Abstract
Metabotropic glutamate (mGlu) receptors are a family of eight GPCRs that are attractive drug discovery targets to modulate glutamate action and response. Here we review the application of computational methods to the study of this family of receptors. X-ray structures of the extracellular and 7-transmembrane domains have played an important role to enable structure-based modeling approaches, whilst we also discuss the successful application of ligand-based methods. We summarize the literature and highlight the areas where modeling and experiment have delivered important understanding for mGlu receptor drug discovery. Finally, we offer suggestions of future areas of opportunity for computational work.
1. Introduction
Glutamate is the major excitatory neurotransmitter in the central nervous system (CNS) and modulates synaptic responses by activating ionotropic glutamate (iGlu) receptors and G protein-coupled receptors (GPCRs) termed metabotropic glutamate (mGlu) receptors. GPCRs are the largest class of membrane proteins in the human genome and are crucial for cell signaling [1]. The metabotropic glutamate (mGlu) receptors are family C GPCRs that participate in the modulation of synaptic transmission and neuronal excitability throughout the CNS. To date, eight mGlu receptor subtypes and multiple splice variants have been cloned and classified into three groups based upon sequence homology, pharmacological profile and preferential signal transduction pathways [2]: Group I mGlu receptors (mGlu1 and mGlu5), group II (mGlu2 and mGlu3) and group III receptors (mGlu4, mGlu6, mGlu7 and mGlu8). They have emerged as interesting drug targets [3] and pharmaceutical research has primarily focused on the pharmacology and development of ligands acting at groups I and II, whereas group III receptors have started to attract attention more recently (Figure 1) [4].
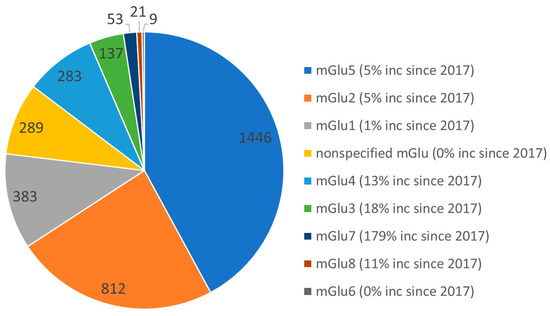
Figure 1.
Pie chart showing reported ligands for mGlu receptors in the Thomson Reuters Integrity database. The most explored are mGlu5, mGlu2, and mGlu1. Extracted on 29 January 2019 and compared with results from March 2017.
Group I receptors, mGlu1 and mGlu5, may be useful targets for psychiatric disorders via their indirect modulation of NMDA receptors [5]. They are coupled to protein kinase C (PKC) via Gq protein and are ubiquitously expressed on postsynaptic excitatory terminals in limbic brain regions that are involved in motivational, emotional and cognitive processes [6]. For instance, mGlu5 and NMDA receptors are co-expressed at GABAergic interneurons, and are not only connected via intracellular signaling pathways but also physically through scaffolding proteins [7,8]. These receptors may have the potential for therapeutic intervention of schizophrenia. Meanwhile, group II mGlu receptors reduce transmission at glutamatergic synapses in brain regions such as the prefrontal cortex and hippocampus where excessive glutamatergic transmission may be implicated in the pathophysiology of anxiety and schizophrenia [9]. It is therefore hypothesized that activation of group II mGlu receptors, mGlu2 and mGlu3, may provide anxiolytic and/or antipsychotic effects [5]. Finally, the group III receptors, mGlu4, mGlu6, mGlu7 and mGlu8, are much less explored but offer huge potential. Ligands of mGlu4 for example have recently shown to be neuroprotective and beneficial in animal models of Parkinson’s disease, pain, and anxiety [10]. Furthermore, ligands of mGlu7 and mGlu8 receptors have possible indications in neurodevelopmental disorders and anxiety, post-traumatic stress, Parkinson’s, and multiple sclerosis [11,12].
The mGlu receptors form cysteine linked dimers. Their structures include a characteristic Venus fly-trap (VFT) extracellular agonist-binding domain, a 7-transmembrane (7TM) domain, and a cysteine rich domain (CRD) that connects the two (Figure 2) [13]. The first structures of a full length mGlu receptor were recently deposited in the protein databank (PDB id 6N51 and 6N52). Solved by electron microscopy they reveal how activation proceeds by compacting the intersubunit VFT dimer interface, bringing the CRDs into proximity, and in-turn enabling the 7TM domains to reposition closer together to initiate signaling [14]. The extracellular loop 2 of the 7TM makes crucial interactions with the CRD. This is a huge breakthrough for the understanding of mGlu receptor function. Future work will likely improve the resolution, and study local interactions and conformational changes, helping to understand the 7TM movements and interactions that occur with activation. Previously, the VFT and 7TM domains have been solved by X-ray crystallography. The crystal structures for the extracellular VFT domains of mGlu’s 1, 2, 3, 5, 7 and 8 reveal the closed agonist-bound active conformations as well as resting and inactive conformations [15]. Meanwhile, the crystal structures of the 7TM domain of mGlu1 and mGlu5 have shown the binding site of allosteric ligands in an analogous position to the orthosteric pocket of the class A receptors. The computational study of class C GPCRs has benefited greatly from the publication of the mGlu1 and mGlu5 7TM crystal structures in 2014 [16,17]. Despite their similarity, the mGlu5 structure was crystallized with a NAM (mavoglurant) binding in a deeper pocket compared to the NAM (FITM) bound to mGlu1 (Figure 2). Two more recent studies provided more mGlu5 7TM X-ray crystallographic information revealing subtle differences in binding modes for different NAMs [18,19].
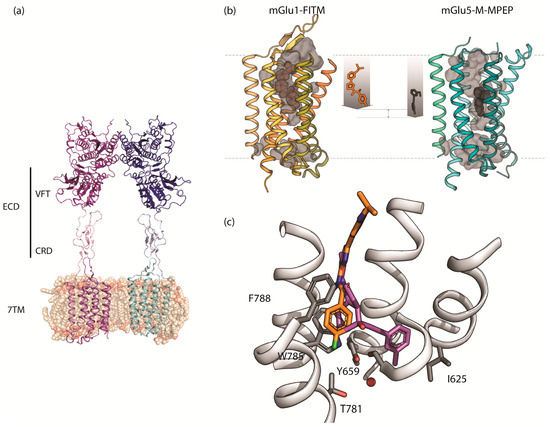
Figure 2.
The structure of mGlu receptors: (a) Structure of the full length apo mGlu5 receptor homodimer showing the VFT, CRD, and 7TM domains (PDB 6N52); (b) X-ray structures of mGlu1 and mGlu5 receptor 7TMs showing the different depth of binding for NAMs. (c) Close up view of the X-ray structures of the 7TM domains showing the overlay of NAMs FITM (orange) and mavoglurant (magenta) bound at mGlu1 and mGlu5 receptors with several amino acids labelled.
As these structures suggest, mGlu receptor drug discovery has focused on targeting both orthosteric and allosteric ligands. Ligands targeting the orthosteric site are often amino-acid like, but reports have shown that group II mGlu receptor ligands can achieve brain penetration and selectivity versus other members of the mGlu family [20]. Despite this, the mGlu receptor family has seen much effort dedicated to allosteric drug discovery which provides ways to modulate GPCRs for potential therapeutic benefit [21]. Allosteric modulators of mGlu receptors avoid amino-acid (glutamate-like) chemistry that can be difficult for brain penetration. The allosteric site is less conserved, helping the identification of selective ligands. Furthermore, allosteric modulators typically only modify receptor response in the presence of the endogenous ligand thus avoiding receptor desensitization or other unwanted-effects of agonist action. Allosteric modulators are classified as either positive allosteric modulators (PAMs) or negative allosteric modulators (NAMs) that either enhance or inhibit the response to glutamate respectively. Silent allosteric modulators (SAMs) are ligands that occupy the allosteric pocket but do not modify activity.
Given the substantial interest in this target class, but the challenges for structure-based drug design, the mGlu receptor field has seen a variety of alternative computational approaches to aid drug discovery. Computationally assisted drug design can offer many varied techniques useful in lead discovery [22], virtual screening (VS) [23] and in lead optimization [24,25,26]. In this manuscript we review the reports of in silico methods applied to this GPCR subfamily. It is found that a wide range of methods have been used, from applying pharmacophores and other tools for VS, modeling and docking to understand binding modes, to molecular dynamics (MD) to understand origins of functional effect of allosteric modulation. Thus, this target family has spurred diverse and creative approaches that can be of interest for future drug discovery target families.
2. Group I mGlu Receptors
2.1. mGlu1 Receptor
The mGlu1 receptor is an attractive target for several psychiatric diseases and neurological disorders [27]. Research on its structure has also benefited the understanding of the features needed to selectively target both orthosteric and allosteric binding sites of this receptor. Many early mutagenesis studies using site-directed mutagenesis, truncations and chimeras of the receptor obtained details on mGlu1 functional behavior. Some papers described the role of C-terminal residues in protein coupling [28] or the ones involved in dimerization [29], but most efforts focused on elucidating the important residues for ligand recognition [30,31,32,33,34,35,36,37].
In the late 1990s the first mGlu1 selective non-competitive antagonists were described, suggesting the existence of an alternative binding site for these compounds. Several mutagenesis studies began to propose mGlu1 residues involved in the recognition of these non-competitive antagonists, such as CPCCOEt (1, Figure 3), suggesting an overlapping binding site in the 7TM domain of the receptor [30,31]. In 2003, Malherbe et al. built a computational homology model of the receptor based on the available bovine rhodopsin crystal structure which allowed to specifically mutate binding pocket facing sidechains and pinpoint a cluster of aromatic residues in TM6 as important for mGlu1 NAM activity [32]. These residues coincided with the non-orthosteric antagonistic binding site noted in the previous mutagenesis studies.
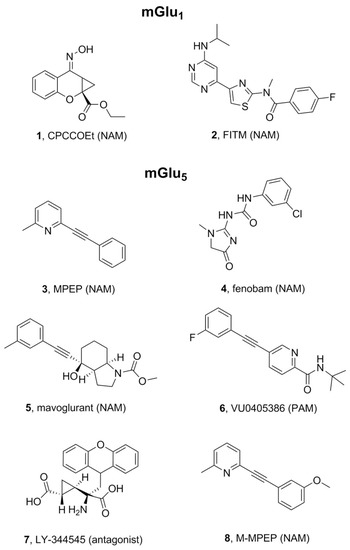
Figure 3.
Chemical structures of selected ligands for mGlu1 and mGlu5 receptors.
In the absence of a solved crystal structure of the full receptor or 7TM, scientists sought other approaches to understand the compound selectivity of mGlu1 receptor antagonists such as the VS performed in 2006 by Noeske et al [38]. They used self-organizing map (SOM) clustering algorithms and a topological pharmacophore descriptor to predict the potential activities of some known NAMs. With this VS strategy, they were able to discriminate between antagonists of mGlu1 and mGlu5 useful for early recognition of potential selectivity concerns in lead discovery. In that same year, researchers performed a docking study using the binding site domain (orthosteric) crystal structure of the mGlu1 receptor. They generated homology models for ECDs of other class C receptors and observed that their VS approach was able to recognize the best ligands in most cases, with a combination of different docking and scoring functions being required for optimal results [39].
