Untargeted Metabolomics Study of the In Vitro Anti-Hepatoma Effect of Saikosaponin d in Combination with NRP-1 Knockdown
Abstract
1. Introduction
2. Results
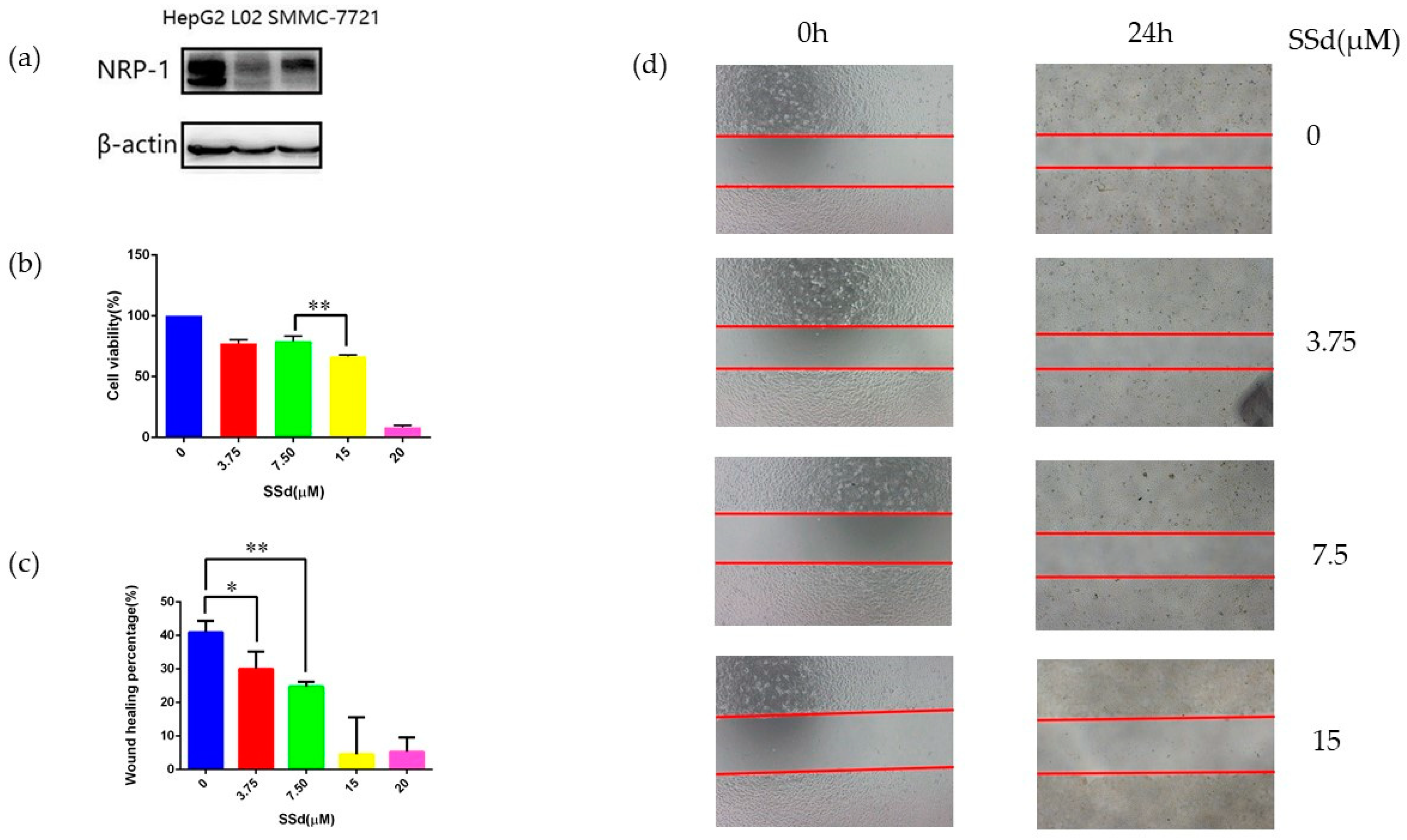
2.1. NRP-1 Was Selected as a Target of SSd
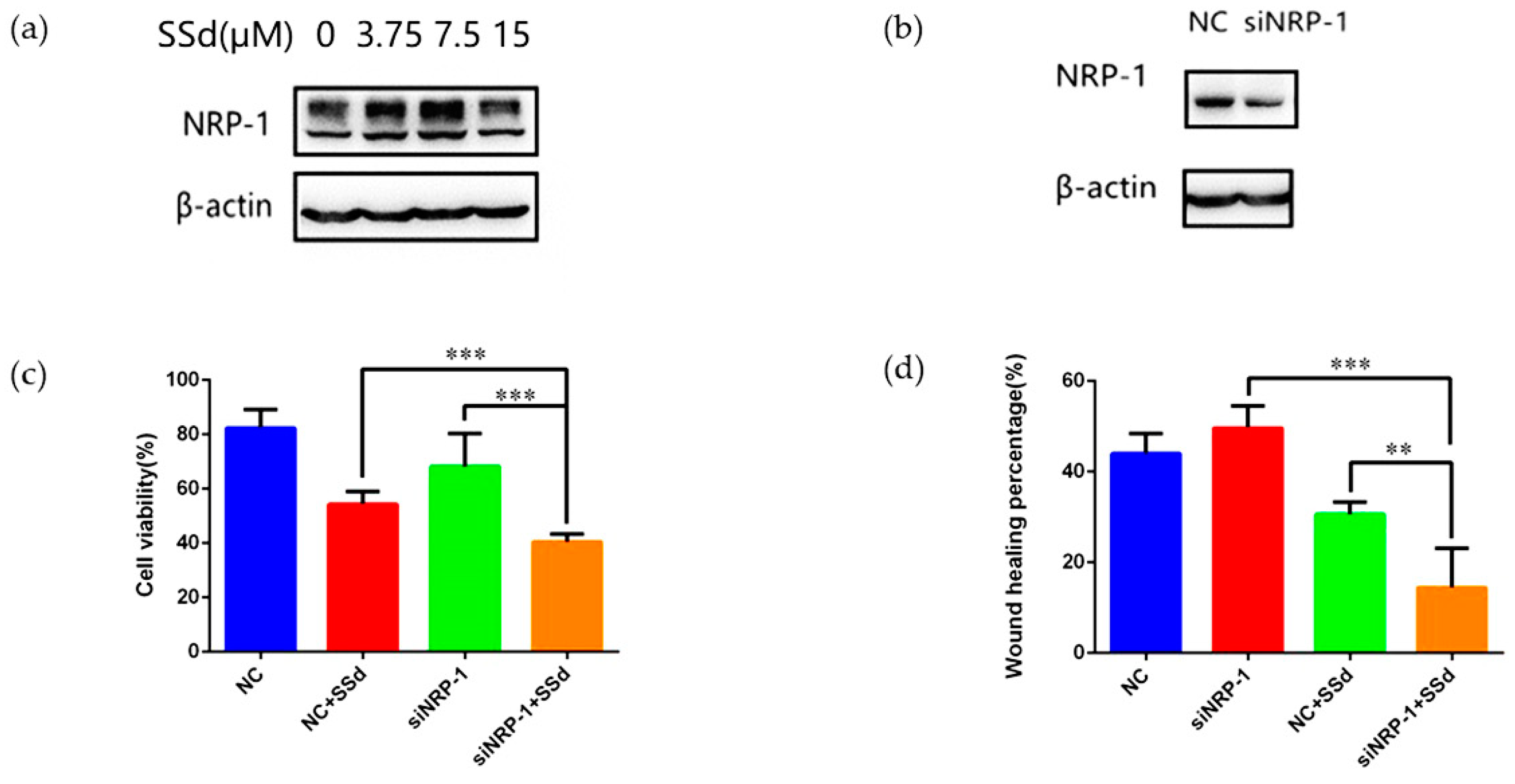
2.2. NRP-1 Knockdown Enhanced the Anti-Hepatoma Effect of SSd
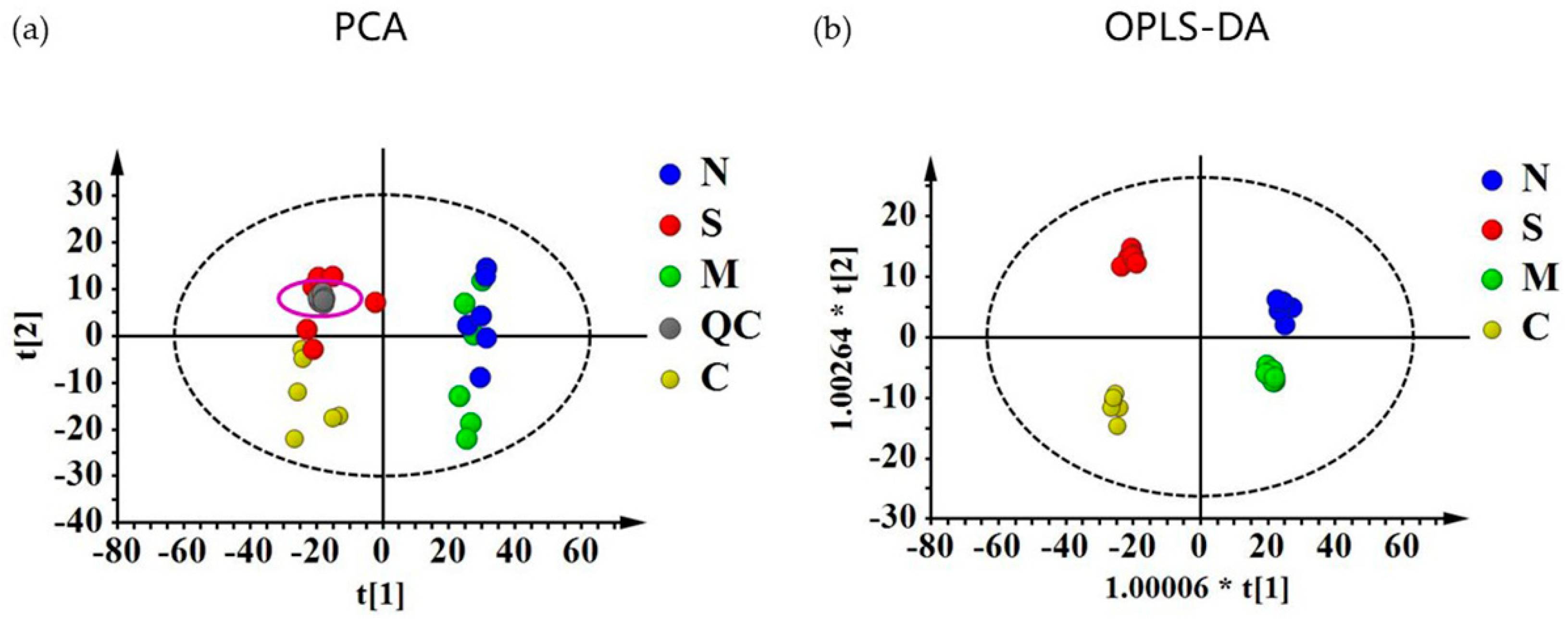
2.3. Quantitative Evaluation of the Effect of NRP-1 Knockdown and SSd by Metabolomics
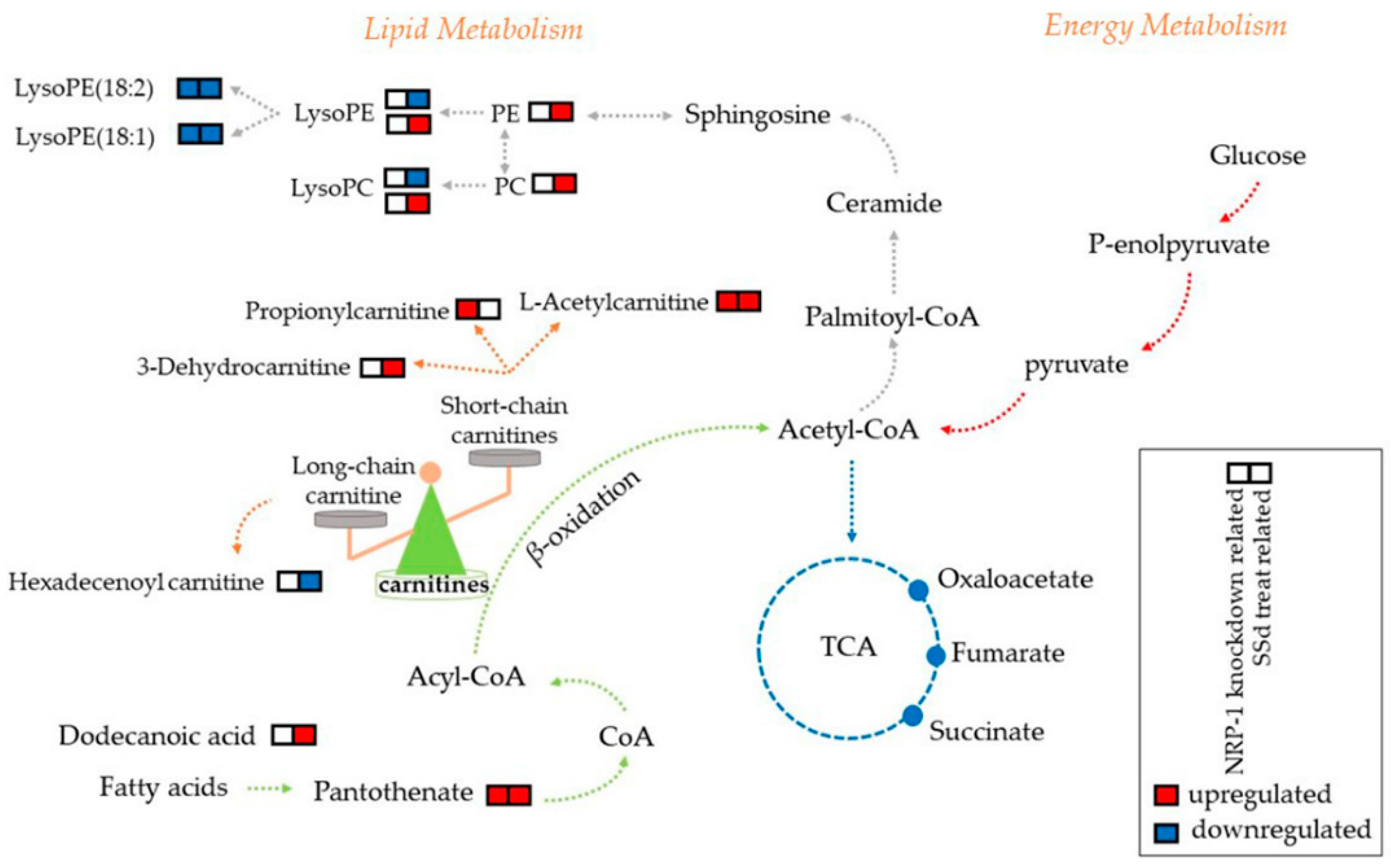
2.4. Carnitines and Phospholipids Were Focalized Based on Metabolomics
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Target Collection and Screening
4.2.1. Target Collection
4.2.2. Target Screening by Molecular Docking
4.3. Verification of the Target on HepG2
4.3.1. Cell Culture
4.3.2. Cell Proliferation Assay
4.3.3. Cell Migration Assay
4.3.4. Western Blot
4.3.5. Knockdown of NRP-1 Using Small Interfering RNA
4.3.6. Effect of SSd on siNRP-1 Cells
4.3.7. Statistical Analysis
4.4. Untargeted Metabolomics Analysis
4.4.1. Sample Collecting and Metabolomic Analysis
4.4.2. Data Preprocessing and Analysis
4.4.3. Quantitative Evaluation of NRP-1 Knockdown and SSd on HepG2 by Metabolites Deregulation Score (MDS)
4.4.4. Differential Metabolites Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dang, S.S.; Wang, B.F.; Cheng, Y.A.; Song, P.; Liu, Z.G.; Li, Z.F. Inhibitory effects of saikosaponin-d on CCl4-induced hepatic fbrogenesis in rats. World J. Gastroenterol. 2007, 13, 557–563. [Google Scholar] [CrossRef]
- Lu, C.N.; Yuan, Z.G.; Zhang, X.L.; Yan, R.; Zhao, Y.Q.; Liao, M.; Chen, J.X. Saikosaponin a and its epimer saikosaponin d exhibit anti-inflammatory activity by suppressing activation of NF-kappaB signaling pathway. Int. Immunopharmacol. 2012, 14, 121–126. [Google Scholar] [CrossRef]
- Wang, B.F.; Dai, Z.J.; Wang, X.J.; Bai, M.H.; Lin, S.; Ma, H.B.; Wang, Y.L.; Song, L.Q.; Ma, X.L.; Zan, Y.; et al. Saikosaponin-d increases the radiosensitivity of smmc-7721 hepatocellular carcinoma cells by adjusting the g0/g1 and g2/m checkpoints of the cell cycle. BMC Complement. Altern. Med. 2013, 13. [Google Scholar] [CrossRef]
