Surface Enhanced Raman Spectroscopy for DNA Biosensors—How Far Are We?
Abstract
1. Introduction
1.1. Mechanism of Surface-Enhanced Raman Spectroscopy
1.2. Currently Developed Methods for Detecting DNA Mutations
1.2.1. Allele-Specific PCR
1.2.2. qPCR Quantitative Polymerase Chain Reaction (qPCR)
1.2.3. PCR-HRM (High Resolution Melt)
1.2.4. Sanger Sequencing
1.2.5. ddPCR
1.2.6. NGS
2. Characteristics of SERS-Based Sensors—General Approach
2.1. Label-Free Sensors
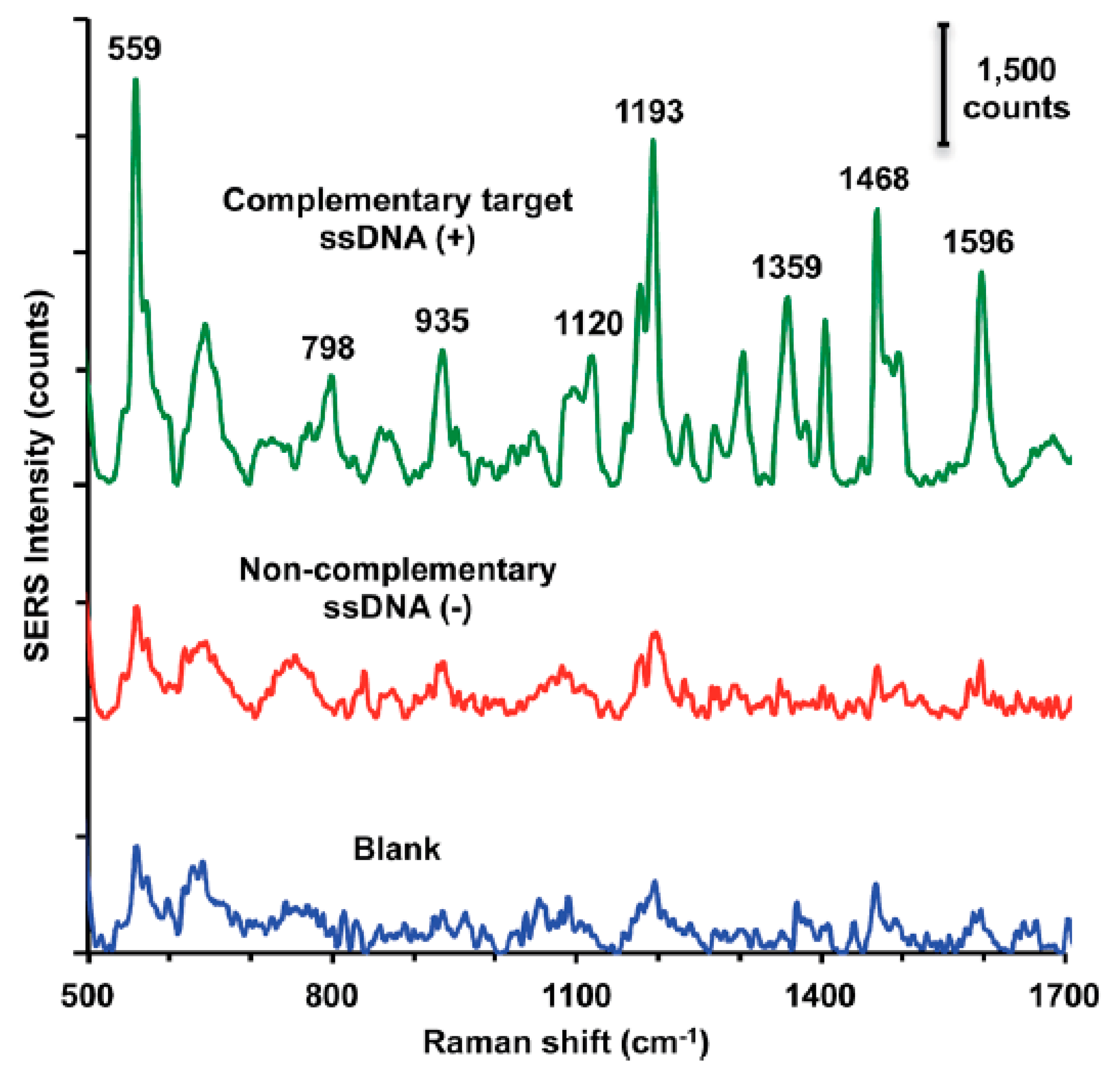
2.2. Typical Sensors with Labels
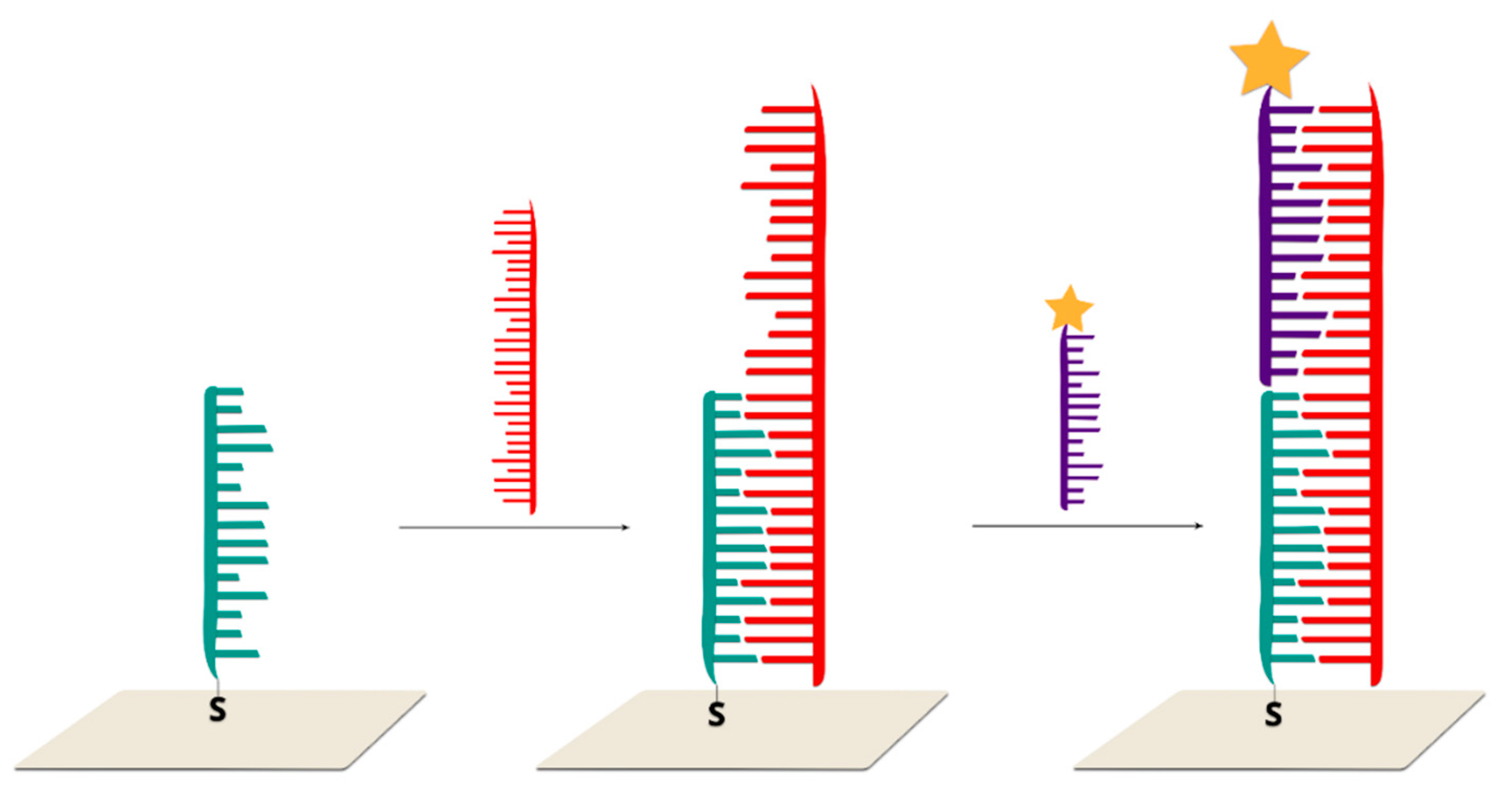
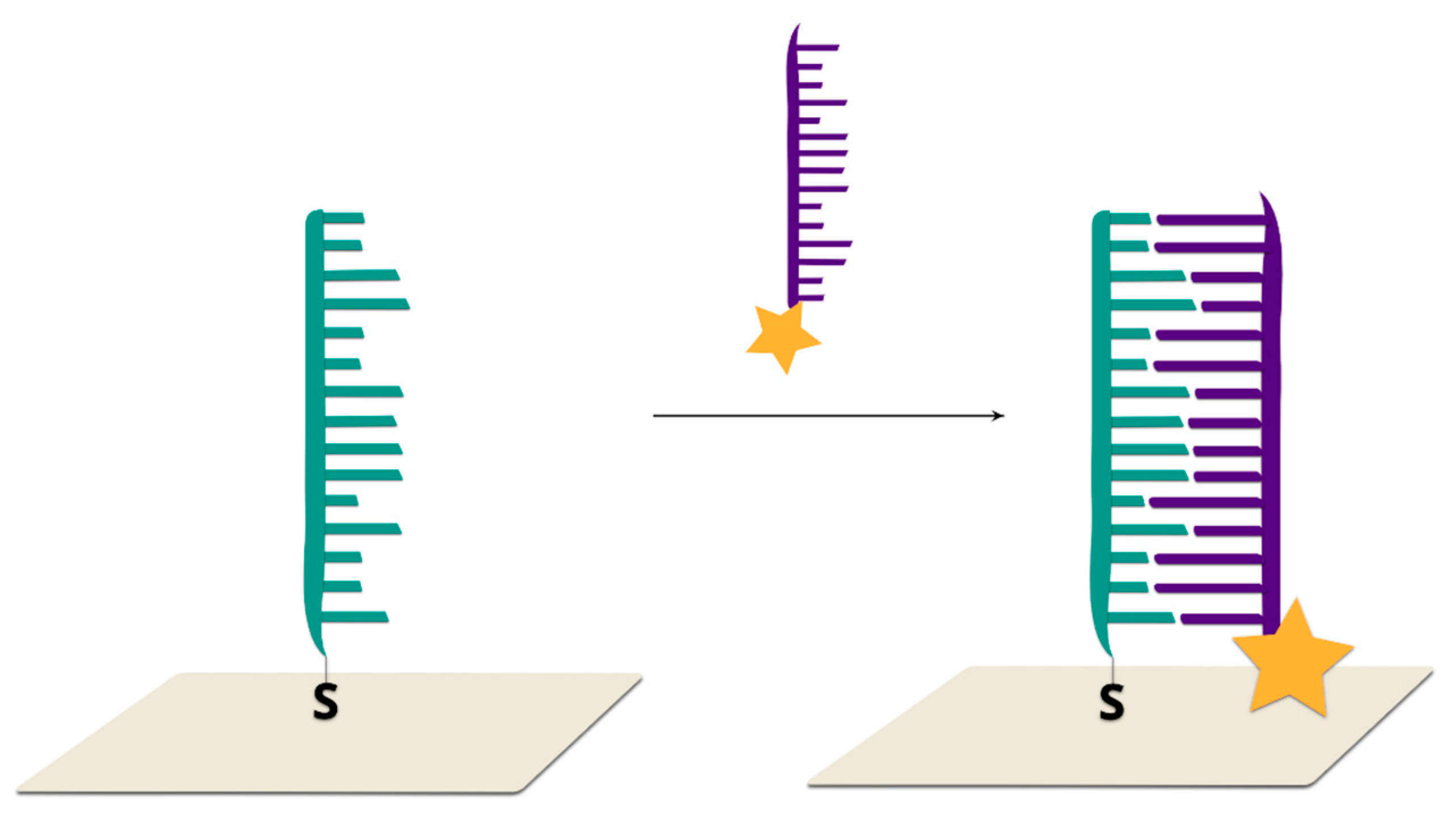
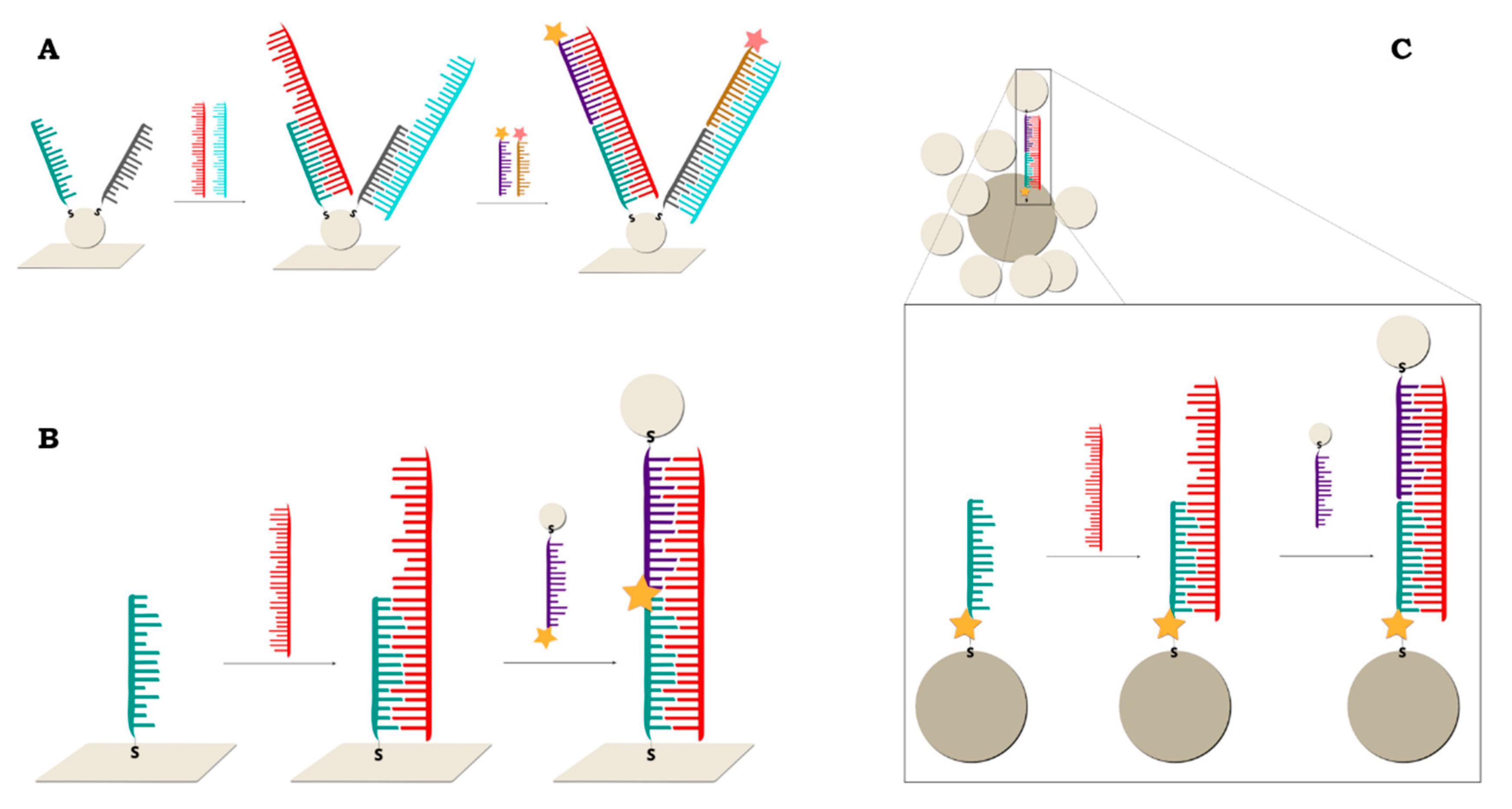
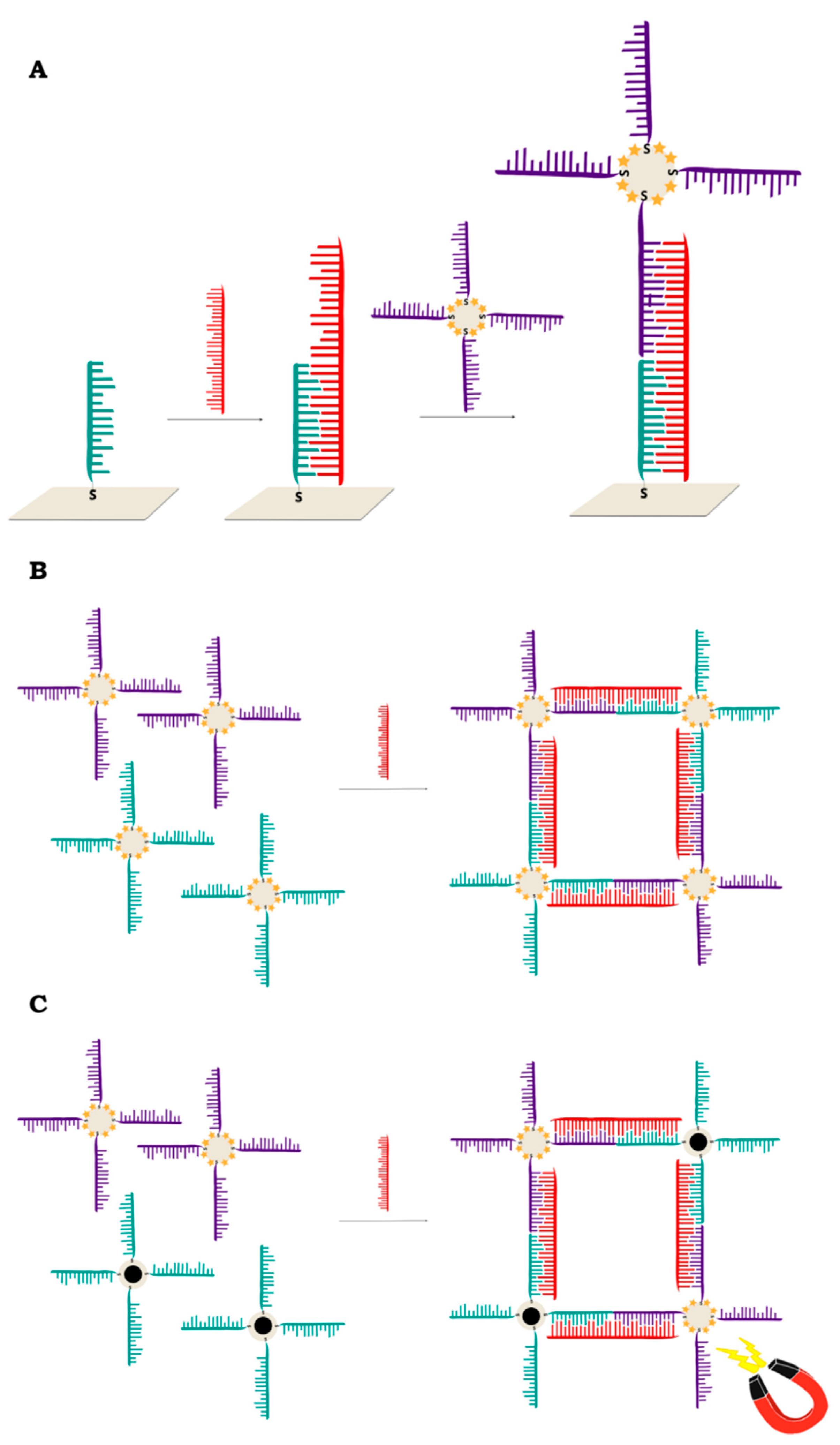
2.2.1. Sandwich Sensors with RR Connected to DNA
2.2.2. Sandwich Sensors with RR Located on Nanoparticles
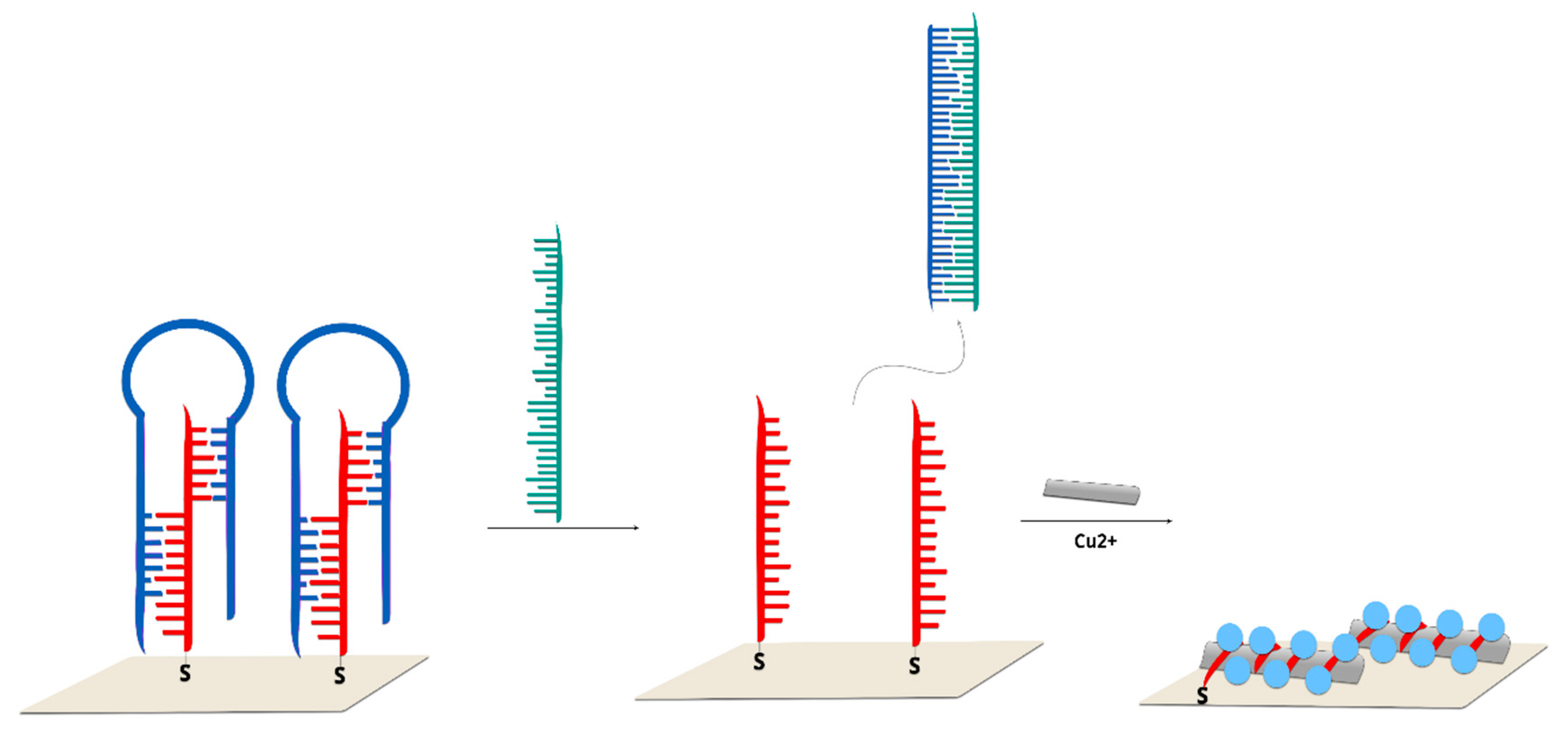
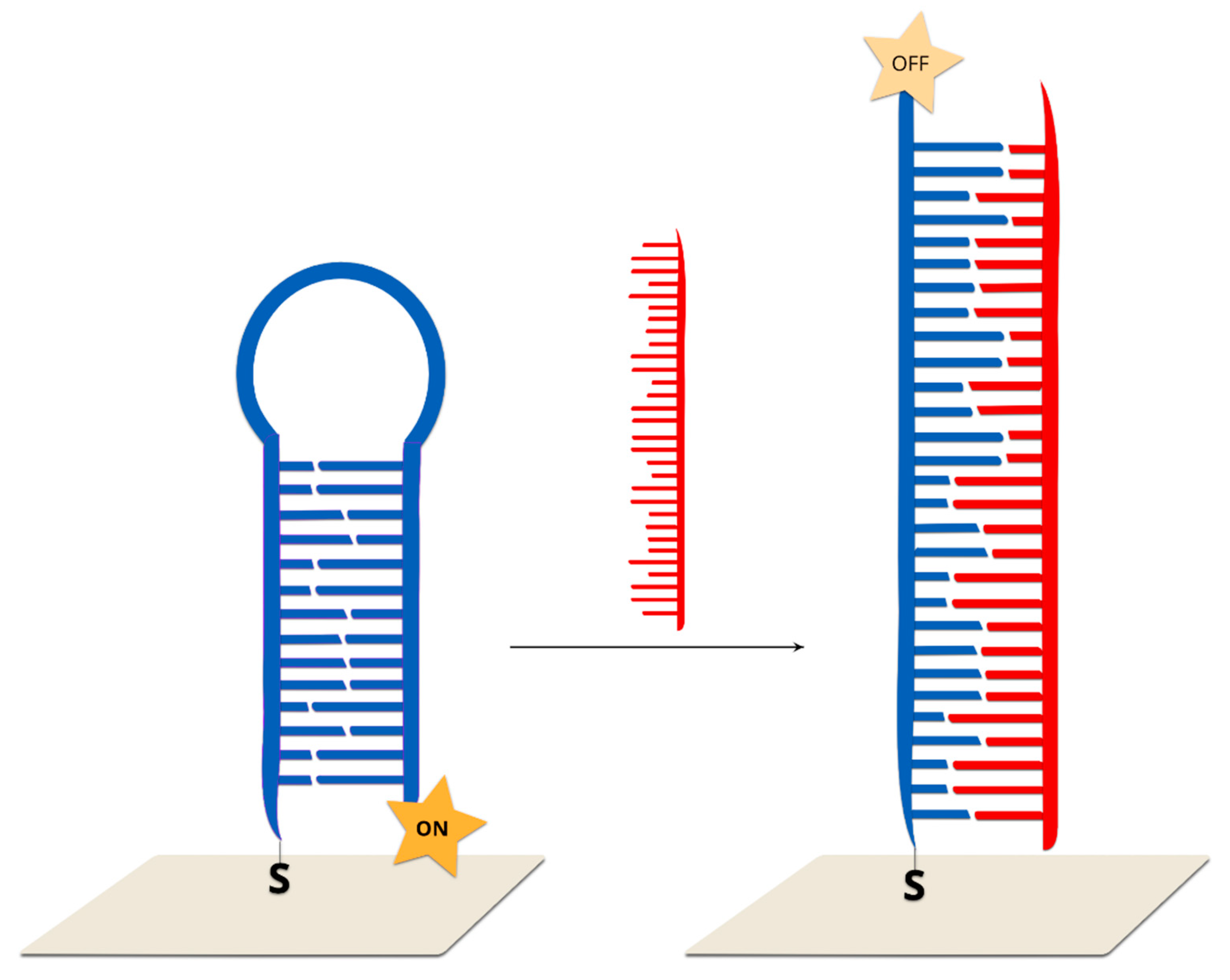
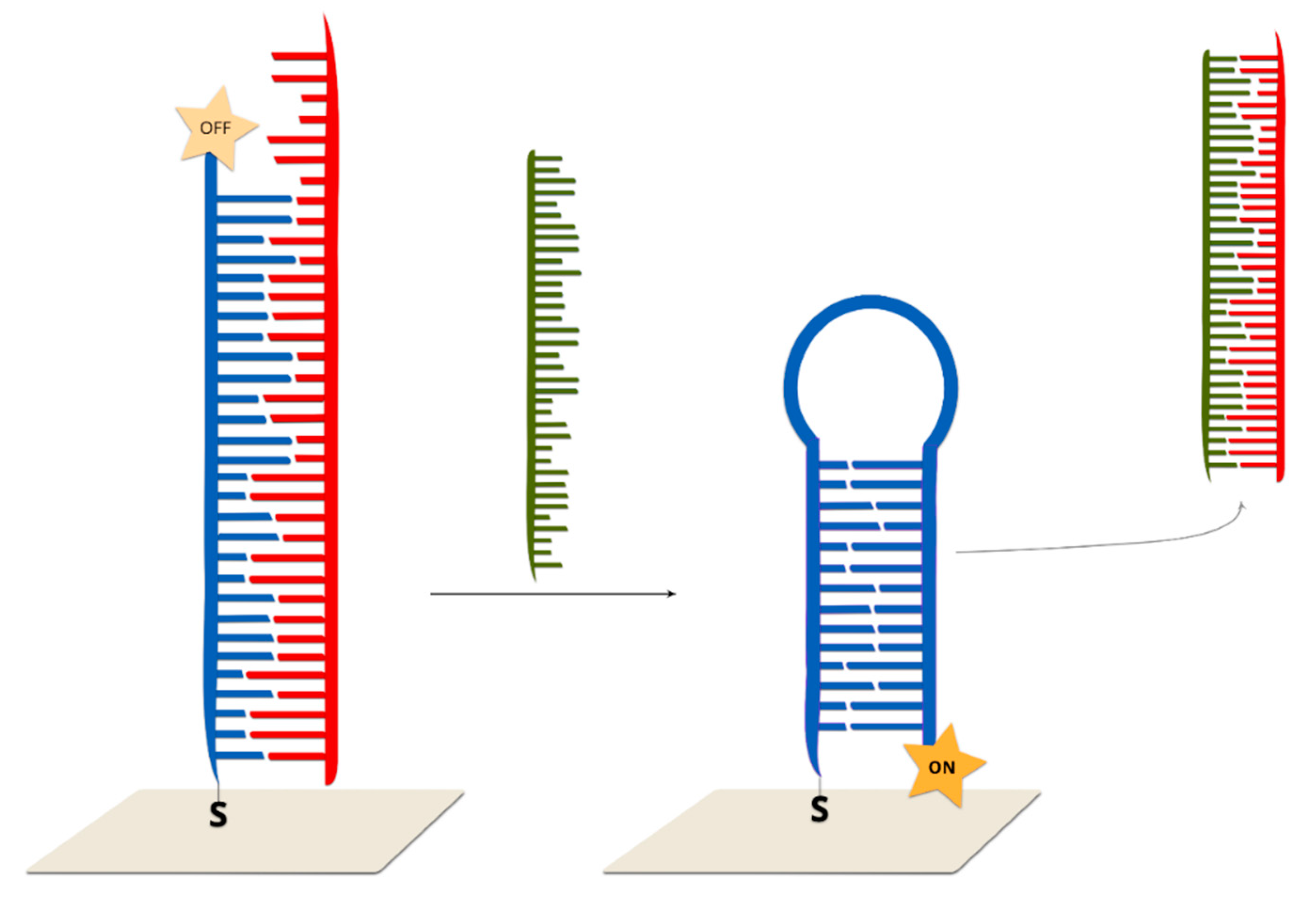
2.2.3. Hairpin On-Off Sensors
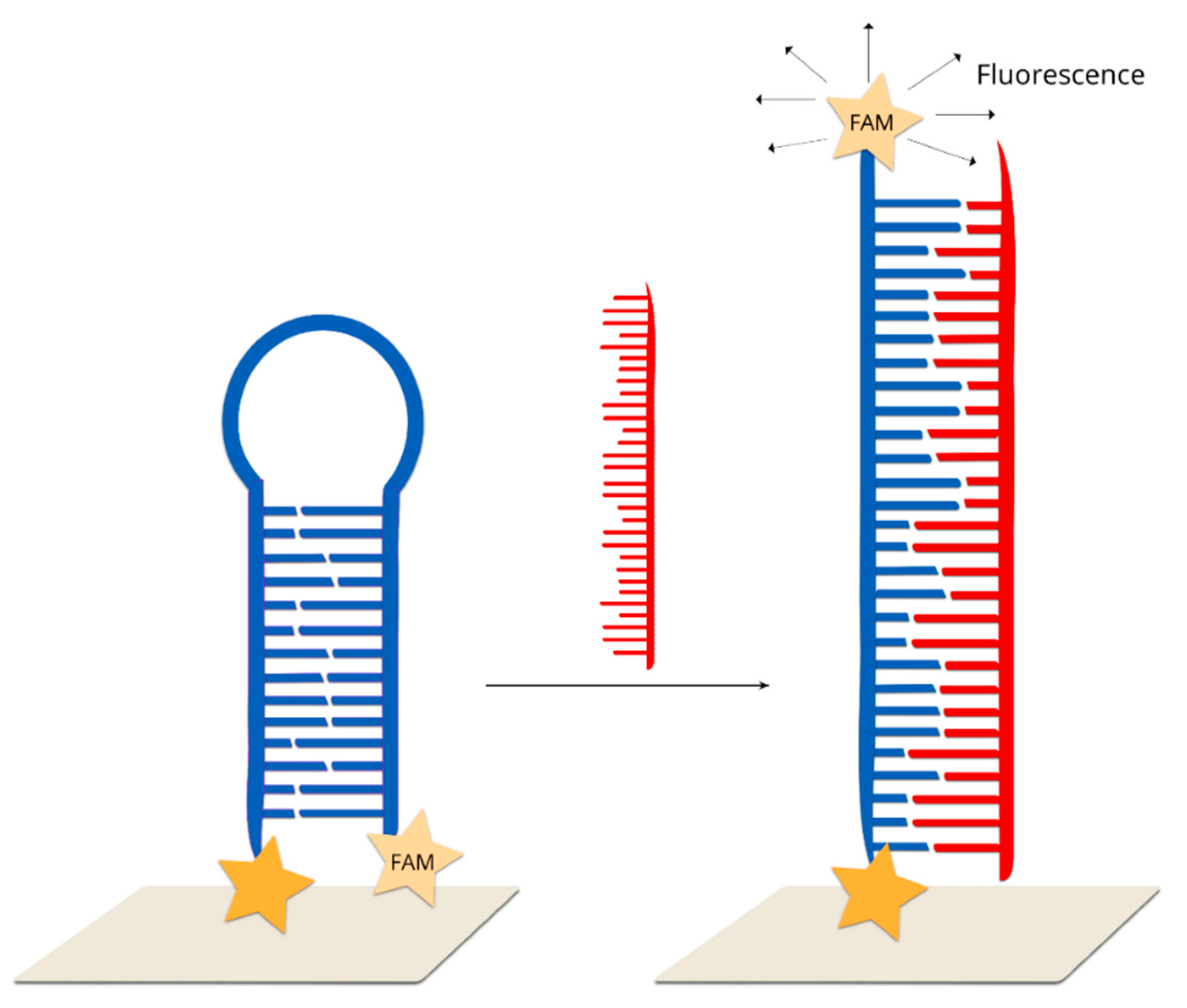
2.2.4. Hairpin Off-On Sensors
2.2.5. Other Examples
3. Summary, Conclusions and Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Syeda, M.M.; Wiggins, J.M.; Corless, B.; Spittle, C.; Karlin-Neumann, G.; Polsky, D. Validation of Circulating Tumor DNA Assays for Detection of Metastatic Melanoma. In Biomarkers for Immunotherapy of Cancer: Methods and Protocols; Thurin, M., Cesano, A., Marincola, F.M., Eds.; Springer: New York, NY, USA, 2020; pp. 155–180. [Google Scholar]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Moskovits, M. Surface-enhanced spectroscopy. Rev. Mod. Phys. 1985, 57, 783–826. [Google Scholar] [CrossRef]
- Albrecht, M.G.; Creighton, J.A. Anomalously intense Raman spectra of pyridine at a silver electrode. J. Am. Chem. Soc. 1977, 99, 5215–5217. [Google Scholar] [CrossRef]
- Jeanmaire, D.L.; Van Duyne, R.P. Surface raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chem. Interfacial Electrochem. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Ni, J.; Lipert, R.J.; Dawson, G.B.; Porter, M.D. Immunoassay Readout Method Using Extrinsic Raman Labels Adsorbed on Immunogold Colloids. Anal. Chem. 1999, 71, 4903–4908. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Su, S.; Guo, Y.; Jiang, X.; Zhong, Y.; Su, Y.; Fan, C.; Lee, S.