Transcriptomic Analysis of Vibrio parahaemolyticus Reveals Different Virulence Gene Expression in Response to Benzyl Isothiocyanate
Abstract
:1. Introduction
2. Results
2.1. Antibacterial Tests
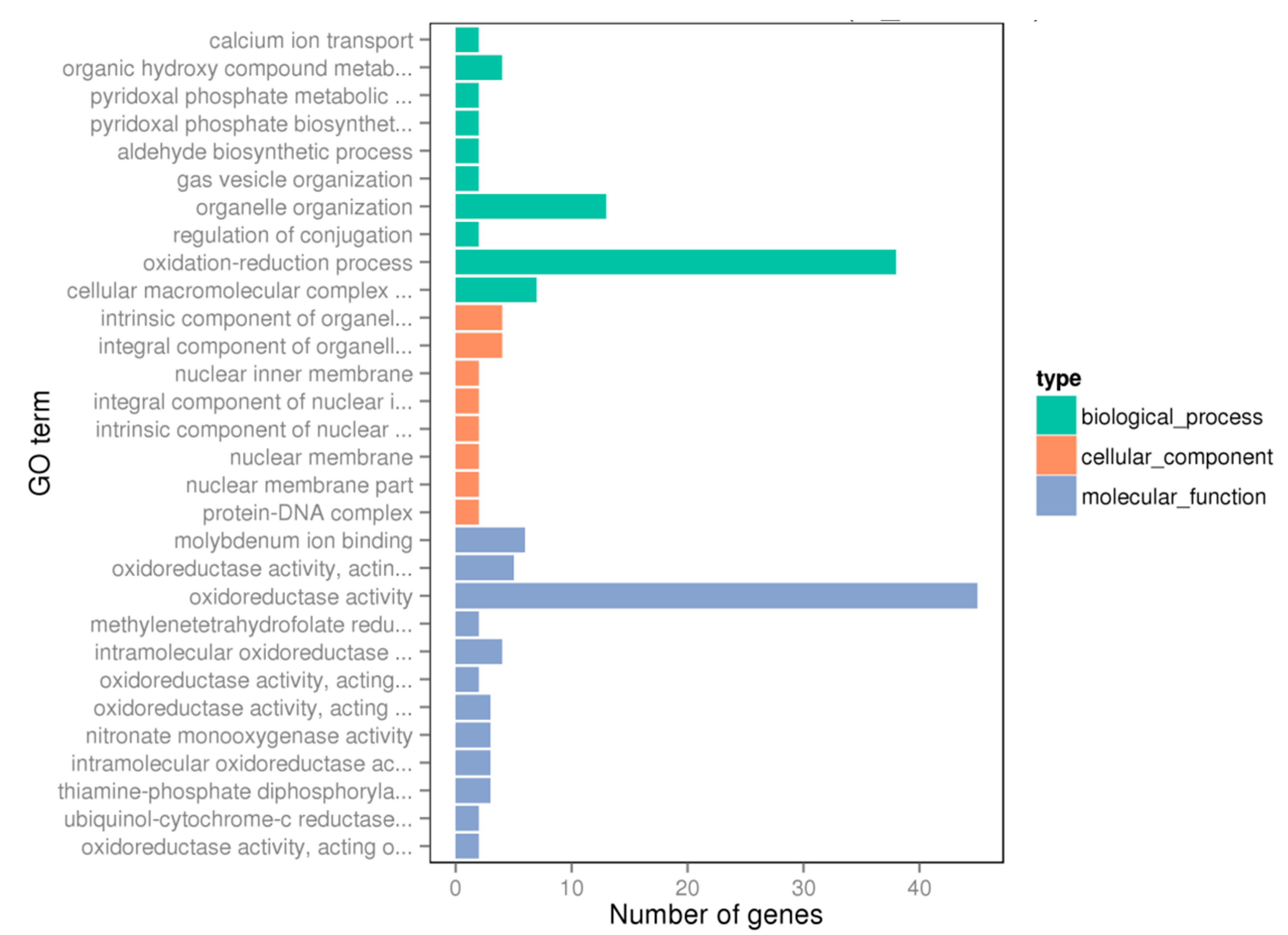
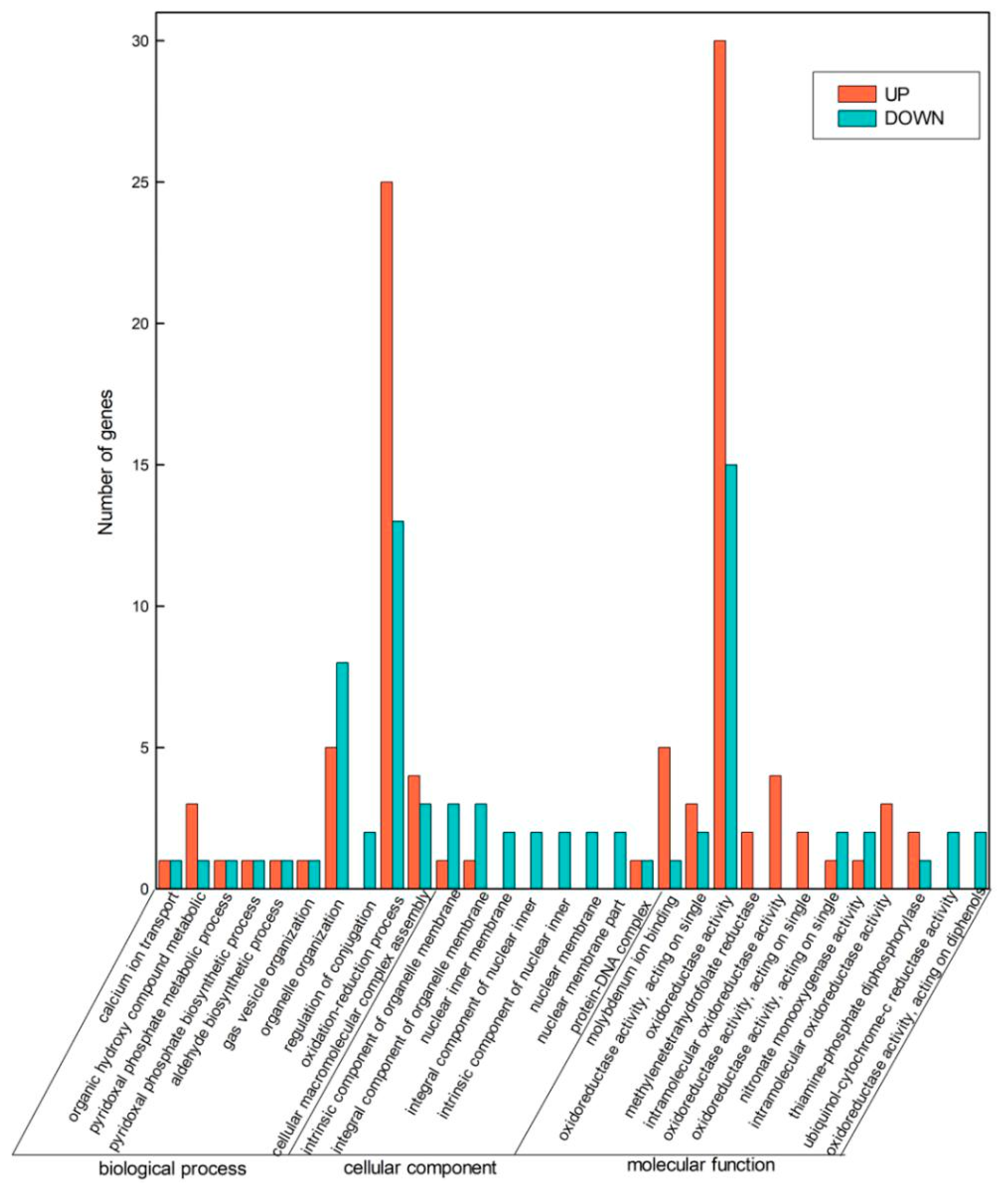
2.2. Global Changes at Transcriptome Level
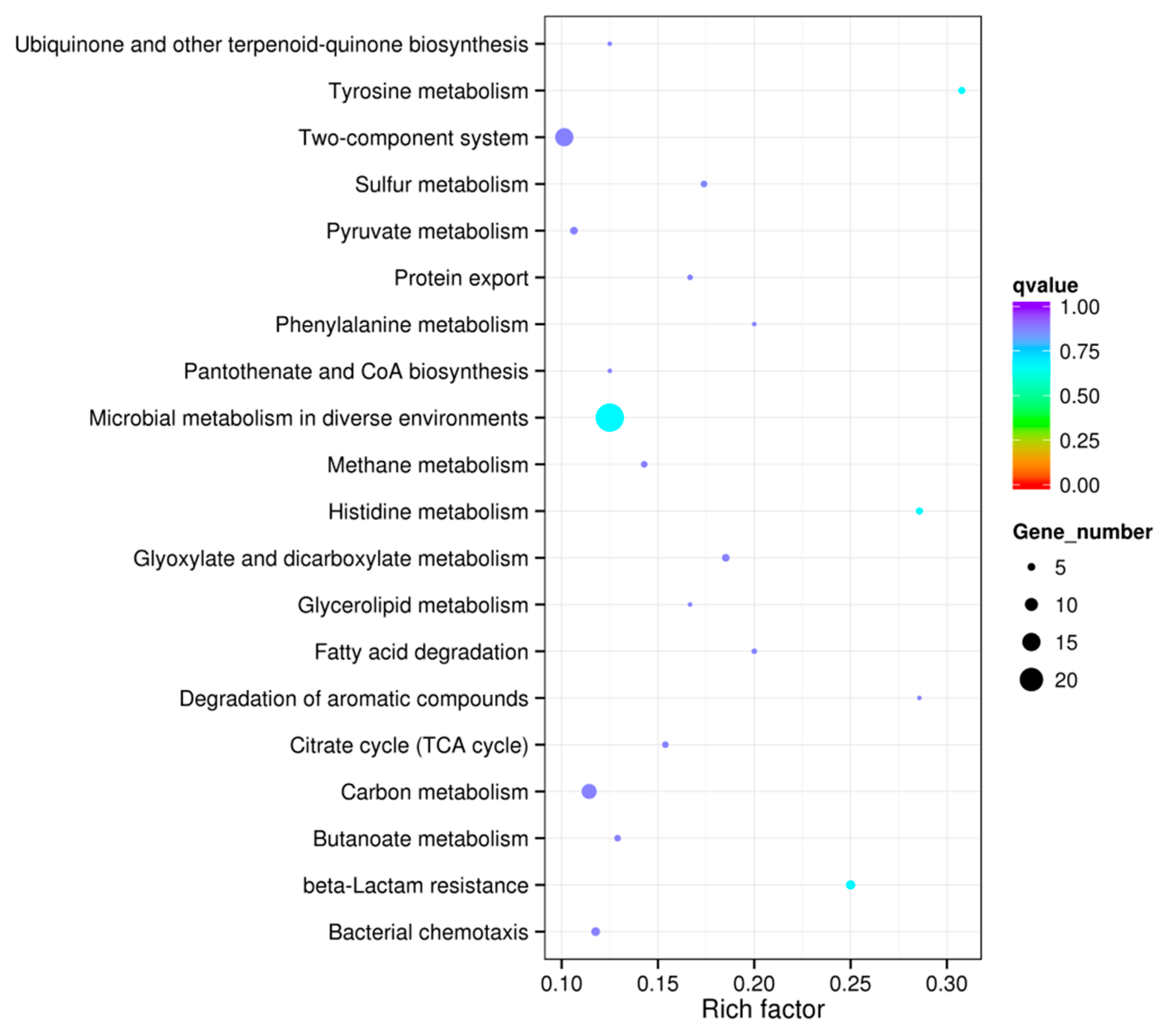
2.3. KEGG Pathway Analysis
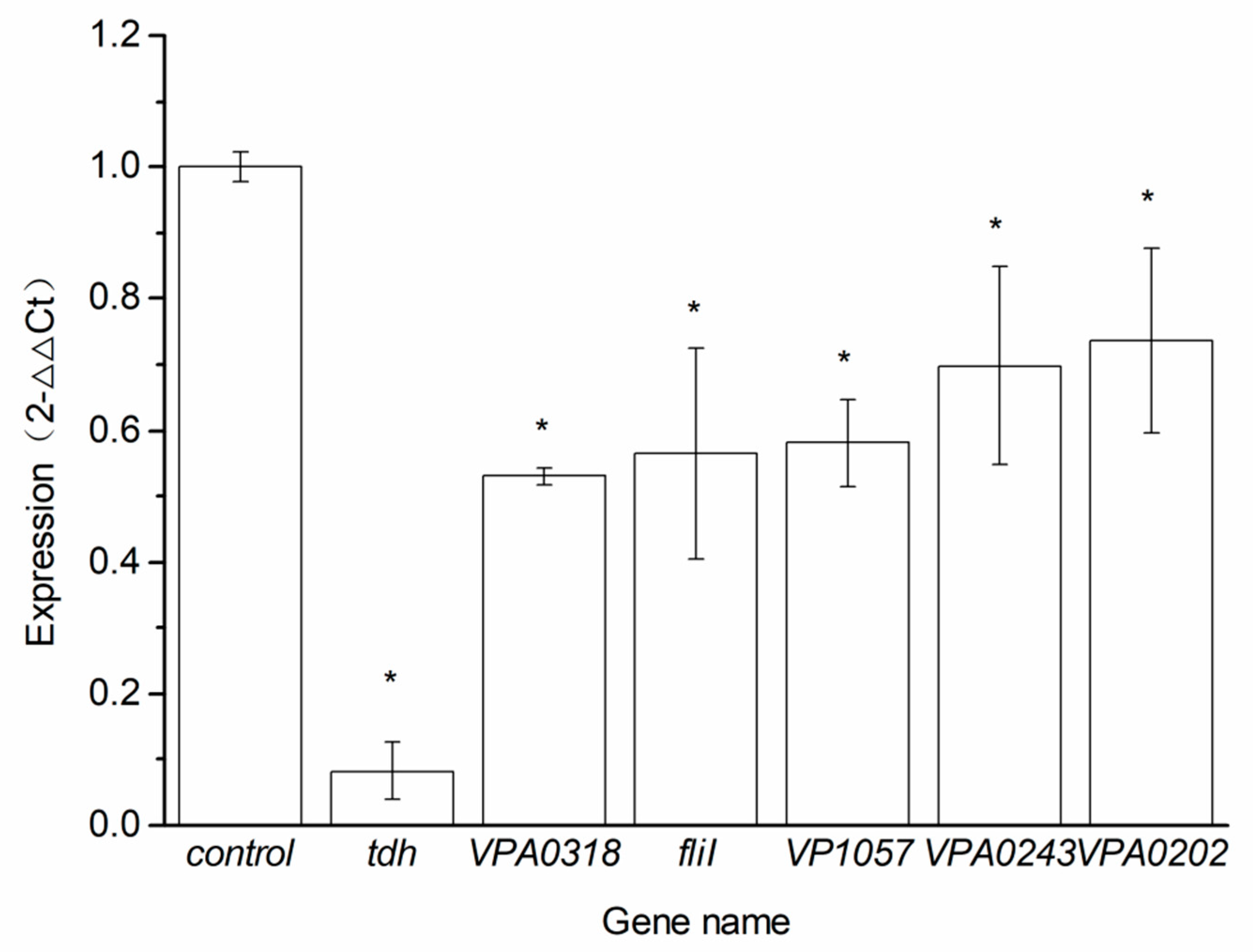
2.4. Genes Related to Virulence and Validation the RNA Sequencing Data by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Growth Conditions
4.2. Antimicrobial Tests
4.3. RNA Extraction
4.4. Library Preparation for Strand-Specific Transcriptome Sequencing
4.5. Bioinformatic Analysis
4.6. qRT-PCR Validation of Differentially Expressed Genes
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Shen, X.; Su, Y.C. Application of grape seed extract in depuration for decontaminating Vibrio parahaemolyticus in Pacific oysters (Crassostrea gigas). Food Control 2016, 73, 601–605. [Google Scholar] [CrossRef]
- Lovell, C.R. Ecological fitness and virulence features of Vibrio parahaemolyticus in estuarine environments. Appl. Microbiol. Biotech. 2017, 101, 1781–1794. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Ma, Y.; Fu, J.; Zhao, A.; Guo, Z.; Malakar, P.K.; Pan, Y.; Zhao, Y. Effect of temperature on pathogenic and non-pathogenic Vibrio parahaemolyticus biofilm formation. Food Contr. 2016, 73, 485–491. [Google Scholar] [CrossRef]