- Wong, V.K.; Zhou, H.; Cheung, S.S.; Li, T.; Liu, L. Mechanistic study of saikosaponin-d (Ssd) on suppression of murine T lymphocyte activation. J. Cell. Biochem. 2009, 107, 303–315. [Google Scholar] [CrossRef]
- Li, Z.Y.; Jiang, Y.M.; Liu, Y.M.; Guo, Z.; Shen, S.N.; Liu, X.M.; Pan, R.L. Saikosaponin D acts against corticosterone-induced apoptosis via regulation of mitochondrial GR translocation and a GR-dependent pathway. Prog. Neuropsychopharmacol. Biol. Psychiatry 2014, 53, 80–89. [Google Scholar] [CrossRef]
- Hao, Y.; Piao, X.; Piao, X. Saikosaponin-d inhibits beta-conglycinin induced activation of rat basophilic leukemia-2H3 cells. Int. Immunopharmacol. 2012, 13, 257–263. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, L.; Jin, H.; Shao, J.; Wu, L.; Lu, Y.; Zheng, S. Activation of Fas death receptor pathway and Bid in hepatocytes is involved in saikosaponin D induction of hepatotoxicity. Environ. Toxicol. Pharmacol. 2016, 41, 8–13. [Google Scholar] [CrossRef]
- Chen, M.F.; Huang, S.J.; Huang, C.C.; Liu, P.S.; Lin, K.I.; Liu, C.W.; Hsieh, W.C.; Shiu, L.Y.; Chen, C.H. Saikosaponin d induces cell death through caspase-3-dependent, caspase-3-independent and mitochondrial pathways in mammalian hepatic stellate cells. BMC Cancer 2016, 16, 532. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, F.; Kong, D.; Zhu, X.; Chen, W.; Wang, A.; Zheng, S. Saikosaponin D disrupts platelet-derived growth factor-beta receptor/p38 pathway leading to mitochondrial apoptosis in human LO2 hepatocyte cells: A potential mechanism of hepatotoxicity. Chem. Biol. Interact. 2013, 206, 76–82. [Google Scholar] [CrossRef]
- Ye, H.; Ye, L.; Kang, H.; Zhang, D.; Tao, L.; Tang, K.; Liu, X.; Zhu, R.; Liu, Q.; Chen, Y.Z.; et al. HIT: Linking herbal active ingredients to targets. Nucleic Acids Res. 2011, 39, D1055–D1059. [Google Scholar] [CrossRef]
- Huang, L.; Xie, D.; Yu, Y.; Liu, H.; Shi, Y.; Shi, T.; Wen, C. TCMID 2.0: A comprehensive resource for TCM. Nucleic Acids Res. 2018, 46, D1117–D1120. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef]