-T.; He, Y. A Molecular Beacon-Based Signal-Off Surface-Enhanced Raman Scattering Strategy for Highly Sensitive, Reproducible, and Multiplexed DNA Detection. Small 2013, 9, 2493–2499. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-N.; Fales, A.M.; Vo-Dinh, T. Plasmonics-based SERS nanobiosensor for homogeneous nucleic acid detection. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 811–814. [Google Scholar] [CrossRef]
- Van Lierop, D.; Faulds, K.; Graham, D. Separation Free DNA Detection Using Surface Enhanced Raman Scattering. Anal. Chem. 2011, 83, 5817–5821. [Google Scholar] [CrossRef]
- Kowalczyk, A.; Krajczewski, J.; Kowalik, A.; Weyher, J.L.; Dzięcielewski, I.; Chłopek, M.; Góźdź, S.; Nowicka, A.M.; Kudelski, A. New strategy for the gene mutation identification using surface enhanced Raman spectroscopy (SERS). Biosens. Bioelectron. 2019, 132, 326–332. [Google Scholar] [CrossRef]
- Lombardi, J.R.; Birke, R.L. A Unified View of Surface-Enhanced Raman Scattering. Acc. Chem. Res. 2009, 42, 734–742. [Google Scholar] [CrossRef]
- Lombardi, J.R.; Birke, R.L. A Unified Approach to Surface-Enhanced Raman Spectroscopy. J. Phys. Chem. C 2008, 112, 5605–5617. [Google Scholar] [CrossRef]
- Pilot, R.; Signorini, R.; Durante, C.; Orian, L.; Bhamidipati, M.; Fabris, L. A Review on Surface-Enhanced Raman Scattering. Biosensors 2019, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Willets, K.A.; Van Duyne, R.P. Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu. Rev. Phys. Chem. 2007, 58, 267–297. [Google Scholar] [CrossRef] [PubMed]
- Stiles, P.L.; Dieringer, J.A.; Shah, N.C.; Van Duyne, R.P. Surface-Enhanced Raman Spectroscopy. Annu. Rev. Anal. Chem. 2008, 1, 601–626. [Google Scholar] [CrossRef] [PubMed]
- Jaworska, A.; Pyrak, E.; Kudelski, A. Comparison of the efficiency of generation of Raman radiation by various Raman reporters connected via DNA linkers to different plasmonic nano-structures. Vib. Spectrosc. 2019, 101, 34–39. [Google Scholar] [CrossRef]
- Ding, S.-Y.; You, E.-M.; Tian, Z.-Q.; Moskovits, M. Electromagnetic theories of surface-enhanced Raman spectroscopy. Chem. Soc. Rev. 2017, 46, 4042–4076. [Google Scholar] [CrossRef]