A similar homology model of mGlu1 was used by Merz Pharmaceuticals’ scientists in studies from 2007 and 2008. The first was a ligand-based VS study, in which they analyzed the binding mode of two representative mGlu1 NAM hits describing potential interaction patterns [38]. In Vanejevs et al. the model was used for the SAR interpretation of novel modulators [40]. They developed a pharmacophoric hypothesis for mGlu1 antagonists based on previously described ligands and used this to perform VS of a large compound database. Lead optimization on the hit scaffold was carried-out, and a small library of compounds was docked into the homology model to predict the binding mode and help SAR understanding.
In 2014, the release of the mGlu1 7TM crystal structure bound to NAM FITM (2, Figure 3) [17] increased the scope of modeling opportunities. In 2015 Harpsøe et al. performed an extensive computational analysis of all the mGlu receptors, merging information on experimental mutagenesis with the new insights from the crystal structures to predict binding modes and discuss common interactions [41]. The work points out several residue hotspots and characteristic features within the allosteric binding pocket of the different mGlu receptors that can explain NAM subtype selectivity. This information has since been incorporated into the GPCR database [42]. With the new crystal structure information more reports began to emerge. For instance, Jiang et al. used pharmacophore models based on known mGlu1 NAM ligands to VS a database of Chinese herbs. The hit compounds were then docked into the mGlu1 crystal structure suggesting new possible allosteric ligands, although this was not confirmed experimentally [43].
A hierarchical VS approach was employed to identify new micromolar mGlu1 receptor NAMs [44]. A homology model of mGlu1 was built based on the D3 receptor and refined using MD simulations. Known mGlu1 antagonists were then used to build a pharmacophore model, as well as a recursive partitioning model, a Bayesian model, and a support vector machine (SVM) model. VS was initially performed using a pharmacophore guided structure-based approach, then hits were classified as likely mGlu1 actives with the remaining models. After applying this approach to commercially available compounds new active mGlu1 allosteric modulators were identified. Retrospective docking validation of the homology model with the released mGlu1 crystal suggested the predicted binding mode of the hits obtained.
Also in 2015, the mGlu1 receptor 7TM crystal structure was used as a test case for studying the functional group affinity patterns for its occluded allosteric binding site [45]. The site identification by ligand competitive saturation (SILCS) methodology uses fragment sampling to map free energy affinity patterns of functional groups at protein surfaces. MD of mGlu1 in a membrane and with different solute fragments was performed and the affinity patterns were obtained based on probability analyses. The method revealed the largely hydrophobic nature of the mGlu1 allosteric binding site consistent with the lipophilic allosteric ligands [46].
Bai et al. combined three computational methods to study the mGlu1 receptor [47]. They performed MD simulations of the receptor wild type and T815M and T805A mutants. Weak interaction analysis and free energy calculations based on the outcome of the simulations provided insights into the dimeric packing and allosteric regulation mechanism of the receptor. Free energy calculations showed that residues T748, C746, K811 and S735 form the secondary binding pocket, where FITM binds before escaping the receptor, suggesting these amino acids are relevant for the dynamic binding of this NAM.
Recently in 2018, quantum mechanical methods were used to study the binding site of FITM in the mGlu1 7TM domain and compare it with twelve close analogs covering a broad range of affinity [48]. The binding site geometries were optimized using the “our own N-layered integrated molecular orbital and molecular mechanics” (ONIOM) method along with the fragment molecular orbital method with energy decomposition analysis (FMO-EDA). The results showed that residues Q6603.28 and/or Y8056.55 were anchor points for all the FITM analogs, while the H-bond with T8157.38 determined only the orientation of very active molecules containing an amino substituent in the pyrimidine moiety (e.g., FITM). The orientation of the other parts of ligands resulted from hydrophobic interactions mainly with L7575.44, F8016.51, or W7986.48. The applied ONIOM/FMO–EDA approach facilitated the study of effects related to very small changes in the ligand structure and led to conclusions regarding the significance of individual interactions in the allosteric binding pocket of mGlu1.
2.2. mGlu5 Receptor
Due to its interest for depression [49], migraine [50], fragile X-syndrome [51] and many other CNS disorders, there has been much effort to understand the mechanism of action and ligand interactions of this receptor.
Since the early 2000s, site-directed mutagenesis has been used frequently to characterize the allosteric binding pocket of the mGlu5 receptor (Figure 4) [52,53,54,55]. However, the mutagenesis data was best interpreted by performing homology modelling. First studies used bovine rhodopsin as the template since it was the only GPCR crystal structure available [32,52]. Later, class A 7TM GPCR structures were used instead, such as the β2 adrenergic receptor or the A2A adenosine receptor [54,56]. These early homology models together with the mutagenesis data were used to hypothesize the binding modes of the mGlu5 ligands. Manual or computed docking of the ligands were carried out for ligand-protein interactions analysis [31]. In this sense, Gregory et al. wanted to understand the molecular determinants dictating allosteric modulator interactions in mGlu5 [57]. Site-directed mutagenesis was performed obtaining six main residues as key for ligand affinity. Many diverse allosteric ligands were docked into their homology model receptor based on β2 adrenergic receptor crystal. They observed a clear overlap in binding modes of the compounds confirming the location of the allosteric pocket in the 7TM domain of the receptor. Overall, the 7TM allosteric binding site defined by experimental mutagenesis and computational studies was later verified by the 7TM crystal structures (Figure 4).
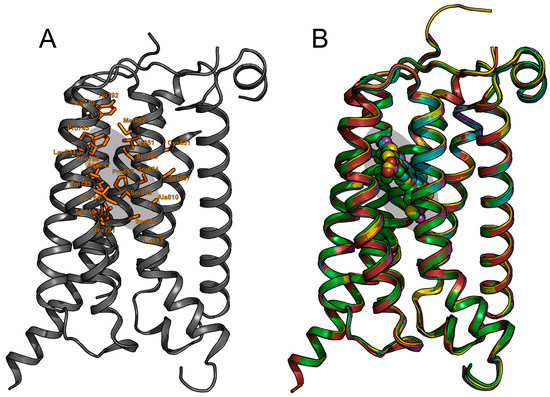
Figure 4.
(a) The mGlu5 7TM showing amino acids identified from experimental mutagenesis as important for allosteric modulator activity, compared with (b) the overlap of the subsequent X-ray crystal structures of multiple mGlu5 receptor NAMs.
Pharmacophore modeling was another computational tool used in the field to identify new allosteric modulators of both mGlu1 and mGlu5 receptors. Coupling of mutagenesis data with a 3D pharmacophoric model of the receptor was performed to elucidate the binding mode of MPEP (3, Figure 3) and fenobam (4, Figure 3) [58]. As mentioned in the mGlu1 section, scientists from Merz Pharmaceuticals built a pharmacophore model for mGlu1 allosteric modulators based on previously described ligands [40,59]. Given the high structural similarity between mGlu1 and mGlu5 allosteric pockets, this model was also used for the design of mGlu5 allosteric modulators. The pharmacophore model was used to screen a wide library of compounds and hit optimization led to highly potent mGlu5 NAMs and PAMs.
Given the difficulties to perform structure-based drug design for mGlu receptors it is understandable that many computational approaches instead relied upon ligand-based methods, especially machine learning methods for discovery of new compounds. One of the first uses of machine learning algorithms in this field was performed by Renner et al. in 2007. They used an artificial neural network (ANN) combined with a 3D pharmacophoric model for discovery of new NAMs of mGlu5. The VS resulted in the identification of three potent mGlu5-specific hits [60]. Later in 2010, a comparative molecular field analysis (CoMFA) 3D-QSAR approach was applied to a set of mGlu5 PAMs [61]. Mueller et al. [62] tried to overcome QSAR limitations by implementing artificial neural networks (ANN) approaches for the identification of new potential mGlu5 modulators. High throughput screening data were used to train an ANN model to predict mGlu5 NAM activity. The model was then applied to identify new NAM chemotypes by VS. The hits led to the identification of a series of allosteric modulators. Further work involved synthesizing MPEP analogues, then docking them into a homology model to understand their selectivity. An mGlu5 receptor model was used based on both β2 adrenergic receptor and A2A receptor crystal structures, validated by previous mutagenesis studies. Docking the compound of interest into the model suggested its binding mode, indicating the relevance of certain residues such as N747, and S809 for NAM binding [63].
In 2014 Dalton et al. built an active state of the 7TM domain by homology modelling. Multiple templates such as β2 adrenergic receptor, sphingosine 1-phosphate receptor 1, dopamine D3 receptor and neurotensin receptor were used for the construction of both the inactive and active state models. Four NAMs and four PAMs previously described in other studies were docked into their respective receptor state model using an induced-fit method. PAM and NAM binding modes were compared and led to identification of amino acids S809, W785, T781 and Y659 as key residues [64].
The 7TM crystal structure of mGlu5 was published in 2014 and proved a turning point for mGlu5 targeted drug design. The previously obtained mutagenesis data and SAR studies could be mapped, (Figure 4). For instance, the above-mentioned study from Harpsøe et al. [41] merged large amounts of experimentally available mutagenesis data with a docking study of several mGlu NAM ligands. Despite many similarities between mGlu1 and mGlu5 structures, their allosteric pockets reveal the determinants of ligand selectivity. A key selectivity residue is the amino acid on TM3 at position 40: 3.40. In mGlu1, S6683.40 together with Thr8157.33 appear to induce selectivity for this receptor by H-bonding to the ligands. On the other hand, mGlu5 has a proline residue at the 3.40 position. The proline occupies less space compared to the bulkier side chains at this position in the other mGlu receptors allowing ligands to pass and bind deeper in mGlu5 (Figure 2). This hypothesis is supported by mutagenesis evidence in which mutations such as Pro655Phe, result in abolishment of NAM effects [54]. The comparison of the first crystal structures of mGlu5 [16] versus mGlu1 [17] confirmed this, all available mGlu receptor 7TM domain structures are summarized in Table 1. Later, Harpsøe et al. used the mGlu5 structure bound to mavoglurant (5, Figure 3) to place ligands in the binding site and establish that subtype selectivity is achieved by reaching the deep pocket of the receptor, and it can be further improved by interacting with S805 [41].

Table 1.
Summary of crystal structures for the 7TM domain of mGlu receptors.
Along the same lines, a medicinal chemistry study by Anighoro et al. [65] used an homology model of mGlu5 based on the crystal structure of mGlu1. Docking of MPEP and fenobam (mGlu5 NAMs) was performed using an induced fit approach. MD simulations of these complexes were performed for refinement of the predicted binding modes, which were confirmed by previous mutagenesis data. The release of the mGlu5 receptor enabled definitive validation of the binding modes described in the work by docking the NAM compounds. They synthesized a new series of NAMs, docked them to the receptor structure and extracted the binding modes and SAR. The obtained insights on interaction modes of NAMs could be useful for the future design of high affinity mGlu5 modulators.