- Mathias, D.; Stefan, G.; Jessica, A.; Burghardt, W.; Robert, P. SuperPred: Drug classification and target prediction. Nucleic Acids Res. 2008, 36, 55–59. [Google Scholar] [CrossRef]
- Nikolai, H.; Jessica, A.; Joachim, V.E.; Mathias, D.; Karel, M.; Andreas, E.; Gilson, M.K.; Bourne, P.E.; Robert, P. SuperTarget goes quantitative: Update on drug-target interactions. Nucleic Acids Res. 2012, 40, 1113–1117. [Google Scholar] [CrossRef]
- Liu, T.; Lin, Y.; Wen, X.; Jorissen, R.N.; Gilson, M.K. BindingDB: A web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007, 35, D198–D201. [Google Scholar] [CrossRef]
- Rognan, D. Chemogenomic approaches to rational drug design. Br. J. Pharmacol. 2007, 152, 38–52. [Google Scholar] [CrossRef]
- Pellet-Many, C.; Frankel, P.; Jia, H.; Zachary, I. Neuropilins: Structure, function and role in disease. Biochem. J. 2008, 411, 211–226. [Google Scholar] [CrossRef]
- Zachary, I.C.; Frankel, P.; Evans, I.M.; Pellet-Many, C. The role of neuropilins in cell signalling. Biochem. Soc. Trans. 2009, 37, 1171–1178. [Google Scholar] [CrossRef]
- Fuh, G.; Garcia, K.C.; de Vos, A.M. The interaction of neuropilin-1 with vascular endothelial growth factor and its receptor flt-1. J. Biol. Chem. 2000, 275, 26690–26695. [Google Scholar] [CrossRef]
- Berge, M.; Allanic, D.; Bonnin, P.; de Montrion, C.; Richard, J.; Suc, M.; Boivin, J.F.; Contreres, J.O.; Lockhart, B.P.; Pocard, M.; et al. Neuropilin-1 is upregulated in hepatocellular carcinoma and contributes to tumour growth and vascular remodelling. J. Hepatol. 2011, 55, 866–875. [Google Scholar] [CrossRef]
- Tse, B.W.C.; Volpert, M.; Ratther, E.; Stylianou, N.; Nouri, M.; McGowan, K.; Lehman, M.L.; McPherson, S.J.; Roshan-Moniri, M.; Butler, M.S.; et al. Neuropilin-1 is upregulated in the adaptive response of prostate tumors to androgen-targeted therapies and is prognostic of metastatic progression and patient mortality. Oncogene 2017, 36, 3417–3427. [Google Scholar] [CrossRef]