- Su, Y.C.; Liu, C. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 2007, 24, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.Y.; Li, W.; Dong, X.; Li, M.S. Benzyl-isothiocyanate induces apoptosis and inhibits migration and invasion of hepatocellular carcinoma cells in vitro. J. Cancer 2017, 8, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Meng, X.; Li, Y.; Zhao, C.N.; Tang, G.Y.; Li, H.B. Antibacterial and antifungal activities of spices. Int. J. Mol. Sci. 2017, 18, 1283–1321. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Zhang, K.; Gao, M.; Shi, C.; Ge, C.; Qu, D.; Zhu, J.; Shi, Y.; Han, J. Inactivation of Vibrio parahaemolyticus by aqueous ozone. J. Microbiol. Biotech. 2018, 28, 1233–1246. [Google Scholar]
- Morse, M.A.; Zu, H.; Galati, A.J.; Schmidt, C.J.; Stoner, G.D. Dose-related inhibition by dietary phenethyl isothiocyanate of esophageal tumorigenesis and DNA methylation induced by N-nitrosomethylbenzylamine in rats. Cancer Lett. 1993, 72, 103–110. [Google Scholar] [CrossRef]
- Smolinska, U.; Morra, M.J.; Knudsen, G.R.; James, R.L. Isothiocyanates produced by Brassicaceae species as inhibitors of Fusarium oxysporum. Plant Disease 2003, 87, 407–412. [Google Scholar] [CrossRef]
- Dufour, V.; Stahl, M.; Baysse, C. The antibacterial properties of isothiocyanates. Microbiology 2015, 161, 229–243. [Google Scholar] [CrossRef] [Green Version]
- Nakano, M.; Takahashi, A.; Su, Z.; Harada, N.; Mawatari, K.; Nakaya, Y. Hfq regulates the expression of the thermostable direct hemolysin gene in Vibrio parahaemolyticus. BMC Microbiol. 2008, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Shirai, H.; Ito, H.; Hirayama, T.; Nakamoto, Y.; Nakabayashi, N.; Kumagai, K.; Nishibuchi, M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 1990, 58, 3568–3573. [Google Scholar] [PubMed]