Feng et al. used the mGlu1 and mGlu5 crystal structures to model the whole family of 1–8 7TM domains and studied the binding modes of multiple allosteric ligands [66]. They compared the different interactions of PAMs and NAMs. They went on to construct a model of a full length mGlu5 receptor (amino acids 26 to 832). However, according to the report, it was only modeled in the monomeric form. This construct was submitted to a 100 ns MD simulation in the presence of an orthosteric antagonist-LY-344545 (6, Figure 3) and the mavoglurant NAM, as well as bound to the agonist glutamate and PAM-VU0405386 (7, Figure 3). They reported large conformational changes of glutamate bound within the orthosteric binding site as well as in the CRD when compared with the simulations performed using the antagonist + NAM. Within the 7TM they observed substantial differences in the behavior of the ionic-lock on the intracellular side of the receptor when comparing the antagonist+NAM with the agonist+PAM simulations. They also witnessed the intracellular opening of TM5 in the presence of PAM and suggested this may facilitate activation.
In 2017 the mGlu5 crystal structure was used to model MPEP activity versus mGlu4 and mGlu5 [67]. The results showed different interaction modes of this ligand with the receptors, which they argued can explain the opposite functionality of this molecule in the different contexts. Dalton et al. suggested two different ligand-receptor interactions given by H-bonding of MPEP with TM3 residues in mGlu4 or with TM7 residues in mGlu5. They also consider the depth of the allosteric pocket a key determinant for the binding modes difference, since 6362.49 residue in mGlu4 receptor is a less bulky G6282.49 in mGlu5 thus allowing the ligand to bind deeper into the allosteric pocket. The authors also proposed the occupancy of the binding pocket by a lipid when no modulator is present. Also in 2017, Bian et al. performed a purely in silico study to generate potential allosteric mGlu5 receptor ligands [68]. They fragmented a database of existing allosteric modulators and used these fragments to recombine and generate new ligands. Docking and scoring identified the top-ranking new molecules which were suggested to be potential new mGlu5 modulators. Later in 2018, Fu et al. [69] performed a computational study of the pharmacophoric binding of several mGlu5 NAMs identifying important amino acids for binding in the allosteric site. They used induced fit docking and MD approaches for their objective, and their results were supported by mutagenesis data.
In 2018, two more crystallographic structures of NAMs, M-MPEP (8, Figure 3) and fenobam, bound to the mGlu5 receptor 7TM domain were solved by Christopher et al [19]. Along with revealing the new structures they also performed a computational study of the role of water in the activity of mGlu5 NAMs. The crystal structures have already been used for some computational studies such as VS using a pharmacophoric model [70]. The pharmacophore was validated by induced-fit docking of the hit compounds, and ensuing MD simulations were performed for analysis of the specific ligand-receptor interactions. The same authors also performed a QSAR study of a thieno[2–b]pyridines series of mGlu5 receptor NAMs [71]. They first built an all-atom QSAR model from the pharmacophore hypothesis of this list of compounds. The model was used for induced-fit docking and binding free energy calculations, MD of the complex were run for stability verification. Again, the results of this paper are thought to contribute useful knowledge for future NAM drug design studies.
A recent study demonstrated for the first time the origin of the functional effect of NAMs and PAMs of the mGlu5 receptor, by studying close analogue molecular switches. A series of MD simulations elucidated the mechanisms by which minor changes in ligand scaffold can elicit completely opposite functional activity [72]. These molecular switches have only been described for mGlu5. An active-like model of the receptor was built using the mGlu5 7TM crystal structure and the β2-Gs active-state complex (PDB ID 3SN6). The Gq protein was included in the model to obtain a fully active-like receptor state. By simulating NAMs bound to the inactive 7TM mGlu5 receptor crystal structures, and PAMs bound to the active-like model of mGlu5 three residues were identified to be crucial for the allosteric activation mechanism of the receptor: S658, Y659 and T781. The hypothesis is consistent with previously described residues in class A and class C receptors that are involved in the activation process, and their relevance is also supported by former mutagenesis studies. Additionally, water analysis of the systems enabled the identification of a key water molecule for stabilization of the inactive state of the receptor. The results can be extrapolated to additional mGlu5 NAM and PAM ligands, to understand how they induce their functional effect. This work contributes to better understanding of the mGlu5 allosterism and activation mechanisms and can be of great help for future drug design studies.
3. Group II mGlu Receptors
3.1. mGlu2 Receptor
Given its links to various neuropsychiatric disorders the mGlu2 receptor has been amongst the most studied from a medicinal chemistry point of view (Figure 1) [73,74,75,76,77,78]. Early computational modeling work involved generating mGlu2 receptor models and docking to support and explain site directed mutagenesis studies. This was particularly relevant to understand the mode of action and identify binding sites for allosteric modulators.
In 2009 and 2011 scientists from Roche (Basel, Switzerland) reported detailed mutagenesis studies of both the orthosteric and allosteric binding sites [79,80]. They modeled an antagonist binding mode in the closed inactive form of the mGlu2 VFT as well as the binding of two NAMs bound in the 7TM. The binding modes could explain the mutant observations but the 7TM modeling preceded the release of the mGlu1 and mGlu5 crystal structures. Instead, human β2 adrenoceptor was used as a structural template which may have led to the binding site being more extracellular facing than later revealed for mGlu receptors.
Subsequently, Janssen (Beerse, Belgium) scientists have also reported detailed experimental mutagenesis studies of mGlu2 PAMs and NAMs that were driven with computational modeling. In the period from 2015 to 2018 this group published several studies that confirmed an overlapping binding site and similar interactions amongst PAMs, and amongst NAMs, but with some interactions being unique to each set [81,82]. These extensive studies involved as many as 12 ligands and 39 mutants. Along with binding displacement studies, and deep analysis of structure activity relationships, this led to relatively confident predictions of the binding mode of the mGlu2 allosteric modulators. Furthermore, the binding hypothesis was confirmed by the computational targeted design of an mGlu2 allosteric covalent ligand [83].
Building upon this relatively solid understanding of the binding mode they went on to consider how the PAMs and NAMs achieve their functional effect. Classical MD simulations were performed for µs timescales using repeats and multiple ligands. It was consistently demonstrated that PAMs and NAMs induce different conformational behavior of amino acids in their direct vicinity (so-called trigger switch), that in turn modifies the conformational behavior of amino acids one turn below in the known transmission switch (Figure 5) [82,84,85]. The amino acids involved in the transmission switch correspond to the same functionally important positions known from class A GPCRs. Based on the modeling they went on to perform additional mutation of amino acids in the transmission switch and confirmed them to be crucial even for functional activity of glutamate alone. Thus, the hypothesis emerges whereby NAMs can block movements in this region, whereas PAMs promote sidechain movement, particularly of amino acids such as threonine 769 in transmembrane helix 6 (T7696.44a.46c) that can induce kinking in helices.
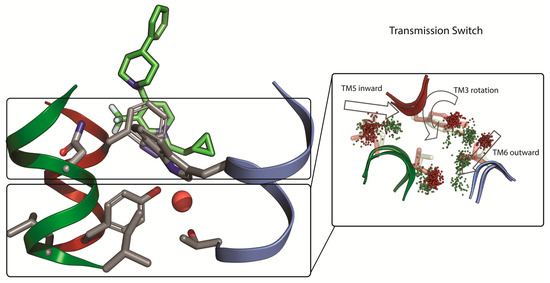
Figure 5.
Origin of the functional effect of mGlu2 PAMs and mGlu2 NAMs. A combined experimental and computational study by Perez-Benito et al. [82] showed that PAMs and NAMs bind above the transmission switch but transmit a different conformational effect. Based on the modelling, amino acids in the transmission switch were proposed for experimental mutagenesis that confirmed their importance for receptor activation. Thus, validating a hypothesis of local allosteric modulator induced conformational changes that promote or block movements of amino acids responsible for functional activity, overall confirming the 7TM analogy with class A GPCRs.
Another interesting modeling study used MD approaches to demonstrate cross talk between a heterodimer of mGlu2 and 5HT2A receptors. Bruno et al. constructed the model and performed classical MD simulations that could explain the allosteric effect of mGlu2 receptor on 5HT2A mediated response [86]. As this study predates the 7TM crystal structures of mGlu1 and mGlu5 receptors, the authors had to use the human β2 adrenergic receptor to model 5HT2A and bovine rhodopsin as a template for mGlu2. Nevertheless, the work suggested the TM4/TM5 interface was likely stable and preferred for initiating the allosteric effect between receptors. Another MD study investigated the conformational behavior of 7TM helix 8 in the mGlu2 receptor [87]. Little is known about the role of helix 8 in class C GPCRs. Extended MD simulations indicated that helix 8 adopts membrane-sensitive conformational states and is dependent on interaction with cholesterol, in short, the more cholesterol present the more stabilized the helix 8 structure.
In 2014 computational modeling of the mGlu2 receptor was used to assist an important study investigating the inter-monomeric rotation during activation of the mGlu2 receptor dimer [88]. Computational models of inactive and active dimeric complexes were built that were able to assist the definition of the cross-linking experiments and aid analysis. The modeling helped to reveal that TM4 and TM5 are mainly cross-linked in the inactive state, whilst TM6 constitutes the interface of the active dimer.
In 2010 scientists from Janssen applied computational ligand-based approaches to help identify new mGlu2 PAMs [89]. They showed how the ligands of several different mGlu2 PAM chemical series shared a common alignment and features (Figure 6). A database of virtual fragments was generated and searched using shape and electrostatic similarity methods to identify alternative scaffolds that mimic the steric hindrance and features but with alternative underlying chemical connectivity. This scaffold-hopping approach led to modification of a pyridone series to a novel imidazopyridine scaffold. Rapid decoration of the new scaffold resulted in ~100 nM mGlu2 PAMs amongst the first synthesized examples and subsequent optimization led to further potent advanced leads [75,84,85,90,91]. This application shows the power of relatively simple, conventional pharmacophore-based approaches to guide medicinal chemistry. More recently a similar approach was applied by Szabo et al [92]. They screened small fragments for allosteric activity versus the mGlu2 receptor and successfully identified a relatively low potency but high ligand efficiency benzotriazine fragment. Comparison with the pharmacophore in the Janssen reports suggested the optimal vectors for substitution to improve activity and more potent ligands were rapidly found.
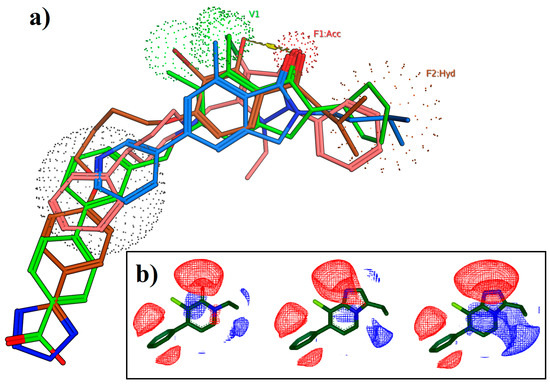
Figure 6.