- Li, L.; Jiang, X.; Zhang, Q.; Dong, X.; Gao, Y.; He, Y.; Qiao, H.; Xie, F.; Xie, X.; Sun, X. Neuropilin-1 is associated with clinicopathology of gastric cancer and contributes to cell proliferation and migration as multifunctional co-receptors. J. Exp. Clin. Cancer Res. 2016, 35, 16. [Google Scholar] [CrossRef]
- Jubb, A.M.; Strickland, L.A.; Liu, S.D.; Mak, J.; Schmidt, M.; Koeppen, H. Neuropilin-1 expression in cancer and development. J. Pathol. 2012, 226, 50–60. [Google Scholar] [CrossRef]
- Casazza, A.; Laoui, D.; Wenes, M.; Rizzolio, S.; Bassani, N.; Mambretti, M.; Deschoemaeker, S.; Van Ginderachter, J.A.; Tamagnone, L.; Mazzone, M. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell 2013, 24, 695–709. [Google Scholar] [CrossRef]
- Tordjman, R.; Lepelletier, Y.; Lemarchandel, V.; Cambot, M.; Gaulard, P.; Hermine, O.; Romeo, P.H. A neuronal receptor, neuropilin-1, is essential for the initiation of the primary immune response. Nat. Immunol. 2002, 3, 477–482. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Connelly, J.; Lindon, J.C.; Holmes, E. Metabonomics: A platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 2002, 1, 153–161. [Google Scholar] [CrossRef]
- Li, Q.; Cheng, T.; Wang, Y.; Bryant, S.H. PubChem as a public resource for drug discovery. Drug Discov. Today 2010, 15, 1052–1057. [Google Scholar] [CrossRef]
- Lu, X.L.; He, S.X.; Ren, M.D.; Wang, Y.L.; Zhang, Y.X.; Liu, E.Q. Chemopreventive effect of saikosaponin-d on diethylinitrosamine-induced hepatocarcinogenesis: Involvement of CCAAT/enhancer binding protein β and cyclooxygenase-2. Mol. Med. Rep. 2012, 5, 637–644. [Google Scholar] [CrossRef]
- Liu, A.; Tanaka, N.; Sun, L.; Guo, B.; Kim, J.H.; Krausz, K.W.; Fang, Z.; Jiang, C.; Yang, J.; Gonzalez, F.J. Saikosaponin d protects against acetaminophen-induced hepatotoxicity by inhibiting NF-κB and STAT3 signaling. Chem. Biol. Int. 2014, 223, 80–86. [Google Scholar] [CrossRef]
- Zu, N.; Li, P.; Li, N.; Choy, P.; Gong, Y. Mechanism of saikosaponin-d in the regulation of rat mesangial cell proliferation and synthesis of extracellular matrix proteins. Biochem. Cell Biol. 2007, 85, 169–174. [Google Scholar] [CrossRef]