- Kennedy, B.J.; Spaeth, S.; Dickey, M.; Carron, K.T. Determination of the Distance Dependence and Experimental Effects for Modified SERS Substrates Based on Self-Assembled Monolayers Formed Using Alkanethiols. J. Phys. Chem. B 1999, 103, 3640–3646. [Google Scholar] [CrossRef]
- Otto, A. On the contribution of charge transfer excitations to SERS. J. Electr. Spectrosc. Relat. Phenom. 1983, 29, 329–342. [Google Scholar] [CrossRef]
- Kudelski, A.; Bukowska, J. Excitation profiles versus potential profiles in the determination of the charge-transfer contribution to SERS of pyridine on a silver electrode. J. Ram. Spectrosc. 1995, 26, 955–958. [Google Scholar] [CrossRef]
- Wang, J. Electrochemical biosensors: Towards point-of-care cancer diagnostics. Biosens. Bioelectron. 2006, 21, 1887–1892. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Wu, X.; Tong, X.; Wang, X.; Chang, Z.; Mao, Y.; Chen, X.; Sun, J.; Wang, Z.; Hong, Z.; et al. Circulating Tumor DNA Mutation Profiling by Targeted Next Generation Sequencing Provides Guidance for Personalized Treatments in Multiple Cancer Types. Sci. Rep. 2017, 7, 583. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Bugawan, T.L.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Analysis of enzymatically amplified β-globin and HLA-DQα DNA with allele-specific oligonucleotide probes. Nature 1986, 324, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Walczyk, A.; Kowalska, A.; Kowalik, A.; Sygut, J.; Wypiórkiewicz, E.; Chodurska, R.; Pięciak, L.; Góźdź, S. The BRAF(V600E) mutation in papillary thyroid microcarcinoma: Does the mutation have an impact on clinical outcome? Clin. Endocrinol. (Oxf) 2014, 80, 899–904. [Google Scholar] [CrossRef]
- Ashida, A.; Uhara, H.; Mikoshiba, A.; Sakaizawa, K.; Kumagai, N.; Koga, H.; Okuyama, R. Melanoma with BRAF Mutation in Circulating Cell-free DNA despite no Mutation in the Primary Lesion: A Case Report. Acta Derm. Venereol. 2016, 96, 128–129. [Google Scholar] [CrossRef]
- Schlücker, S. Surface-Enhanced Raman Spectroscopy: Concepts and Chemical Applications. Angew. Chem. Int. Ed. 2014, 53, 4756–4795. [Google Scholar] [CrossRef]