Ligand-based approaches leading to identification of new mGlu2 PAMs. (a) The structural alignment and pharmacophore of selected mGlu2 receptor PAMs. (b) The electrostatic fields showing the similarity between pyridone and subsequent imidazopyridine and triazopyridine scaffolds.
Also based on the consistent ligand-based alignment hypothesis, scientists at Janssen demonstrated how computational QSAR and machine learning methods could be used to generate models to predict new potent substituents in a series of mGlu2 PAMs [93]. The newly identified compounds were highly potent in nearly all cases being better than 100 nM mGlu2 PAMs. The two different models had relatively low errors suggesting they were extremely valuable for compound activity prediction. The SVM derived model showed less tendency of overtraining compared to the 3D QSAR approach and gave better predictions over the duration of the project.
3.2. mGlu3 Receptor
The second group II mGlu receptor is mGlu3, and similar to mGlu2, it is also implicated in neuropsychiatric disorders such as schizophrenia [94]. Furthermore, the evolution of research around mGlu3 is closely intertwined with mGlu2 because the earliest small molecule synthetic orthosteric ligands designed for group II receptors suffered from limited selectivity [95]. Nowadays selective ligands exist and due to the single nucleotide polymorphisms in the GRM3 gene associated with schizophrenia, interest in the target is increasing.
Compared to mGlu2 there have been relatively few computational modeling studies of this receptor. Apart from the reports that have studied the whole mGlu family, specific focus on mGlu3 has involved work such as Yao et al. who built a homology model of the extra-cellular VFT and used it to study the binding mode of agonists [96]. Experimental mutagenesis revealed the important amino acids for [3H]DCG-IV binding and the model allowed visualization of the ligand in the binding site showing its important interactions.
The chloride ion has been shown to act as an agonist of group II and III mGlu receptors, and in particular it is an agonist of mGlu3 but not mGlu2 [97]. Mutagenesis and modeling showed the ion to bind in a cavity near the orthosteric binding site in the VFT domain. This small pocket is unavailable in mGlu2 receptors due to sequence differences. The phenomena of chloride ion activation were studied in more detail very recently in 2018 by Tora et al. [98]. They modeled the VFT domain and went on to show that Cl− ions stabilize the glutamate-induced active state of the extracellular domain of the mGlu3 receptor and the large basal activity that had for many years been seen was due to chloride mediated positive allosteric modulation effect on low levels of residual glutamate concentrations. This is consistent with other recent reports of mGlu receptor activation being sensitive to residual levels of glutamate [99].
Finally, scientists at Lilly (Indianapolis, Indiana) have extensive experience in the field of orthosteric group II mGlu receptor ligands. Ever since the beginning of their work, computational methods have featured, such as with the design of conformational restrained glutamate analogs [100]. More up to date, they have released several reports of structure-based drug design efforts for group II agonists [101,102]. They used X-ray crystallography and docking and conformational modeling during the lead optimization of group II agonists to reach selective mGlu3 ligands [20].
4. Group III mGlu Receptors
4.1. mGlu4 Receptor
The mGlu4 receptor has emerged as an exciting therapeutic target for multiple indications such as Parkinson’s disease (PD) [103]. Agonists of group III mGlu receptors have shown robust efficacy in rodent models of PD including a highly selective mGlu4 receptor PAM, PHCCC. In addition to providing symptomatic relief for PD patients, several studies suggest that mGlu4 activation could provide a neuroprotective effect and slow disease progression.
In 2010 Slevam et al. identified new mGlu4 receptor agonists based on VS. Using their validated homology models of the mGlu4 binding pocket, they ran a VS campaign with the idea of finding new compounds that could reach other parts of the orthosteric pocket of mGlu4 to gain subtype selectivity. They examined if longer chain analogues of glutamate could still act as mGlu4 agonists, being able to reach less conserved regions outside the glutamate binding pocket [104]. Five new compounds were identified to activate mGlu4, but only (R)-PCEP was chosen for further chemical optimization revealing a noticeable number of new group III mGlu receptor agonists.
In 2014 Rovira et al. studied ligands of mGlu4 receptor with the assistance of computational modeling and docking [105]. The modeling and docking studies identified two overlapping 7TM binding pockets as follows: a first site homologous to the pocket of natural agonists of class A GPCRs linked to allosteric agonism and a second one pointing toward a site topographically homologous to the Na+ binding pocket of class A GPCRs, occupied by PAMs exhibiting the strongest cooperativity. These results reveal that intrinsic efficacy and cooperativity of mGlu4 PAMs are correlated with their binding mode, and vice versa. Finally, as mentioned above, Dalton et al. studied a ligand with dual mGlu4 and mGlu5 functional activity using MD methods [67].
4.2. mGlu7 Receptor
Among the Group III mGlu receptors, mGlu7 is receiving increased attention. It is reported to be a potential target for numerous neurological and psychiatric disorders, such as: schizophrenia, epilepsy, anxiety and depression. It is differentiated from other members of group III due to its low affinity for glutamate binding. Amongst allosteric modulators, AMN082 was identified as the first selective allosteric-agonist of mGlu7, nevertheless the biology of mGlu7 is still only partially explored due to the difficulties to find potent selective tool compounds [11,106,107].
In 2017, scientists from Janssen reported a VS approach to find hits for mGlu7 positive allosteric modulation [108]. They used a method known as proteochemometrics that combines molecule and protein features into a single descriptor for each bioactivity point. The method is well suited to finding new hits for targets that are less explored, but where other protein family members have lots of associated screening data. The large numbers of bioactivity points available for molecules tested versus mGlu1, mGlu2 and mGlu5 for instance could be incorporated into a single model, and used to predict likelihood of activity versus mGlu7. The VS led to the identification of new mGlu7 allosteric modulators from several series. One of these was used as the starting point for a medicinal chemistry optimization program that was assisted with docking into a homology modeling of the mGlu7 receptor [109]. The compounds were rapidly optimized to nanomolar levels of activity validating the binding mode hypothesis and providing a promising series of mGlu7 PAMs.
4.3. mGlu8 Receptor
The mGlu8 receptor has been indicated in drug addiction [110] or anxiety [111], among a number of CNS disorders [112]. The structural opening and closing of the extracellular VFT domain have been discussed. Various studies that have included computational modeling have focused on elucidating the conformational changes of the extracellular domain of mGlu8 and its relation to receptor-specificity of orthosteric ligands targeting the receptor.
As early as 2002, Bessis et al. tried to elucidate the role of two antagonists in the closing of the VFT domain [113]. They modeled the mGlu8 receptor using the available mGlu1 crystal structure and then docked two antagonists (ACPT-II and MAP4) into the model. The antagonists were supposed to prevent the closing of the extracellular domain, hence hindering the activation of the receptor. They identified the key role of D309 and Y227 in the steric clash responsible for the inactivating action of ACPT-II and MAP4, further confirming the conformational changes in VFT needed for receptor activation.
Very recently, in 2018, three mGlu8 VFT domain crystal structures were solved. Schkeryantz et al. were the first to release the mGlu8 crystal in complex with l-glu and l-AP4 [114]. The binding mode of l-glu in this receptor differed from that previously described in mGlu1 crystals. In this work, they showed A154, A177 and E399 to be responsible for the differences observed with other mGlus. They also point to the key role played by a phosphate ion of l-AP4 in agonist potency and selectivity.
The last mGlu8 crystal structure was also released in 2018 by the same group., the mGlu8-(S)DCPG complex [112]. This structure differs from the previously solved (l-glu- and l-AP4 bound) by showing a larger opening angle between the two lobes of the domain. They also generated a homology model of mGlu8 using the mGlu1–l-glu crystal structure as template and docked (S)DCPG into it. Comparison between their crystallized complex, the previous structures, and the model uncovered the relevance of lobe opening angles, which may be considered when studying bulky agonists in future reports.
Feng et al. also built a homology model of the mGlu8 7TM domain based on the mGlu5 crystal structure in their attempt to characterize the allosteric binding pocket of the non-crystallized mGlu receptors [66]. They docked NAM and PAM compounds to the model in order to identify residues involved in selectivity. Two different poses for AZ12216052 in mGlu8 were selected. Finally, they performed a 30 ns MD simulation of mGlu8 bound to AZ12216052 PAM to validate the stability of the complex and the ligand-receptor interactions implicated. Results showed a binding pose of this ligand that involved S663 and S829 forming key H-bonds with the PAM. With the recent release of extracellular domain crystal structures of mGlu8 bound to three different agonists, new opportunities to better understand the orthosteric pocket of the receptor arise. However, mGlu8 allosteric regulation still represents a challenge and there has been relatively little computational work in the area.
5. Conclusions
The mGlu1 and mGlu5 receptors have been studied in a relatively similar manner with computational approaches. Before their crystal structures were known most studies relied on ligand-based approaches to better understand these receptors focusing on virtual screening and pharmacophore building approaches. Homology models based on class A GPCR templates available at the time were built to map the mutagenesis data. With the solving of the 7TM domain structures of mGlu1 and mGlu5 in 2014, the data collected up to then could be validated. Moreover, the confirmation of the allosteric pocket location in the 7TM domain opened new horizons for structure-based drug design studies targeting mGlu receptors. Also, the release of four more NAM bound crystallographic 7TM structures of mGlu5 provided further insight in the binding mode of different scaffolds to this receptor. However, the activation mechanism and PAM SARs still represent a challenge since no available structure of an active state of the receptor had been released but modeling work on molecular switches has suggested new insights. The new dimeric full length structures of mGlu5 open great opportunities for better understanding of these aspects.
In group II, the mGlu2 receptor has been widely studied. An array of ligand and structure-based approaches have been used. The use of ligand-based approaches has been shown to have a significant impact on medicinal chemistry by identifying new hits from VS and scaffolds for lead series. Meanwhile, the experimental mutagenesis was supported with molecular modeling to confirm the allosteric binding site. A series of detailed MD studies followed that have demonstrated a hypothesis for the origins of PAM versus NAM activity. The PAMs generate increased movement amongst amino acid sidechains of the transmission switch, whilst the NAMs generally stabilize amino acids in the inactive-like state, showing less movement, consistent with MD simulations and analyses on mGlu5. Such results can offer great promise if they can be extrapolated to the design of new compounds.
Group III mGlu receptors remain the least studied of the family, yet they offer great therapeutic opportunity. Recently, VS studies led to identifying promising compounds targeting mGlu4, mGlu7 and mGlu8 receptors. To better understand the selectivity of these compounds for each receptor, modelling and docking approaches were applied. Nevertheless, this group of mGlu receptors still needs to be further characterized.
MD has been used in several studies to understand details of functional activity [72,82,84,85]. As the new structure of the full length mGlu5 receptor has revealed the large-scale motions that accompany activation, it will be fascinating to see in later studies how more local conformational changes affect functional activity. The binding site of class A orthosteric ligands overlaps with class C allosteric modulators. However, the allosteric ligands are not able to activate the receptor alone. The microswitch hypothesis proposes that certain groupings of amino acids which are spatially co-located can translate ligand induced effects into knock-on movements that stabilize active states, promoting more activation of the receptor population and that this can be relevant for mGlu receptor allosteric modulation [72,82]. Therefore, will the hypothesis of translating features for class A activation to class C allosteric ligands hold true? In terms of future computational approaches, there has been increased interest in both machine learning and physics based computational modeling. The former is well suited to GPCRs as it relies upon using available data to make future predictions. Such methods have already shown promise for mGlu receptors [60,109]. On the other hand, there are no reports on the application of physics-based methods such as free energy perturbation for mGlu receptors. It is expected that the future will provide significant opportunities for more quantitative mGlu receptor drug design, building on early studies demonstrating the feasibility of these approaches for GPCRs in general [115].