- Banu, S.F.; Rubini, D.; Murugan, R.; Vadivel, V.; Gowrishankar, S.; Nithyanand, P. Exploring the antivirulent and sea food preservation efficacy of essential oil combined with DNase on Vibrio parahaemolyticus. LWT 2018, 95, 107–115. [Google Scholar] [CrossRef]
- Tan, X.J.; Qin, N.; Wu, C.Y. Transcriptome analysis of the biofilm formed by methicillinsusceptible Staphylococcus aureus. Sci. Rep. 2015, 5, 11997. [Google Scholar] [CrossRef] [PubMed]
- Pieta, L.; Escudero, F.L.G.; Jacobus, A.P. Comparative transcriptomic analysis of Listeria monocytogenes reveals upregulation of stress genes and downregulation of virulence genes in response to essential oil extracted from Baccharis psiadioides. Ann. Microbiol. 2017, 67, 479–490. [Google Scholar] [CrossRef]
- Aagesen, A.M.; Phuvasate, S.; Su, Y.C. Characterizing the adherence profiles of virulent Vibrio parahaemolyticus isolates. Microb. Ecol. 2018, 75, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Z.; Zhong, Y.F.; Gu, X.S. The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Front. Microbiol. 2015, 6, 144. [Google Scholar] [CrossRef] [PubMed]
- Sofrata, A.; Santangelo, E.M.; Azeem, M.; Gustafsson, A. Benzyl isothiocyanate, a major component from the roots of Salvadora persica is highly active against gram-negative bacteria. PLoS ONE 2011, 6, e23045. [Google Scholar] [CrossRef] [PubMed]
- Hong, L. Analysis of the collective food poisoning events in Shanghai from 1990 to 2000. Chinese J. Nat. Med. 2003, 5, 17–20. [Google Scholar]