- Garcia-Rico, E.; Alvarez-Puebla, R.A.; Guerrini, L. Direct surface-enhanced Raman scattering (SERS) spectroscopy of nucleic acids: From fundamental studies to real-life applications. Chem. Soc. Rev. 2018, 47, 4909–4923. [Google Scholar] [CrossRef]
- Morla-Folch, J.; Gisbert-Quilis, P.; Masetti, M.; Garcia-Rico, E.; Alvarez-Puebla, R.A.; Guerrini, L. Conformational SERS Classification of K-Ras Point Mutations for Cancer Diagnostics. Angew. Chem. Int. Ed. 2017, 56, 2381–2385. [Google Scholar] [CrossRef]
- Ngo, H.T.; Wang, H.-N.; Fales, A.M.; Vo-Dinh, T. Label-Free DNA Biosensor Based on SERS Molecular Sentinel on Nanowave Chip. Anal. Chem. 2013, 85, 6378–6383. [Google Scholar] [CrossRef]
- Barhoumi, A.; Halas, N.J. Label-Free Detection of DNA Hybridization Using Surface Enhanced Raman Spectroscopy. J. Am. Chem. Soc. 2010, 132, 12792–12793. [Google Scholar] [CrossRef]
- Papadopoulou, E.; Bell, S.E.J. DNA reorientation on Au nanoparticles: Label-free detection of hybridization by surface enhanced Raman spectroscopy. Chem. Commun. 2011, 47, 10966–10968. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, E.; Bell, S.E.J. Label-Free Detection of Single-Base Mismatches in DNA by Surface-Enhanced Raman Spectroscopy. Angew. Chem. Int. Ed. 2011, 50, 9058–9061. [Google Scholar] [CrossRef] [PubMed]
- Torres-Nuñez, A.; Faulds, K.; Graham, D.; Alvarez-Puebla, R.A.; Guerrini, L. Silver colloids as plasmonic substrates for direct label-free surface-enhanced Raman scattering analysis of DNA. Analyst 2016, 141, 5170–5180. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gao, T.; Xu, G.; Xiang, X.; Zhao, B.; Han, X.X.; Guo, X. Direct Approach toward Label-Free DNA Detection by Surface-Enhanced Raman Spectroscopy: Discrimination of a Single-Base Mutation in 50 Base-Paired Double Helixes. Anal. Chem. 2019, 91, 7980–7984. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Kanchanapally, R.; Ray, P.C. Hybrid Graphene Oxide Based Ultrasensitive SERS Probe for Label-Free Biosensing. J. Phys. Chem. Lett. 2013, 4, 3813–3818. [Google Scholar] [CrossRef]
- Gao, F.; Lei, J.; Ju, H. Label-Free Surface-Enhanced Raman Spectroscopy for Sensitive DNA Detection by DNA-Mediated Silver Nanoparticle Growth. Anal. Chem. 2013, 85, 11788–11793. [Google Scholar] [CrossRef] [PubMed]