- Hsu, Y.L.; Kuo, P.L.; Lin, C.C. The proliferative inhibition and apoptotic mechanism of Saikosaponin D in human non-small cell lung cancer A549 cells. Life Sci. 2004, 75, 1231–1242. [Google Scholar] [CrossRef]
- Lin, X.; Wu, S.; Wang, Q.; Shi, Y.; Liu, G.; Zhi, J.; Wang, F. Saikosaponin-D Reduces H2O2-Induced PC12 Cell Apoptosis by Removing ROS and Blocking MAPK-Dependent Oxidative Damage. Cell. Mol. Neurobiol. 2016, 36, 1–11. [Google Scholar] [CrossRef]
- Fan, J.; Li, X.; Li, P.; Li, N.; Wang, T.; Shen, H.; Siow, Y.; Choy, P.; Gong, Y. Saikosaponin-d attenuates the development of liver fibrosis by preventing hepatocyte injury. Biochem. Cell Biol. 2007, 85, 189–195. [Google Scholar] [CrossRef]
- Chiang, L.C.; Ng LTLiu, L.T.; Shieh, D.E.; Lin, C.C. Cytotoxicity and anti-hepatitis B virus activities of saikosaponins from bupleurum species. Planta Med. 2003, 69, 705–709. [Google Scholar] [CrossRef]
- Hsu, M.J.; Cheng, J.S.; Huang, H.C. Effect of saikosaponin, a triterpene saponin, on apoptosis in lymphocytes: Association with c-myc, p53, and bcl-2 mRNA. Br. J. Pharmacol. 2000, 131, 1285–1293. [Google Scholar] [CrossRef]
- Xu, J.; Xia, J. NRP-1 silencing suppresses hepatocellular carcinoma cell growth in vitro and in vivo. Exp. Ther. Med. 2013, 5, 150–154. [Google Scholar] [CrossRef]
- Pan, Q.; Chanthery, Y.; Liang, W.C.; Stawicki, S.; Mak, J.; Rathore, N.; Tong, R.K.; Kowalski, J.; Yee, S.F.; Pacheco, G.; et al. Blocking neuropilin-1 function has an additive effect with anti-VEGF to inhibit tumor growth. Cancer Cell 2007, 11, 53–67. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, P.; Jiang, Y.; Dou, X.; Yan, J.; Ma, C.; Fan, Q.; Wang, W.; Su, F.; Tang, H.; et al. High Expression of Neuropilin-1 Associates with Unfavorable Clinicopathological Features in Hepatocellular Carcinoma. Pathol. Oncol. Res. 2016, 22, 367–375. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef]
- Luo, M.; Hou, L.; Li, J.; Shao, S.; Huang, S.; Meng, D.; Liu, L.; Feng, L.; Xia, P.; Qin, T.; et al. VEGF/NRP-1axis promotes progression of breast cancer via enhancement of epithelial-mesenchymal transition and activation of NF-kappaB and beta-catenin. Cancer Lett. 2016, 373, 1–11. [Google Scholar] [CrossRef]