Author Contributions
This manuscript was written, reviewed and edited by all authors.
Funding
LPB received funding support from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 675451 (CompBioMed project). CLT is recipient of an FPI fellowship (BES-2017-081872).
Acknowledgments
We thank Leonardo Pardo for insight and discussion.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Fagerberg, L.; Jonasson, K.; von Heijne, G.; Uhlén, M.; Berglund, L. Prediction of the human membrane proteome. Proteomics 2010, 10, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Pin, J.-P.; Galvez, T.; Prézeau, L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol. Ther. 2003, 98, 325–354. [Google Scholar] [CrossRef]
- Niswender, C.M.; Conn, P.J. Metabotropic Glutamate Receptors: Physiology, Pharmacology, and Disease. Annu. Rev. Pharmacool. Toxicol. 2010, 50, 295–322. [Google Scholar] [CrossRef] [PubMed]
- Lavreysen, H.; Dautzenberg, F.M. Therapeutic Potential of Group III Metabotropic Glutamate Receptors. Curr. Med. Chem. 2008, 15, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Conn, P.J.; Lindsley, C.W.; Jones, C.K. Activation of metabotropic glutamate receptors as a novel approach for the treatment of schizophrenia. Trends Pharmacol. Sci. 2009, 30, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Shigemoto, R.; Mizuno, N. Chapter III Metabotropic glutamate receptors—Immunocytochemical and in situ hybridization analyses. In Handbook of Chemical Neuroanatomy; Ottersen, O.P., Storm-Mathisen, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2000; Volume 18, pp. 63–98. [Google Scholar]
- Cauli, B.; Porter, J.T.; Tsuzuki, K.; Lambolez, B.; Rossier, J.; Quenet, B.; Audinat, E. Classification of fusiform neocortical interneurons based on unsupervised clustering. Proc. Natl. Acad. Sci. USA 2000, 97, 6144. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-M.; Jia, Z.; Janus, C.; Henderson, J.T.; Gerlai, R.; Wojtowicz, J.M.; Roder, J.C. Mice Lacking Metabotropic Glutamate Receptor 5 Show Impaired Learning and Reduced CA1 Long-Term Potentiation (LTP) But Normal CA3 LTP. J. Neurosci. 1997, 17, 5196. [Google Scholar] [CrossRef]
- Wright, R.A.; Johnson, B.G.; Zhang, C.; Salhoff, C.; Kingston, A.E.; Calligaro, D.O.; Monn, J.A.; Schoepp, D.D.; Marek, G.J. CNS distribution of metabotropic glutamate 2 and 3 receptors: Transgenic mice and [3H]LY459477 autoradiography. Neuropharmacology 2013, 66, 89–98. [Google Scholar] [CrossRef]
- Mercier, M.S.; Lodge, D. Group III Metabotropic Glutamate Receptors: Pharmacology, Physiology and Therapeutic Potential. Neurochem. Res. 2014, 39, 1876–1894. [Google Scholar] [CrossRef]
- Fisher, N.M.; Seto, M.; Lindsley, C.W.; Niswender, C.M. Metabotropic Glutamate Receptor 7: A New Therapeutic Target in Neurodevelopmental Disorders. Front. Mol. Neurosci. 2018, 11, 387. [Google Scholar] [CrossRef]
- Raber, J.; Duvoisin, R.M. Novel metabotropic glutamate receptor 4 and glutamate receptor 8 therapeutics for the treatment of anxiety. Expert Opin. Investig. Drugs 2015, 24, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Congreve, M.; Oswald, C.; Marshall, F.H. Applying Structure-Based Drug Design Approaches to Allosteric Modulators of GPCRs. Trends Pharmacol. Sci. 2017, 38, 837–847. [Google Scholar] [CrossRef]
- Koehl, A.; Hu, H.; Feng, D.; Sun, B.; Zhang, Y.; Robertson, M.J.; Chu, M.; Kobilka, T.S.; Laermans, T.; Steyaert, J.; et al. Structural insights into the activation of metabotropic glutamate receptors. Nature 2019, 566, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Kunishima, N.; Shimada, Y.; Tsuji, Y.; Sato, T.; Yamamoto, M.; Kumasaka, T.; Nakanishi, S.; Jingami, H.; Morikawa, K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 2000, 407, 971. [Google Scholar] [CrossRef] [PubMed]
- Doré, A.S.; Okrasa, K.; Patel, J.C.; Serrano-Vega, M.; Bennett, K.; Cooke, R.M.; Errey, J.C.; Jazayeri, A.; Khan, S.; Tehan, B.; et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 2014, 511, 557. [Google Scholar] [CrossRef]
- Wu, H.; Wang, C.; Gregory, K.J.; Han, G.W.; Cho, H.P.; Xia, Y.; Niswender, C.M.; Katritch, V.; Meiler, J.; Cherezov, V.; et al. Structure of a Class C GPCR Metabotropic Glutamate Receptor 1 Bound to an Allosteric Modulator. Science 2014, 344, 58. [Google Scholar] [CrossRef]
- Christopher, J.A.; Aves, S.J.; Bennett, K.A.; Doré, A.S.; Errey, J.C.; Jazayeri, A.; Marshall, F.H.; Okrasa, K.; Serrano-Vega, M.J.; Tehan, B.G.; et al. Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile). J. Med. Chem. 2015, 58, 6653–6664. [Google Scholar] [CrossRef]
- Christopher, J.A.; Orgován, Z.; Congreve, M.; Doré, A.S.; Errey, J.C.; Marshall, F.H.; Mason, J.S.; Okrasa, K.; Rucktooa, P.; Serrano-Vega, M.J.; et al. Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures. J. Med. Chem. 2018, 62, 207–222. [Google Scholar] [CrossRef]
- Monn, J.A.; Henry, S.S.; Massey, S.M.; Clawson, D.K.; Chen, Q.; Diseroad, B.A.; Bhardwaj, R.M.; Atwell, S.; Lu, F.; Wang, J.; et al. Synthesis and Pharmacological Characterization of C4β-Amide-Substituted 2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1S,2S,4S,5R,6S)-2-Amino-4-[(3-methoxybenzoyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2794193), a Highly Potent and Selective mGlu3 Receptor Agonist. J. Med. Chem. 2018, 61, 2303–2328. [Google Scholar]
- Lindsley, C.W.; Emmitte, K.A.; Hopkins, C.R.; Bridges, T.M.; Gregory, K.J.; Niswender, C.M.; Conn, P.J. Practical Strategies and Concepts in GPCR Allosteric Modulator Discovery: Recent Advances with Metabotropic Glutamate Receptors. Chem. Rev. 2016, 116, 6707–6741. [Google Scholar] [CrossRef]
- Tresadern, G.; Rombouts, F.J.R.; Oehlrich, D.; Macdonald, G.; Trabanco, A.A. Industrial medicinal chemistry insights: Neuroscience hit generation at Janssen. Drug Discov. Today 2017, 22, 1478–1488. [Google Scholar] [CrossRef]
- Scior, T.; Bender, A.; Tresadern, G.; Medina-Franco, J.L.; Martínez-Mayorga, K.; Langer, T.; Cuanalo-Contreras, K.; Agrafiotis, D.K. Recognizing Pitfalls in Virtual Screening: A Critical Review. J. Chem. Inf. Model. 2012, 52, 867–881. [Google Scholar] [CrossRef]
- Ciordia, M.; Pérez-Benito, L.; Delgado, F.; Trabanco, A.S.A.; Tresadern, G. Application of free energy perturbation for the design of BACE1 inhibitors. J. Chem. Inf. Model. 2016, 56, 1856–1871. [Google Scholar] [CrossRef]
- Keränen, H.; Pérez-Benito, L.; Ciordia, M.; Delgado, F.; Steinbrecher, T.B.; Oehlrich, D.; van Vlijmen, H.W.; Trabanco, A.S.A.; Tresadern, G. Acylguanidine beta secretase 1 inhibitors: A combined experimental and free energy perturbation study. J. Chem. Theory Comput. 2017, 13, 1439–1453. [Google Scholar] [CrossRef]
- Pérez-Benito, L.; Keränen, H.; van Vlijmen, H.; Tresadern, G. Predicting Binding Free Energies of PDE2 Inhibitors. The Difficulties of Protein Conformation. Sci. Rep. 2018, 8, 4883. [Google Scholar] [CrossRef]
- Nicoletti, F.; Bruno, V.; Ngomba, R.T.; Gradini, R.; Battaglia, G. Metabotropic glutamate receptors as drug targets: What’s new? Curr. Opin. Pharmacol. 2015, 20, 89–94. [Google Scholar] [CrossRef]
- Mary, S.; Gomeza, J.; Prézeau, L.; Bockaert, J.; Pin, J.-P. A Cluster of Basic Residues in the Carboxyl-terminal Tail of the Short Metabotropic Glutamate Receptor 1 Variants Impairs Their Coupling to Phospholipase C. J. Biol. Chem. 1998, 273, 425–432. [Google Scholar] [CrossRef]
- Ray, K.; Hauschild, B.C. Cys-140 Is Critical for Metabotropic Glutamate Receptor-1 Dimerization. J. Biol. Chem. 2000, 275, 34245–34251. [Google Scholar] [CrossRef]
- Litschig, S.; Gasparini, F.; Rueegg, D.; Stoehr, N.; Flor, P.J.; Vranesic, I.; Prézeau, L.; Pin, J.-P.; Thomsen, C.; Kuhn, R. CPCCOEt, a Noncompetitive Metabotropic Glutamate Receptor 1 Antagonist, Inhibits Receptor Signaling Without Affecting Glutamate Binding. Mol. Pharmacol. 1999, 55, 453. [Google Scholar]
- Pagano, A.; Rüegg, D.; Litschig, S.; Stoehr, N.; Stierlin, C.; Heinrich, M.; Floersheim, P.; Prezèau, L.; Carroll, F.; Pin, J.-P.; et al. The Non-competitive Antagonists 2-Methyl-6-(phenylethynyl)pyridine and 7-Hydroxyiminocyclopropan[b]chromen-1a-carboxylic Acid Ethyl Ester Interact with Overlapping Binding Pockets in the Transmembrane Region of Group I Metabotropic Glutamate Receptors. J. Biol. Chem. 2000, 275, 33750–33758. [Google Scholar] [CrossRef]