- Guselnikova, O.; Postnikov, P.; Pershina, A.; Svorcik, V.; Lyutakov, O. Express and portable label-free DNA detection and recognition with SERS platform based on functional Au grating. Appl. Surf. Sci. 2019, 470, 219–227. [Google Scholar] [CrossRef]
- Dick, S.; Bell, S.E.J. Quantitative surface-enhanced Raman spectroscopy of single bases in oligodeoxynucleotides. Faraday Discuss. 2017, 205, 517–536. [Google Scholar] [CrossRef]
- Papadopoulou, E.; Bell, S.E.J. Label-Free Detection of Nanomolar Unmodified Single- and Double-Stranded DNA by Using Surface-Enhanced Raman Spectroscopy on Ag and Au Colloids. Chem. Eur. J. 2012, 18, 5394–5400. [Google Scholar] [CrossRef]
- Harper, M.M.; Dougan, J.A.; Shand, N.C.; Graham, D.; Faulds, K. Detection of SERS active labelled DNA based on surface affinity to silver nanoparticles. Analyst 2012, 137, 2063–2068. [Google Scholar] [CrossRef]
- Pyrak, E.; Jaworska, A.; Kudelski, A. SERS Studies of Adsorption on Gold Surfaces of Mononucleotides with Attached Hexanethiol Moiety: Comparison with Selected Single-Stranded Thiolated DNA Fragments. Molecules 2019, 24, 3921. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Xiao, X.; Chen, K.; Yin, W.; Li, Q.; Wang, P.; Lu, Z.; Ma, J.; Han, H. Ultrasensitive SERS detection of Bacillus thuringiensis special gene based on Au@Ag NRs and magnetic beads. Biosens. Bioelectron. 2017, 92, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, T.; Smith, W.E.; Faulds, K.; Graham, D. Silver and magnetic nanoparticles for sensitive DNA detection by SERS. Chem. Commun. 2014, 50, 12907–12910. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-M.; Ma, W.-F.; You, L.-J.; Guo, J.; Hu, J.; Wang, C.-C. Highly sensitive detection of target ssDNA based on SERS liquid chip using suspended magnetic nanospheres as capturing substrates. Langmuir 2013, 29, 6147–6155. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; He, Y.; Wang, X.; Yuan, R. A SERS biosensor with magnetic substrate CoFe2O4@Ag for sensitive detection of Hg2+. Appl. Surf. Sci. 2017, 416, 581–586. [Google Scholar] [CrossRef]