- Cui, D.N.; Wang, X.; Chen, J.Q.; Lv, B.; Zhang, P.; Zhang, W.; Zhang, Z.J.; Xu, F.G. Quantitative Evaluation of the Compatibility Effects of Huangqin Decoction on the Treatment of Irinotecan-Induced Gastrointestinal Toxicity Using Untargeted Metabolomics. Front. Pharmacol. 2017, 8, 211. [Google Scholar] [CrossRef]
- Drier, Y.; Sheffer, M.; Domany, E. Pathway-based personalized analysis of cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 6388–6393. [Google Scholar] [CrossRef]
- Huang, S.; Chong, N.; Lewis, N.E.; Jia, W.; Xie, G.; Garmire, L.X. Novel personalized pathway-based metabolomics models reveal key metabolic pathways for breast cancer diagnosis. Genome Med. 2016, 8, 34. [Google Scholar] [CrossRef]
- Liang, Q.; Liu, H.; Zhang, T.; Jiang, Y.; Xing, H.; Zhang, A.-h. Metabolomics-based screening of salivary biomarkers for early diagnosis of Alzheimer’s disease. RSC Adv. 2015, 5, 96074–96079. [Google Scholar] [CrossRef]
- Hoppel, C. The role of carnitine in normal and altered fatty acid metabolism. Am. J. Kidney Dis. 2003, 41, S4–S12. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Q.; Yin, P.; Xing, W.; Wu, Z.; Chen, S.; Lu, X.; Zhang, Y.; Lin, X.; Xu, G. Serum metabolomics reveals the deregulation of fatty acids metabolism in hepatocellular carcinoma and chronic liver diseases. Anal. Bioanal. Chem. 2012, 403, 203–213. [Google Scholar] [CrossRef]
- Lu, Y.; Li, N.; Gao, L.; Xu, Y.J.; Huang, C.; Yu, K.; Ling, Q.; Cheng, Q.; Chen, S.; Zhu, M.; et al. Acetylcarnitine Is a Candidate Diagnostic and Prognostic Biomarker of Hepatocellular Carcinoma. Cancer Res. 2016, 76, 2912–2920. [Google Scholar] [CrossRef]
- Ren, C.; Liu, J.; Zhou, J.; Liang, H.; Wang, Y.; Sun, Y.; Ma, B.; Yin, Y. Lipidomic analysis of serum samples from migraine patients. Lipids Health Dis. 2018, 17, 22. [Google Scholar] [CrossRef]
- Guo, Q.; Zhang, Q.-Q.; Chen, J.-Q.; Zhang, W.; Qiu, H.-C.; Zhang, Z.-J.; Liu, B.-M.; Xu, F.-G. Liver metabolomics study reveals protective function of Phyllanthus urinaria against CCl 4 -induced liver injury. Chin. J. Nat. Med. 2017, 15, 525–533. [Google Scholar] [CrossRef]