- Romocastillo, M.; Andrade, A.; Espinosa, N. EscO, a functional and structural analog of the flagellar FliJ protein, is a positive regulator of EscN ATPase activity of the enteropathogenic Escherichia coli injectisome. J. Bacteriol. 2014, 196, 2227–2241. [Google Scholar] [CrossRef] [PubMed]
- Claret, L.; Calder, S.R.; Higgins, M.; Hughes, C. Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol. Microbiol. 2010, 48, 1349–1355. [Google Scholar] [CrossRef]
- Cornelis, G.R. The type III secretion injectisome. Nat. Rev. Microbiol. 2006, 4, 811–825. [Google Scholar] [CrossRef] [PubMed]
- Haiko, J.; Westerlund-Wikström, B. The role of the bacterial flagellum in adhesion and virulence. Biology 2013, 2, 1242–1267. [Google Scholar] [CrossRef] [PubMed]
- Detweiler, C.S.; Monack, D.M.; Brodsky, I.E.; Mathew, H.; Falkow, S. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 2010, 48, 385–400. [Google Scholar] [CrossRef]
- Spencer, H.; Karavolos, M.H.; Bulmer, D.M.; Aldridge, P.; Chhabra, S.R.; Winzer, K.; Williams, P.; Khan, C.M.A. Genome-wide transposon mutagenesis identifies a role for host neuroendocrine stress hormones in regulating the expression of virulence genes in Salmonella. J. Bacteriol. 2010, 192, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.M.; Moreira, L.M.; Ferro, J.A.; Soares, M.R.; Laia, M.L.; Varani, A.M.; de Oliveira, J.C.; Ferro, M.I. Unravelling potential virulence factor candidates in Xanthomonas citri. subsp. citri by secretome analysis. Peer J. 2016, 4, e1734. [Google Scholar] [CrossRef]
- Gabriela, T.P.; Lucia, C.D.A.; Humberto, L.M.; Ken, T.; Fernando, N.G. VirK is a periplasmic protein required for efficient secretion of plasmid-encoded toxin from enteroaggregative Escherichia coli. Infect. Immun. 2012, 80, 2276–2285. [Google Scholar]