- Liang, Y.; Gong, J.-L.; Huang, Y.; Zheng, Y.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Biocompatible core-shell nanoparticle-based surface-enhanced Raman scattering probes for detection of DNA related to HIV gene using silica-coated magnetic nanoparticles as separation tools. Talanta 2007, 72, 443–449. [Google Scholar] [CrossRef]
- Ngo, H.T.; Gandra, N.; Fales, A.M.; Taylor, S.M.; Vo-Dinh, T. Sensitive DNA detection and SNP discrimination using ultrabright SERS nanorattles and magnetic beads for malaria diagnostics. Biosens. Bioelectron. 2016, 81, 8–14. [Google Scholar] [CrossRef]
- Yu, J.; Jeon, J.; Choi, N.; Lee, J.O.; Kim, Y.-P.; Choo, J. SERS-based genetic assay for amplification-free detection of prostate cancer specific PCA3 mimic DNA. Sens. Actuators B Chem. 2017, 251, 302–309. [Google Scholar] [CrossRef]
- Strelau, K.K.; Brinker, A.; Schnee, C.; Weber, K.; Möller, R.; Popp, J. Detection of PCR products amplified from DNA of epizootic pathogens using magnetic nanoparticles and SERS. J. Ram. Spectrosc. 2011, 42, 243–250. [Google Scholar] [CrossRef]
- Duan, B.; Zhou, J.; Fang, Z.; Wang, C.; Wang, X.; Hemond, H.F.; Chan-Park, M.B.; Duan, H. Surface enhanced Raman scattering by graphene-nanosheet-gapped plasmonic nanoparticle arrays for multiplexed DNA detection. Nanoscale 2015, 7, 12606–12613. [Google Scholar] [CrossRef]
- Barhoumi, A.; Zhang, D.; Tam, F.; Halas, N.J. Surface-Enhanced Raman Spectroscopy of DNA. J. Am. Chem. Soc. 2008, 130, 5523–5529. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, E.; Bell, S.E.J. Surface-Enhanced Raman Evidence of Protonation, Reorientation, and Ag+ Complexation of Deoxyadenosine and Deoxyadenosine-5′-Monophosphate (dAMP) on Ag and Au Surfaces. J. Phys. Chem. C 2011, 115, 14228–14235. [Google Scholar] [CrossRef]
- Guerrini, L.; Krpetić, Ž.; van Lierop, D.; Alvarez-Puebla, R.A. Graham Duncan Direct Surface-Enhanced Raman Scattering Analysis of DNA Duplexes. Angew. Chem. Int. Ed. 2015, 54, 1144–1148. [Google Scholar] [CrossRef] [PubMed]
- Morla-Folch, J.; Alvarez-Puebla, R.A.; Guerrini, L. Direct Quantification of DNA Base Composition by Surface-Enhanced Raman Scattering Spectroscopy. J. Phys. Chem. Lett. 2016, 7, 3037–3041. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Rao, Y.; Tao, Q.; Dong, J.; Su, T.; Liu, F.; Qian, W. Fabrication of large-scale gold nanoplate films as highly active SERS substrates for label-free DNA detection. Biosens. Bioelectron. 2013, 43, 193–199. [Google Scholar] [CrossRef]