- Malherbe, P.; Kratochwil, N.; Zenner, M.-T.; Piussi, J.; Diener, C.; Kratzeisen, C.; Fischer, C.; Porter, R.H.P. Mutational Analysis and Molecular Modeling of the Binding Pocket of the Metabotropic Glutamate 5 Receptor Negative Modulator 2-Methyl-6-(phenylethynyl)-pyridine. Mol. Pharmacol. 2003, 64, 823. [Google Scholar] [CrossRef]
- Knoflach, F.; Mutel, V.; Jolidon, S.; Kew, J.N.C.; Malherbe, P.; Vieira, E.; Wichmann, J.; Kemp, J.A. Positive allosteric modulators of metabotropic glutamate 1 receptor: Characterization, mechanism of action, and binding site. Proc. Natl. Acad. Sci. USA 2001, 98, 13402. [Google Scholar] [CrossRef]
- Hemstapat, K.; de Paulis, T.; Chen, Y.; Brady, A.E.; Grover, V.K.; Alagille, D.; Tamagnan, G.D.; Conn, P.J. A Novel Class of Positive Allosteric Modulators of Metabotropic Glutamate Receptor Subtype 1 Interact with a Site Distinct from That of Negative Allosteric Modulators. Mol. Pharmacol. 2006, 70, 616. [Google Scholar] [CrossRef]
- Fukuda, J.; Suzuki, G.; Kimura, T.; Nagatomi, Y.; Ito, S.; Kawamoto, H.; Ozaki, S.; Ohta, H. Identification of a novel transmembrane domain involved in the negative modulation of mGluR1 using a newly discovered allosteric mGluR1 antagonist, 3-cyclohexyl-5-fluoro-6-methyl-7-(2-morpholin-4-ylethoxy)-4H-chromen-4-one. Neuropharmacology 2009, 57, 438–445. [Google Scholar] [CrossRef]
- Chen, Y.; Goudet, C.; Pin, J.-P.; Conn, P.J. N-{4-Chloro-2-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-2-hydroxybenzamide (CPPHA) Acts through a Novel Site as a Positive Allosteric Modulator of Group 1 Metabotropic Glutamate Receptors. Mol. Pharmacol. 2008, 73, 909. [Google Scholar] [CrossRef]
- Suzuki, G.; Kimura, T.; Satow, A.; Kaneko, N.; Fukuda, J.; Hikichi, H.; Sakai, N.; Maehara, S.; Kawagoe-Takaki, H.; Hata, M.; et al. Pharmacological characterization of a new, orally active and potent allosteric metabotropic glutamate receptor 1 antagonist, 4-[1-(2-fluoropyridin-3-yl)-5-methyl-1H-1,2,3-triazol-4-yl]-N-isopropyl-N-methyl-3,6-dihydropyridine-1(2H)-carboxamide (FTIDC). J. Pharmacol. Exp. Ther. 2007, 321, 1144. [Google Scholar] [CrossRef]
- Noeske, T.; Jirgensons, A.; Starchenkovs, I.; Renner, S.; Jaunzeme, I.; Trifanova, D.; Hechenberger, M.; Bauer, T.; Kauss, V.; Parsons, C.G.; et al. Virtual Screening for Selective Allosteric mGluR1 Antagonists and Structure-Activity Relationship Investigations for Coumarine Derivatives. ChemMedChem 2007, 2, 1763–1773. [Google Scholar] [CrossRef]
- Wang, M.; Hampson, D.R. An evaluation of automated in silico ligand docking of amino acid ligands to Family C G-protein coupled receptors. Biorg. Med. Chem. 2006, 14, 2032–2039. [Google Scholar] [CrossRef]
- Vanejevs, M.; Jatzke, C.; Renner, S.; Müller, S.; Hechenberger, M.; Bauer, T.; Klochkova, A.; Pyatkin, I.; Kazyulkin, D.; Aksenova, E.; et al. Positive and Negative Modulation of Group I Metabotropic Glutamate Receptors. J. Med. Chem. 2008, 51, 634–647. [Google Scholar] [CrossRef]
- Harpsøe, K.; Isberg, V.; Tehan, B.G.; Weiss, D.; Arsova, A.; Marshall, F.H.; Bräuner-Osborne, H.; Gloriam, D.E. Selective Negative Allosteric Modulation Of Metabotropic Glutamate Receptors—A Structural Perspective of Ligands and Mutants. Sci. Rep. 2015, 5, 13869. [Google Scholar] [CrossRef]
- Munk, C.; Harpsøe, K.; Hauser, A.S.; Isberg, V.; Gloriam, D.E. Integrating structural and mutagenesis data to elucidate GPCR ligand binding. Curr. Opin. Pharmacol. 2016, 30, 51–58. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, X.; Chen, X.; He, Y.; Qiao, L.; Zhang, Y.; Li, G.; Xiang, Y. Virtual Screening and Molecular Dynamics Study of Potential Negative Allosteric Modulators of mGluR1 from Chinese Herbs. Molecules 2015, 20, 12769–12786. [Google Scholar] [CrossRef]
- Jang, J.W.; Cho, N.-C.; Min, S.-J.; Cho, Y.S.; Park, K.D.; Seo, S.H.; No, K.T.; Pae, A.N. Novel Scaffold Identification of mGlu1 Receptor Negative Allosteric Modulators Using a Hierarchical Virtual Screening Approach. Chem. Biol. Drug Des. 2016, 87, 239–256. [Google Scholar] [CrossRef]
- Lakkaraju, S.K.; Yu, W.; Raman, E.P.; Hershfeld, A.V.; Fang, L.; Deshpande, D.A.; MacKerell, A.D. Mapping Functional Group Free Energy Patterns at Protein Occluded Sites: Nuclear Receptors and G-Protein Coupled Receptors. J. Chem. Inf. Model. 2015, 55, 700–708. [Google Scholar] [CrossRef]
- van Westen, G.J.P.; Gaulton, A.; Overington, J.P. Chemical, Target, and Bioactive Properties of Allosteric Modulation. PLoS Comput. Biol. 2014, 10, e1003559. [Google Scholar] [CrossRef]
- Bai, Q.; Yao, X. Investigation of allosteric modulation mechanism of metabotropic glutamate receptor 1 by molecular dynamics simulations, free energy and weak interaction analysis. Sci. Rep. 2016, 6, 21763. [Google Scholar] [CrossRef]
- Śliwa, P.; Kurczab, R.; Bojarski, A.J. ONIOM and FMO-EDA study of metabotropic glutamate receptor 1: Quantum insights into the allosteric binding site. Int. J. Quantum. Chem. 2018, 118, e25617. [Google Scholar] [CrossRef]
- Borroto-Escuela, D.O.; Fuxe, K. Basimglurant for treatment of major depressive disorder: A novel negative allosteric modulator of metabotropic glutamate receptor 5. Expert Opin. Investig. Drugs 2015, 24, 1247–1260. [Google Scholar]
- Waung, M.W.; Akerman, S.; Wakefield, M.; Keywood, C.; Goadsby, P.J. Metabotropic glutamate receptor 5: A target for migraine therapy. Ann. Clin. Trans. Neurol. 2016, 3, 560–571. [Google Scholar] [CrossRef]
- Scharf, S.H.; Jaeschke, G.; Wettstein, J.G.; Lindemann, L. Metabotropic glutamate receptor 5 as drug target for Fragile X syndrome. Curr. Opin. Pharmacol. 2015, 20, 124–134. [Google Scholar] [CrossRef]
- Mühlemann, A.; Ward, N.A.; Kratochwil, N.; Diener, C.; Fischer, C.; Stucki, A.; Jaeschke, G.; Malherbe, P.; Porter, R.H.P. Determination of key amino acids implicated in the actions of allosteric modulation by 3,3′-difluorobenzaldazine on rat mGlu5 receptors. Eur. J. Pharmacol. 2006, 529, 95–104. [Google Scholar] [CrossRef]
- Chen, Y.; Nong, Y.; Goudet, C.; Hemstapat, K.; de Paulis, T.; Pin, J.-P.; Conn, P.J. Interaction of Novel Positive Allosteric Modulators of Metabotropic Glutamate Receptor 5 with the Negative Allosteric Antagonist Site Is Required for Potentiation of Receptor Responses. Mol. Pharmacol. 2007, 71, 1389. [Google Scholar] [CrossRef]
- Gregory, K.J.; Nguyen, E.D.; Reiff, S.D.; Squire, E.F.; Stauffer, S.R.; Lindsley, C.W.; Meiler, J.; Conn, P.J. Probing the Metabotropic Glutamate Receptor 5 (mGlu5) Positive Allosteric Modulator (PAM) Binding Pocket: Discovery of Point Mutations That Engender a “Molecular Switch” in PAM Pharmacology. Mol. Pharmacol. 2013, 83, 991. [Google Scholar] [CrossRef]
- Gregory, K.J.; Noetzel, M.J.; Rook, J.M.; Vinson, P.N.; Stauffer, S.R.; Rodriguez, A.L.; Emmitte, K.A.; Zhou, Y.; Chun, A.C.; Felts, A.S.; et al. Investigating Metabotropic Glutamate Receptor 5 Allosteric Modulator Cooperativity, Affinity, and Agonism: Enriching Structure-Function Studies and Structure-Activity Relationships. Mol. Pharmacol. 2012, 82, 860. [Google Scholar] [CrossRef]
- Turlington, M.; Noetzel, M.J.; Chun, A.; Zhou, Y.; Gogliotti, R.D.; Nguyen, E.D.; Gregory, K.J.; Vinson, P.N.; Rook, J.M.; Gogi, K.K.; et al. Exploration of Allosteric Agonism Structure–Activity Relationships within an Acetylene Series of Metabotropic Glutamate Receptor 5 (mGlu5) Positive Allosteric Modulators (PAMs): Discovery of 5-((3-Fluorophenyl)ethynyl)-N-(3-methyloxetan-3-yl)picolinamide (ML254). J. Med. Chem. 2013, 56, 7976–7996. [Google Scholar]
- Gregory, K.J.; Nguyen, E.D.; Malosh, C.; Mendenhall, J.L.; Zic, J.Z.; Bates, B.S.; Noetzel, M.J.; Squire, E.F.; Turner, E.M.; Rook, J.M.; et al. Identification of Specific Ligand-Receptor Interactions That Govern Binding and Cooperativity of Diverse Modulators to a Common Metabotropic Glutamate Receptor 5 Allosteric Site. ACS Chem. Neurosci. 2014, 5, 282–295. [Google Scholar] [CrossRef]
- Malherbe, P.; Kratochwil, N.; Mühlemann, A.; Zenner, M.-T.; Fischer, C.; Stahl, M.; Gerber, P.R.; Jaeschke, G.; Porter, R.H.P. Comparison of the binding pockets of two chemically unrelated allosteric antagonists of the mGlu5 receptor and identification of crucial residues involved in the inverse agonism of MPEP. J. Neurochem. 2006, 98, 601–615. [Google Scholar] [CrossRef]
- Noeske, T.; Trifanova, D.; Kauss, V.; Renner, S.; Parsons, C.G.; Schneider, G.; Weil, T. Synergism of virtual screening and medicinal chemistry: Identification and optimization of allosteric antagonists of metabotropic glutamate receptor 1. Biorg. Med. Chem. 2009, 17, 5708–5715. [Google Scholar] [CrossRef]