- Abdelhamed, H.; Lu, J.; Lawrence, M.L.; Karsi, A. Involvement of tolQ and tolR genes in Edwardsiella ictaluri virulence. Microb. Pathogenesis 2016, 100, 90–94. [Google Scholar] [CrossRef]
- Heilpern, A.J.; Waldor, M.K. CTXphi infection of Vibrio cholerae requires the tolQRA gene products. J. Bacteriol. 2000, 182, 1739–1747. [Google Scholar] [CrossRef]
- Bowe, F.; Lipps, C.J.; Tsolis, R.M.; Groisman, E.; Heffron, F.; Kusters, J.G. At least four percent of the Salmonella typhimurium genome is required for fatal infection of mice. Infect. Immun. 1998, 66, 3372–3377. [Google Scholar]
- Judith, H.; Jr, R.J.; Meganm, T.; Jennifere, A.; Hshaw, W. Bacterial peptidoglycan-associated lipoprotein is released into the bloodstream in gram-negative sepsis and causes inflammation and death in mice. J. Biol. Chem. 2002, 277, 14274–14280. [Google Scholar]
- Tamayo, R.; Ryan, S.S.; Mccoy, A.J. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar typhimurium. Infect. Immun. 2002, 70, 6770–6778. [Google Scholar] [CrossRef] [PubMed]
- Paterson, G.K.; Northen, H.; Cone, D.B.; Willers, C.; Peters, S.E.; Maskell, D.J. Deletion of tolA in Salmonella Typhimurium generates an attenuated strain with vaccine potential. Microbiology 2009, 155, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Mccarter, L.L. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J. Bacteriol. 2007, 189, 4094–4107. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Beyhan, S.; Yildiz, F.H. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J. Bacteriol. 2007, 189, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.P.; Wang, C.; Ye, M.Z. Differentially expressed outer membrane proteins of Vibrio alginolyticusin response to six types of antibiotics. Mar. Biotech. 2010, 12, 686–695. [Google Scholar] [CrossRef]