- Pierre, M.C.S.; Haes, A.J. Purification Implications on SERS Activity of Silica Coated Gold Nanospheres. Anal. Chem. 2012, 84, 7906–7911. [Google Scholar] [CrossRef]
- He, L.; Langlet, M.; Stambouli, V. Role of the external NH2 linker on the conformation of surface immobilized single strand DNA probes and their SERS detection. Appl. Surf. Sci. 2017, 399, 702–710. [Google Scholar] [CrossRef]
- Han, B.; Zhang, Y.-L.; Zhu, L.; Chen, X.-H.; Ma, Z.-C.; Zhang, X.-L.; Wang, J.-N.; Wang, W.; Liu, Y.-Q.; Chen, Q.-D.; et al. Direct laser scribing of AgNPs@RGO biochip as a reusable SERS sensor for DNA detection. Sens. Actuators B Chem. 2018, 270, 500–507. [Google Scholar] [CrossRef]
- Zhang, J.; Joshi, P.; Zhou, Y.; Ding, R.; Zhang, P. Quantitative SERS-based DNA detection assisted by magnetic microspheres. Chem. Commun. 2015, 51, 15284–15286. [Google Scholar] [CrossRef]
- Zhou, Q.; Zheng, J.; Qing, Z.; Zheng, M.; Yang, J.; Yang, S.; Ying, L.; Yang, R. Detection of Circulating Tumor DNA in Human Blood via DNA-Mediated Surface-Enhanced Raman Spectroscopy of Single-Walled Carbon Nanotubes. Anal. Chem. 2016, 88, 4759–4765. [Google Scholar] [CrossRef]
- Neumann, O.; Zhang, D.; Tam, F.; Lal, S.; Wittung-Stafshede, P.; Halas, N.J. Direct optical detection of aptamer conformational changes induced by target molecules. Anal. Chem. 2009, 81, 10002–10006. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Neumann, O.; McClain, M.J.; Yang, X.; Zhou, L.; Zhang, C.; Nordlander, P.; Halas, N.J. Aluminum Nanocrystals: A Sustainable Substrate for Quantitative SERS-Based DNA Detection. Nano Lett. 2017, 17, 5071–5077. [Google Scholar] [CrossRef] [PubMed]
- Marotta, N.E.; Beavers, K.R.; Bottomley, L.A. Limitations of Surface Enhanced Raman Scattering in Sensing DNA Hybridization Demonstrated by Label-Free DNA Oligos as Molecular Rulers of Distance-Dependent Enhancement. Anal. Chem. 2013, 85, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Van Lierop, D.; Larmour, I.A.; Faulds, K.; Graham, D. SERS Primers and Their Mode of Action for Pathogen DNA Detection. Anal. Chem. 2013, 85, 1408–1414. [Google Scholar] [CrossRef]