- Wang, X.; Cui, D.N.; Dai, X.M.; Wang, J.; Zhang, W.; Zhang, Z.J.; Xu, F.G. HuangQin Decoction Attenuates CPT-11-Induced Gastrointestinal Toxicity by Regulating Bile Acids Metabolism Homeostasis. Front. Pharmacol. 2017, 8, 156. [Google Scholar] [CrossRef]
- Zhang, M.-H.; Chen, J.-Q.; Guo, H.-M.; Li, R.-T.; Gao, Y.-Q.; Tian, Y.; Zhang, Z.-J.; Huang, Y. Combination of LC/MS and GC/MS based metabolomics to study the hepatotoxic effect of realgar nanoparticles in rats. Chin. J. Nat. Med. 2017, 15, 684–694. [Google Scholar] [CrossRef]
- Wong, V.K.W.; Molly Miao, Z.; Hua, Z.; Lam, K.Y.C.; Po Ling, C.; Law, C.K.M.; Yue, P.Y.K.; Liang, L. Saikosaponin-d Enhances the Anticancer Potency of TNF-α via Overcoming Its Undesirable Response of Activating NF-Kappa B Signalling in Cancer Cells. Evid.-Based Complement. Altern. Med. 2013, 745295. [Google Scholar] [CrossRef]
- He, S.; Lu, G.; Hou, H.; Zhao, Z.; Zhu, Z.; Lu, X.; Chen, J.; Wang, Z. Saikosaponin-d suppresses the expression of cyclooxygenase-2 through the phospho-signal transducer and activator of transcription 3/hypoxia-inducible factor-1α pathway in hepatocellular carcinoma cells. Mol. Med. Rep. 2014, 10, 2556. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

| Number | PDB ID | Gene Name | Uniprot ID | Target Name | Docking Score | Validated or Not |
|---|---|---|---|---|---|---|
| 1 | 5F19 | PTGS2 | P35354 | Prostaglandin G/H synthase 2 | −6.67 | Validated [28] |
| 2 | 2QQI | NRP1 | O14786 | Neuropilin−1 | −6.469 | Unvalidated |
| 3 | 1NFI | RELA | Q04206 | Transcription factor p65 | −5.585 | Validated [1] |
| 4 | 2W96 | CDK4 | P11802 | Cell division protein kinase 4 | −5.577 | Validated [30] |
| 5 | 1IKN | NFKBIA | P25963 | NF-kappa-B inhibitor alpha | −5.346 | Validated [1] |
| 6 | 5JFD | F2 | P00734 | Prothrombin | −5.325 | Unvalidated |
| 7 | 4AGN | P53 | P04637 | Cellular tumor antigen p53 | −5.205 | Validated [31] |
| 8 | 4MAN | BCL-2 | P10415 | Apoptosis regulator Bcl-2 | −4.857 | Validated [7] |
| 9 | 5TBE | MAPK14 | Q16539 | Mitogen-activated protein kinase 14 | −4.792 | Validated [32] |