- KIM, M.S.; JIN, J.W.; JUNG, S.H. Genetic variations of outer membrane protein genes of Vibrio harveyi isolated in Korea and immunogenicity of OmpW in olive flounder, Paralichthys olivaceus. J. Fish. Mar. Sci. Educ. 2015, 27, 1508–1521. [Google Scholar]
- Taylor, R.K.; Manoil, C.; Mekalanos, J.J. Broad-host-range vectors for delivery of TnphoA: Use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J. Bacteriol. 1989, 171, 1870–1878. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

| Gene | Primer | Sequence (5′→3′) |
|---|---|---|
| 16S rRNA | 16S rRNA-F | TATCCTTGTTTGCCAGCGAG |
| 16S rRNA-R | CTACGACGCACTTTTTGGGA | |
| tdh | tdh-F | GGCATTTGGATGACCGAAGTA |
| tdh-R | CTGACCAATCGCAACCACTTC | |
| VPA0318 | VPA0318-F | AGGTTACTTAGCGGGTGCG |
| VPA0318-R | TTCACGGTCTTTGATGCC | |
| fliI | fliI-F | TGCGGAACCCATCAACCC |
| fliI-R | CGTCCGTCTTCGCCCAAA | |
| VP1057 | VP1057-F | CGGTTCAATCAGCCCATAC |
| VP1057-R | AACGCTTCTGCGATACCTG | |
| VPA0243 | VPA0243-F | AACGCTTCTGCGATACCTG |
| VPA0243-R | TTGCCATAGTGCGTCGTAGTCG | |
| VPA0202 | VPA0202-F | CGAAGAAGTGATGGTGGTG |
| VPA0202-R | CTCGCATTGGTGAGTTGACG |
| Sample Name | Raw Reads | Clean Reads | Clean Bases (Gb) | Error (%) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| C_BITC1 | 11,409,316 | 10,480,246 | 1.57 | 0.02 | 97.13 | 92.51 | 48.29 |
| C_BITC2 | 12,152,830 | 11,176,478 | 1.68 | 0.02 | 96.93 | 92.11 | 48.39 |
| C_BITC3 | 11,803,196 | 11,105,878 | 1.67 | 0.02 | 97.00 | 92.28 | 48.35 |
