, K.; Osaka, I.; Arakawa, R. ph-Dependent Synthesis of Pepsin-Mediated Gold Nanoclusters with Blue Green and Red Fluorescent Emission. Adv. Funct. Mater. 2011, 21, 3508–3515. [Google Scholar] [CrossRef]
- Ma, X.; Wen, X.; Toh, Y.R.; Huang, K.Y.; Tang, J.; Yu, P. Dynamic study on the transformation process of gold nanoclusters. Nanotechnology 2014, 25, 445705. [Google Scholar] [CrossRef]
- Wen, X.; Yu, P.; Toh, Y.-R.; Tang, J. Quantum Confined Stark Effect in Au8 and Au25 Nanoclusters. J. Phys. Chem. C 2013, 117, 3621–3626. [Google Scholar] [CrossRef]
- Russell, B.A.; Kubiak-Ossowska, K.; Mulheran, P.A.; Birch, D.J.; Chen, Y. Locating the nucleation sites for protein encapsulated gold nanoclusters: A molecular dynamics and fluorescence study. Phys. Chem. Chem. Phys. 2015, 17, 21935–21941. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Shi, M.; Zhou, R.; Chen, X.; Chen, H. Blending of HAuCl4 and histidine in aqueous solution: A simple approach to the Au10 cluster. Nanoscale 2011, 3, 2596–2601. [Google Scholar] [CrossRef]
- Das, T.; Ghosh, P.; Shanavas, M.S.; Maity, A.; Mondal, S.; Purkayastha, P. Protein-templated gold nanoclusters: Size dependent inversion of fluorescence emission in the presence of molecular oxygen. Nanoscale 2012, 4, 6018–6024. [Google Scholar] [CrossRef]
- Schaaff, T.G.; Whetten, R.L. Giant gold− glutathione cluster compounds: Intense optical activity in metal-based transitions. J. Phys. Chem. B 2000, 104, 2630–2641. [Google Scholar] [CrossRef]
- Le Guevel, X.; Daum, N.; Schneider, M. Synthesis and characterization of human transferrin-stabilized gold nanoclusters. Nanotechnology 2011, 22, 275103. [Google Scholar] [CrossRef] [PubMed]
- Arnold, R.J.; Reilly, J.P. High-resolution time-of-flight mass spectra of alkanethiolate-coated gold nanocrystals. J. Am. Chem. Soc. 1998, 120, 1528–1532. [Google Scholar] [CrossRef]
- Tan, Y.N.; Lee, J.Y.; Wang, D.I. Uncovering the design rules for peptide synthesis of metal nanoparticles. J. Am. Chem. Soc. 2010, 132, 5677–5686. [Google Scholar] [CrossRef]
- Jhan, S.Y.; Huang, L.J.; Wang, T.F.; Chou, H.H.; Chen, S.H. Dimethyl Labeling Coupled with Mass Spectrometry for Topographical Characterization of Primary Amines on Monoclonal Antibodies. Anal. Chem. 2017, 89, 4255–4263. [Google Scholar] [CrossRef] [PubMed]
- Majorek, K.A.; Porebski, P.J.; Dayal, A.; Zimmerman, M.D.; Jablonska, K.; Stewart, A.J.; Chruszcz, M.; Minor, W. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol. Immunol. 2012, 52, 174–182. [Google Scholar] [CrossRef]
- Peters, T.J. All About Albumins; Academic Press: San Diego, CA, USA, 1996. [Google Scholar]
- Christodoulou, J.; Sadler, P.J.; Tucker, A. A new structural transition of serum albumin dependent on the state of Cys34: Detection by 1H-NMR spectroscopy. Eur. J. Biochem. 1994, 225, 363–368. [Google Scholar] [CrossRef]
- Peters, T., Jr.; Stewart, A.J. Albumin research in the 21st century. Biochim. Biophys. Acta 2013, 1830, 5351–5353. [Google Scholar] [CrossRef]
- Anand, U.; Mukherjee, S. Binding, unfolding and refolding dynamics of serum albumins. Biochim. Biophys. Acta 2013, 1830, 5394–5404. [Google Scholar] [CrossRef]
- Tsai, C.Y.; Li, C.W.; Li, J.R.; Jang, B.H.; Chen, S.H. Steroid Probes Conjugated with Protein-Protected Gold Nanocluster: Specific and Rapid Fluorescence Imaging of Steroid Receptors in Target Cells. J. Fluoresc. 2016, 26, 1239–1248. [Google Scholar] [CrossRef]
- Accessible Surface Area and Accessibility Calculation for Protein. Available online: http://cib.cf.ocha.ac.jp/bitool/ASA/ (accessed on 23 April 2019).
Sample Availability: Samples of the compounds are available from the authors. |

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, Y.-C.; Hung, M.-J.; Chen, Y.-A.; Wang, T.-F.; Ou, Y.-R.; Chen, S.-H. Identifying Reducing and Capping Sites of Protein-Encapsulated Gold Nanoclusters. Molecules 2019, 24, 1630. https://doi.org/10.3390/molecules24081630
Hsu Y-C, Hung M-J, Chen Y-A, Wang T-F, Ou Y-R, Chen S-H. Identifying Reducing and Capping Sites of Protein-Encapsulated Gold Nanoclusters. Molecules. 2019; 24(8):1630. https://doi.org/10.3390/molecules24081630
Chicago/Turabian StyleHsu, Yu-Chen, Mei-Jou Hung, Yi-An Chen, Tsu-Fan Wang, Ying-Ru Ou, and Shu-Hui Chen. 2019. "Identifying Reducing and Capping Sites of Protein-Encapsulated Gold Nanoclusters" Molecules 24, no. 8: 1630. https://doi.org/10.3390/molecules24081630
APA StyleHsu, Y.-C., Hung, M.-J., Chen, Y.-A., Wang, T.-F., Ou, Y.-R., & Chen, S.-H. (2019). Identifying Reducing and Capping Sites of Protein-Encapsulated Gold Nanoclusters. Molecules, 24(8), 1630. https://doi.org/10.3390/molecules24081630