| 10 | 1IL6 | IL6 | P05231 | Interleukin-6 | −4.666 | Validated [1] |
| 11 | 3V3K | CASP9 | P55211 | Caspase-9 | −4.49 | Validated [8] |
| 12 | 4PRY | CASP3 | P42574 | Caspase-3 | −4.443 | Validated [8] |
| 13 | 5T46 | EIF4G1 | Q04637 | Eukaryotic translation initiation factor 4 | −4.414 | Unvalidated |
| 14 | 4LXO | FN1 | P02751 | Fibronectin | −4.303 | Unvalidated |
| 15 | 2DBF | NFKB1 | P19838 | Nuclear factor NF-kappa-B p105 | −3.902 | Unvalidated |
| 16 | 5FF0 | TGFB1 | P01137 | Transforming growth factor beta-1 | −3.642 | Validated [33] |
| 17 | 5T01 | JUN | P05412 | Transcription factor AP-1 | −3.611 | Validated [32] |
| 18 | 4S0O | BAX | Q07812 | Apoptosis regulator BAX | −3.412 | Validated [8] |
| 19 | 4FDL | CASP7 | P55210 | Caspase-7 | −3.328 | Validated [34] |
| 20 | 5I4Z | MYC | P01106 | Myc proto-oncogene protein | −2.997 | Validated [35] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, Y.; Hou, X.; Zhang, Q.; Li, R.; Xu, L.; Chen, Y.; Tian, Y.; Sun, R.; Zhang, Z.; Xu, F. Untargeted Metabolomics Study of the In Vitro Anti-Hepatoma Effect of Saikosaponin d in Combination with NRP-1 Knockdown. Molecules 2019, 24, 1423. https://doi.org/10.3390/molecules24071423
Lv Y, Hou X, Zhang Q, Li R, Xu L, Chen Y, Tian Y, Sun R, Zhang Z, Xu F. Untargeted Metabolomics Study of the In Vitro Anti-Hepatoma Effect of Saikosaponin d in Combination with NRP-1 Knockdown. Molecules. 2019; 24(7):1423. https://doi.org/10.3390/molecules24071423
Chicago/Turabian StyleLv, Yingtong, Xiaoying Hou, Qianqian Zhang, Ruiting Li, Lei Xu, Yadong Chen, Yuan Tian, Rong Sun, Zunjian Zhang, and Fengguo Xu. 2019. "Untargeted Metabolomics Study of the In Vitro Anti-Hepatoma Effect of Saikosaponin d in Combination with NRP-1 Knockdown" Molecules 24, no. 7: 1423. https://doi.org/10.3390/molecules24071423
APA StyleLv, Y., Hou, X., Zhang, Q., Li, R., Xu, L., Chen, Y., Tian, Y., Sun, R., Zhang, Z., & Xu, F. (2019). Untargeted Metabolomics Study of the In Vitro Anti-Hepatoma Effect of Saikosaponin d in Combination with NRP-1 Knockdown. Molecules, 24(7), 1423. https://doi.org/10.3390/molecules24071423