| Q_BITC1 | 12,650,370 | 11,915,336 | 1.79 | 0.02 | 97.06 | 92.39 | 48.20 |
| Q_BITC2 | 9,990,268 | 9,170,324 | 1.38 | 0.02 | 97.05 | 92.36 | 48.01 |
| Q_BITC3 | 12,204,690 | 11,470,672 | 1.72 | 0.02 | 97.14 | 92.53 | 48.08 |
| Gene_ID | Gene Name | log2 Fold Change (Q_BITC vs. C_BITC) | Pval (Q_BITC vs. C_BITC) | Padj (Q_BITC vs. C_BITC) | Significant (Q_BITC vs. C_BITC) |
|---|---|---|---|---|---|
| VPA1509 | tdh | −0.32858 | 0.04469 | 0.62131 | DOWN |
| VPA0318 | - | −0.95983 | 8.44E−10 | 7.18E−07 | DOWN |
| VP2246 | fliI | −0.3946 | 0.029624 | 0.57503 | DOWN |
| VP1057 | - | −0.38624 | 0.049817 | 0.63785 | DOWN |
| VPA0243 | - | −0.49488 | 0.0045398 | 0.19316 | DOWN |
| VPA0202 | - | −0.4573 | 0.030909 | 0.58184 | DOWN |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Hou, H.-M.; Wu, H.-Y.; Li, K.-X.; Wang, Y.; Zhou, Q.-Q.; Zhang, G.-L. Transcriptomic Analysis of Vibrio parahaemolyticus Reveals Different Virulence Gene Expression in Response to Benzyl Isothiocyanate. Molecules 2019, 24, 761. https://doi.org/10.3390/molecules24040761
Song J, Hou H-M, Wu H-Y, Li K-X, Wang Y, Zhou Q-Q, Zhang G-L. Transcriptomic Analysis of Vibrio parahaemolyticus Reveals Different Virulence Gene Expression in Response to Benzyl Isothiocyanate. Molecules. 2019; 24(4):761. https://doi.org/10.3390/molecules24040761
Chicago/Turabian StyleSong, Jie, Hong-Man Hou, Hong-Yan Wu, Ke-Xin Li, Yan Wang, Qian-Qian Zhou, and Gong-Liang Zhang. 2019. "Transcriptomic Analysis of Vibrio parahaemolyticus Reveals Different Virulence Gene Expression in Response to Benzyl Isothiocyanate" Molecules 24, no. 4: 761. https://doi.org/10.3390/molecules24040761