- Renner, S.; Hechenberger, M.; Noeske, T.; Böcker, A.; Jatzke, C.; Schmuker, M.; Parsons, C.G.; Weil, T.; Schneider, G. Searching for Drug Scaffolds with 3D Pharmacophores and Neural Network Ensembles. Angew. Chem. Int. Ed. 2007, 46, 5336–5339. [Google Scholar] [CrossRef]
- Lowe, E.W.; Ferrebee, A.; Rodriguez, A.L.; Conn, P.J.; Meiler, J. 3D-QSAR CoMFA study of benzoxazepine derivatives as mGluR5 positive allosteric modulators. Bioorg. Med. Chem. Lett. 2010, 20, 5922–5924. [Google Scholar] [CrossRef]
- Mueller, R.; Dawson, E.S.; Meiler, J.; Rodriguez, A.L.; Chauder, B.A.; Bates, B.S.; Felts, A.S.; Lamb, J.P.; Menon, U.N.; Jadhav, S.B.; et al. Discovery of 2-(2-Benzoxazoyl amino)-4-Aryl-5-Cyanopyrimidine as Negative Allosteric Modulators (NAMs) of Metabotropic Glutamate Receptor 5 (mGlu5): From an Artificial Neural Network Virtual Screen to an In Vivo Tool Compound. ChemMedChem 2012, 7, 406–414. [Google Scholar] [CrossRef]
- Kaae, B.H.; Harpsøe, K.; Kvist, T.; Mathiesen, J.M.; Mølck, C.; Gloriam, D.; Jimenez, H.N.; Uberti, M.A.; Nielsen, S.M.; Nielsen, B.; et al. Structure-Activity Relationships for Negative Allosteric mGluR5 Modulators. ChemMedChem 2012, 7, 440–451. [Google Scholar] [CrossRef]
- Dalton, J.A.R.; Gómez-Santacana, X.; Llebaria, A.; Giraldo, J. Computational Analysis of Negative and Positive Allosteric Modulator Binding and Function in Metabotropic Glutamate Receptor 5 (In)Activation. J. Chem. Inf. Model. 2014, 54, 1476–1487. [Google Scholar] [CrossRef]
- Anighoro, A.; Graziani, D.; Bettinelli, I.; Cilia, A.; De Toma, C.; Longhi, M.; Mangiarotti, F.; Menegon, S.; Pirona, L.; Poggesi, E.; et al. Insights into the interaction of negative allosteric modulators with the metabotropic glutamate receptor 5: Discovery and computational modeling of a new series of ligands with nanomolar affinity. Biorg. Med. Chem. 2015, 23, 3040–3058. [Google Scholar] [CrossRef]
- Feng, Z.; Ma, S.; Hu, G.; Xie, X.-Q. Allosteric Binding Site and Activation Mechanism of Class C G-Protein Coupled Receptors: Metabotropic Glutamate Receptor Family. AAPS J. 2015, 17, 737–753. [Google Scholar] [CrossRef]
- Dalton, J.A.R.; Pin, J.-P.; Giraldo, J. Analysis of positive and negative allosteric modulation in metabotropic glutamate receptors 4 and 5 with a dual ligand. Sci. Rep. 2017, 7, 4944. [Google Scholar] [CrossRef]
- Bian, Y.; Feng, Z.; Yang, P.; Xie, X.-Q. Integrated In Silico Fragment-Based Drug Design: Case Study with Allosteric Modulators on Metabotropic Glutamate Receptor 5. AAPS J. 2017, 19, 1235–1248. [Google Scholar] [CrossRef]
- Fu, T.; Zheng, G.; Tu, G.; Yang, F.; Chen, Y.; Yao, X.; Li, X.; Xue, W.; Zhu, F. Exploring the Binding Mechanism of Metabotropic Glutamate Receptor 5 Negative Allosteric Modulators in Clinical Trials by Molecular Dynamics Simulations. ACS Chem. Neurosci. 2018, 9, 1492–1502. [Google Scholar] [CrossRef]
- Vijaya Prabhu, S.; Singh, S.K. E-pharmacophore-based screening of mGluR5 negative allosteric modulators for central nervous system disorder. Comput. Biol. Chem. 2018, 78, 414–423. [Google Scholar] [CrossRef]
- Vijaya Prabhu, S.; Singh, S.K. Atom-based 3D-QSAR, induced fit docking, and molecular dynamics simulations study of thieno[2–b]pyridines negative allosteric modulators of mGluR5. J. Recept. Signal Transduct. 2018, 38, 225–239. [Google Scholar] [CrossRef]
- Llinas del Torrent, C.; Casajuana-Martin, N.; Pardo, L.; Tresadern, G.; Pérez-Benito, L. Mechanisms Underlying Allosteric Molecular Switches of Metabotropic Glutamate Receptor 5. J. Chem. Inf. Model. 2019. [Google Scholar] [CrossRef] [PubMed]
- Trabanco, A.A.; Cid, J.M.; Lavreysen, H.; Macdonald, G.J.; Tresadern, G. Progress in the Developement of Positive Allosteric Modulators of the Metabotropic Glutamate Receptor 2. Curr. Med. Chem. 2011, 18, 47–68. [Google Scholar] [CrossRef] [PubMed]
- Trabanco, A.A.; Cid, J.M. mGluR2 positive allosteric modulators: A patent review (2009–present). Expert Opin. Ther. Patents 2013, 23, 629–647. [Google Scholar] [CrossRef]
- Trabanco, A.A.; Tresadern, G.; Macdonald, G.J.; Vega, J.A.; de Lucas, A.I.; Matesanz, E.; García, A.; Linares, M.L.; Alonso de Diego, S.A.; Alonso, J.M.; et al. Imidazo[1,2-a]pyridines: Orally Active Positive Allosteric Modulators of the Metabotropic Glutamate 2 Receptor. J. Med. Chem. 2012, 55, 2688–2701. [Google Scholar] [CrossRef]
- Cid, J.M.; Duvey, G.; Cluzeau, P.; Nhem, V.; Macary, K.; Raux, A.; Poirier, N.; Muller, J.; Boléa, C.; Finn, T.; et al. Discovery of 1,5-Disubstituted Pyridones: A New Class of Positive Allosteric Modulators of the Metabotropic Glutamate 2 Receptor. ACS Chem. Neurosci. 2010, 1, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Cid, J.M.; Tresadern, G.; Duvey, G.; Lütjens, R.; Finn, T.; Rocher, J.-P.; Poli, S.; Vega, J.A.; de Lucas, A.I.; Matesanz, E.; et al. Discovery of 1-Butyl-3-chloro-4-(4-phenyl-1-piperidinyl)-(1H)-pyridone (JNJ-40411813): A Novel Positive Allosteric Modulator of the Metabotropic Glutamate 2 Receptor. J. Med. Chem. 2014, 57, 6495–6512. [Google Scholar] [CrossRef] [PubMed]
- Cid, J.M.; Duvey, G.; Tresadern, G.; Nhem, V.; Furnari, R.; Cluzeau, P.; Vega, J.A.; de Lucas, A.I.; Matesanz, E.; Alonso, J.M.; et al. Discovery of 1,4-Disubstituted 3-Cyano-2-pyridones: A New Class of Positive Allosteric Modulators of the Metabotropic Glutamate 2 Receptor. J. Med. Chem. 2012, 55, 2388–2405. [Google Scholar] [CrossRef] [PubMed]
- Lundström, L.; Bissantz, C.; Beck, J.; Wettstein, J.G.; Woltering, T.J.; Wichmann, J.; Gatti, S. Structural determinants of allosteric antagonism at metabotropic glutamate receptor 2: Mechanistic studies with new potent negative allosteric modulators. Br. J. Pharmacol. 2011, 164, 521–537. [Google Scholar] [CrossRef]
- Lundström, L.; Kuhn, B.; Beck, J.; Borroni, E.; Wettstein, J.G.; Woltering, T.J.; Gatti, S. Mutagenesis and Molecular Modeling of the Orthosteric Binding Site of the mGlu2 Receptor Determining Interactions of the Group II Receptor Antagonist 3H-HYDIA. ChemMedChem 2009, 4, 1086–1094. [Google Scholar] [CrossRef]
- Farinha, A.; Lavreysen, H.; Peeters, L.; Russo, B.; Masure, S.; Trabanco, A.A.; Cid, J.; Tresadern, G. Molecular determinants of positive allosteric modulation of the human metabotropic glutamate receptor 2. Br. J. Pharmacol. 2015, 172, 2383–2396. [Google Scholar] [CrossRef]
- Pérez-Benito, L.; Doornbos, M.L.J.; Cordomí, A.; Peeters, L.; Lavreysen, H.; Pardo, L.; Tresadern, G. Molecular Switches of Allosteric Modulation of the Metabotropic Glutamate 2 Receptor. Structure 2017, 25, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Doornbos, M.L.J.; Wang, X.; Vermond, S.C.; Peeters, L.; Pérez-Benito, L.; Trabanco, A.A.; Lavreysen, H.; Cid, J.M.; Heitman, L.H.; Tresadern, G.; et al. Covalent Allosteric Probe for the Metabotropic Glutamate Receptor 2: Design, Synthesis, and Pharmacological Characterization. J. Med. Chem. 2018, 62, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Doornbos, M.L.J.; Pérez-Benito, L.; Tresadern, G.; Mulder-Krieger, T.; Biesmans, I.; Trabanco, A.A.; Cid, J.M.; Lavreysen, H.; Ijzerman, A.P.; Heitman, L.H. Molecular mechanism of positive allosteric modulation of the metabotropic glutamate receptor 2 by JNJ-46281222. Br. J. Pharmacol. 2015, 173, 588–600. [Google Scholar] [CrossRef] [PubMed]
- Cid, J.M.; Tresadern, G.; Vega, J.A.; de Lucas, A.I.; del Cerro, A.; Matesanz, E.; Linares, M.L.; García, A.; Iturrino, L.; Pérez-Benito, L.; et al. Discovery of 8-Trifluoromethyl-3-cyclopropylmethyl-7-[(4-(2,4-difluorophenyl)-1-piperazinyl)methyl]-1,2,4-triazolo[4,3-a]pyridine (JNJ-46356479), a Selective and Orally Bioavailable mGlu2 Receptor Positive Allosteric Modulator (PAM). J. Med. Chem. 2016, 59, 8495–8507. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.; Guadix, A.E.; Costantino, G. Molecular Dynamics Simulation of the Heterodimeric mGluR2/5HT2A Complex. An Atomistic Resolution Study of a Potential New Target in Psychiatric Conditions. J. Chem. Inf. Model. 2009, 49, 1602–1616. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.; Costantino, G.; de Fabritiis, G.; Pastor, M.; Selent, J. Membrane-Sensitive Conformational States of Helix 8 in the Metabotropic Glu2 Receptor, a Class C GPCR. PLoS ONE 2012, 7, e42023. [Google Scholar] [CrossRef]
- Xue, L.; Rovira, X.; Scholler, P.; Zhao, H.; Liu, J.; Pin, J.-P.; Rondard, P. Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat. Chem. Biol. 2014, 11, 134. [Google Scholar] [CrossRef]
- Tresadern, G.; Cid, J.M.; Macdonald, G.J.; Vega, J.A.; de Lucas, A.I.; García, A.; Matesanz, E.; Linares, M.L.; Oehlrich, D.; Lavreysen, H.; et al. Scaffold hopping from pyridones to imidazo[1,2-a]pyridines. New positive allosteric modulators of metabotropic glutamate 2 receptor. Bioorg. Med. Chem. Lett. 2010, 20, 175–179. [Google Scholar] [CrossRef]
- Cid, J.M.; Tresadern, G.; Vega, J.A.; de Lucas, A.I.; Matesanz, E.; Iturrino, L.; Linares, M.L.; Garcia, A.; Andrés, J.I.; Macdonald, G.J.; et al. Discovery of 3-Cyclopropylmethyl-7-(4-phenylpiperidin-1-yl)-8-trifluoromethyl[1,2,4]triazolo[4,3-a]pyridine (JNJ-42153605): A Positive Allosteric Modulator of the Metabotropic Glutamate 2 Receptor. J. Med. Chem. 2012, 55, 8770–8789. [Google Scholar] [CrossRef]
- Ahnaou, A.; Lavreysen, H.; Tresadern, G.; Cid, J.M.; Drinkenburg, W.H. mGlu2 Receptor Agonism, but Not Positive Allosteric Modulation, Elicits Rapid Tolerance towards Their Primary Efficacy on Sleep Measures in Rats. PLoS ONE 2015, 10, e0144017. [Google Scholar] [CrossRef]
- Szabó, G.; Túrós, G.I.; Kolok, S.; Vastag, M.; Sánta, Z.; Dékány, M.; Lévay, G.I.; Greiner, I.; Natsumi, M.; Tatsuya, W.; et al. Fragment Based Optimization of Metabotropic Glutamate Receptor 2 (mGluR2) Positive Allosteric Modulators in the Absence of Structural Information. J. Med. Chem. 2019, 62, 234–246. [Google Scholar] [CrossRef]
- Tresadern, G.; Cid, J.-M.; Trabanco, A.A. QSAR design of triazolopyridine mGlu2 receptor positive allosteric modulators. J. Mol. Graph. Model. 2014, 53, 82–91. [Google Scholar] [CrossRef]
- Harrison, P.J.; Lyon, L.; Sartorius, L.J.; Burnet, P.; Lane, T.A. Review: The group II metabotropic glutamate receptor 3 (mGluR3, mGlu3, GRM3): Expression, function and involvement in schizophrenia. J. Psychopharmacol. 2008, 22, 308–322. [Google Scholar] [CrossRef]
- Maksymetz, J.; Moran, S.P.; Conn, P.J. Targeting metabotropic glutamate receptors for novel treatments of schizophrenia. Mol. Brain 2017, 10, 15. [Google Scholar] [CrossRef]
- Yao, Y.; Pattabiraman, N.; Michne, W.F.; Huang, X.-P.; Hampson, D.R. Molecular modeling and mutagenesis of the ligand-binding pocket of the mGlu3 subtype of metabotropic glutamate receptor. J. Neurochem. 2003, 86, 947–957. [Google Scholar] [CrossRef]
- DiRaddo, J.O.; Miller, E.J.; Bowman-Dalley, C.; Wroblewska, B.; Javidnia, M.; Grajkowska, E.; Wolfe, B.B.; Liotta, D.C.; Wroblewski, J.T. Chloride is an Agonist of Group II and III Metabotropic Glutamate Receptors. Mol. Pharmacol. 2015, 88, 450–459. [Google Scholar] [CrossRef]
- Tora, A.S.; Rovira, X.; Cao, A.-M.; Cabayé, A.; Olofsson, L.; Malhaire, F.; Scholler, P.; Baik, H.; Van Eeckhaut, A.; Smolders, I.; et al. Chloride ions stabilize the glutamate-induced active state of the metabotropic glutamate receptor 3. Neuropharmacology 2018, 140, 275–286. [Google Scholar] [CrossRef]
- Doornbos, M.L.J.; Van der Linden, I.; Vereyken, L.; Tresadern, G.; Ijzerman, A.P.; Lavreysen, H.; Heitman, L.H. Constitutive activity of the metabotropic glutamate receptor 2 explored with a whole-cell label-free biosensor. Biochem. Pharmacol. 2018, 152, 201–210. [Google Scholar] [CrossRef]
- Monn, J.A.; Valli, M.J.; Massey, S.M.; Wright, R.A.; Salhoff, C.R.; Johnson, B.G.; Howe, T.; Alt, C.A.; Rhodes, G.A.; Robey, R.L.; et al. Design, Synthesis, and Pharmacological Characterization of (+)-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY354740): A Potent, Selective, and Orally Active Group 2 Metabotropic Glutamate Receptor Agonist Possessing Anticonvulsant and Anxiolytic Properties. J. Med. Chem. 1997, 40, 528–537. [Google Scholar]
- Monn, J.A.; Massey, S.M.; Valli, M.J.; Henry, S.S.; Stephenson, G.A.; Bures, M.; Hérin, M.; Catlow, J.; Giera, D.; Wright, R.A.; et al. Synthesis and Metabotropic Glutamate Receptor Activity of S-Oxidized Variants of (−)-4-Amino-2-thiabicyclo-[3.1.0]hexane-4,6-dicarboxylate: Identification of Potent, Selective, and Orally Bioavailable Agonists for mGlu2/3 Receptors. J. Med. Chem. 2007, 50, 233–240. [Google Scholar] [CrossRef]
- Monn, J.A.; Prieto, L.; Taboada, L.; Pedregal, C.; Hao, J.; Reinhard, M.R.; Henry, S.S.; Goldsmith, P.J.; Beadle, C.D.; Walton, L.; et al. Synthesis and Pharmacological Characterization of C4-Disubstituted Analogs of 1S,2S,5R,6S-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a Potent, Selective Metabotropic Glutamate Receptor Agonist and Determination of Agonist-Bound Human mGlu2 and mGlu3 Amino Terminal Domain Structures. J. Med. Chem. 2015, 58, 1776–1794. [Google Scholar] [PubMed]
- Hopkins, C.R.; Lindsley, C.W.; Niswender, C.M. mGluR4-positive allosteric modulation as potential treatment for Parkinson’s disease. Future Med. Chem. 2009, 1, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Selvam, C.; Oueslati, N.; Lemasson, I.A.; Brabet, I.; Rigault, D.; Courtiol, T.; Cesarini, S.; Triballeau, N.; Bertrand, H.-O.; Goudet, C.; et al. A Virtual Screening Hit Reveals New Possibilities for Developing Group III Metabotropic Glutamate Receptor Agonists. J. Med. Chem. 2010, 53, 2797–2813. [Google Scholar] [CrossRef] [PubMed]
- Rovira, X.; Malhaire, F.; Scholler, P.; Rodrigo, J.; Gonzalez-Bulnes, P.; Llebaria, A.; Pin, J.-P.; Giraldo, J.; Goudet, C. Overlapping binding sites drive allosteric agonism and positive cooperativity in type 4 metabotropic glutamate receptors. FASEB J. 2014, 29, 116–130. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Villa, H.; Trabanco, A.A. Progress toward allosteric ligands of metabotropic glutamate 7 (mGlu7) receptor: 2008–present. MedChemComm 2019, 10, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Reed, C.W.; Yohn, S.E.; Washecheck, J.P.; Roenfanz, H.F.; Quitalig, M.C.; Luscombe, V.B.; Jenkins, M.T.; Rodriguez, A.L.; Engers, D.W.; Blobaum, A.L.; et al. Discovery of an Orally Bioavailable and Central Nervous System (CNS) Penetrant mGlu7 Negative Allosteric Modulator (NAM) in Vivo Tool Compound: N-(2-(1H-1,2,4-triazol-1-yl)-5-(trifluoromethoxy)phenyl)-4-(cyclopropylmethoxy)-3-methoxybenzamide (VU6012962). J. Med. Chem. 2019. [Google Scholar] [CrossRef] [PubMed]
- Tresadern, G.; Trabanco, A.A.; Pérez-Benito, L.; Overington, J.P.; van Vlijmen, H.W.T.; van Westen, G.J.P. Identification of Allosteric Modulators of Metabotropic Glutamate 7 Receptor Using Proteochemometric Modeling. J. Chem. Inf. Model. 2017, 57, 2976–2985. [Google Scholar] [CrossRef]
- Cid, J.M.; Lavreysen, H.; Tresadern, G.; Pérez-Benito, L.; Tovar, F.; Fontana, A.; Trabanco, A.A. Computationally Guided Identification of Allosteric Agonists of the Metabotropic Glutamate 7 Receptor. ACS Chem. Neurosci. 2018. [Google Scholar] [CrossRef]
- Cleva, R.M.; Olive, M.F. Metabotropic glutamate receptors and drug addiction. Wiley Interdiscip.Rev. Membr. Transp. Signal. 2012, 1, 281–295. [Google Scholar] [CrossRef]
- Duvoisin, R.M.; Pfankuch, T.; Wilson, J.M.; Grabell, J.; Chhajlani, V.; Brown, D.G.; Johnson, E.; Raber, J. Acute pharmacological modulation of mGluR8 reduces measures of anxiety. Behav. Brain Res. 2010, 212, 168–173. [Google Scholar] [CrossRef]
- Chen, Q.; Ho, J.D.; Ashok, S.; Vargas, M.C.; Wang, J.; Atwell, S.; Bures, M.; Schkeryantz, J.M.; Monn, J.A.; Hao, J. Structural Basis for (S)-3,4-Dicarboxyphenylglycine (DCPG) As a Potent and Subtype Selective Agonist of the mGlu8 Receptor. J. Med. Chem. 2018, 61, 10040–10052. [Google Scholar] [CrossRef]
- Bessis, A.-S.; Rondard, P.; Gaven, F.; Brabet, I.; Triballeau, N.; Prézeau, L.; Acher, F.; Pin, J.-P. Closure of the Venus flytrap module of mGlu8 receptor and the activation process: Insights from mutations converting antagonists into agonists. Proc. Natl. Acad. Sci. USA 2002, 99, 11097. [Google Scholar] [CrossRef]
- Schkeryantz, J.M.; Chen, Q.; Ho, J.D.; Atwell, S.; Zhang, A.; Vargas, M.C.; Wang, J.; Monn, J.A.; Hao, J. Determination of L-AP4-bound human mGlu8 receptor amino terminal domain structure and the molecular basis for L-AP4′s group III mGlu receptor functional potency and selectivity. Bioorg. Med. Chem. Lett. 2018, 28, 612–617. [Google Scholar] [CrossRef]
- Lenselink, E.B.; Louvel, J.; Forti, A.F.; van Veldhoven, J.P.D.; de Vries, H.; Mulder-Krieger, T.; McRobb, F.M.; Negri, A.; Goose, J.; Abel, R.; et al. Predicting Binding Affinities for GPCR Ligands Using Free-Energy Perturbation. ACS Omega 2016, 1, 293–304. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).