- Culha, M.; Stokes, D.; Allain, L.R.; Vo-Dinh, T. Surface-Enhanced Raman Scattering Substrate Based on a Self-Assembled Monolayer for Use in Gene Diagnostics. Anal. Chem. 2003, 75, 6196–6201. [Google Scholar] [CrossRef]
- Culha, M.; Stokes, D.; Vo-Dinh, T. Surface-enhanced Raman scattering for cancer diagnostics: Detection of the BCL2 gene. Expert Rev. Mol. Diagn. 2003, 3, 669–675. [Google Scholar] [CrossRef]
- Lee, S.; Ongko, A.; Kim, H.Y.; Yim, S.-G.; Jeon, G.; Jeong, H.J.; Lee, S.; Kwak, M.; Yang, S.Y. Sub-100 nm gold nanohole-enhanced Raman scattering on flexible PDMS sheets. Nanotechnology 2016, 27, 315301. [Google Scholar] [CrossRef]
- Yuan, W.; Ho, H.P.; Lee, R.K.Y.; Kong, S.K. Surface-enhanced Raman scattering biosensor for DNA detection on nanoparticle island substrates. Appl. Opt. 2009, 48, 4329–4337. [Google Scholar] [CrossRef]
- Harpster, M.H.; Zhang, H.; Sankara-Warrier, A.K.; Ray, B.H.; Ward, T.R.; Kollmar, J.P.; Carron, K.T.; Mecham, J.O.; Corcoran, R.C.; Wilson, W.C.; et al. SERS detection of indirect viral DNA capture using colloidal gold and methylene blue as a Raman label. Biosens. Bioelectron. 2009, 25, 674–681. [Google Scholar] [CrossRef]
- Vo-Dinh, T.; Allain, L.R.; Stokes, D.L. Cancer gene detection using surface-enhanced Raman scattering (SERS). J. Ram. Spectrosc. 2002, 33, 511–516. [Google Scholar] [CrossRef]
- Cao, Y.C.; Jin, R.; Mirkin, C.A. Nanoparticles with Raman Spectroscopic Fingerprints for DNA and RNA Detection. Science 2002, 297, 1536–1540. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Cao, Y.C.; Thaxton, C.S.; Mirkin, C.A. Glass-Bead-Based Parallel Detection of DNA Using Composite Raman Labels. Small 2006, 2, 375–380. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Su, S.; Xu, T.; Zhong, Y.; Zapien, J.A.; Li, J.; Fan, C.; Lee, S.-T. Silicon nanowires-based highly-efficient SERS-active platform for ultrasensitive DNA detection. Nano Today 2011, 6, 122–130. [Google Scholar] [CrossRef]
- Zengin, A.; Tamer, U.; Caykara, T. SERS detection of hepatitis B virus DNA in a temperature-responsive sandwich-hybridization assay. J. Ram. Spectrosc. 2017, 48, 668–672. [Google Scholar] [CrossRef]
- Jiang, Z.Y.; Jiang, X.X.; Su, S.; Wei, X.P.; Lee, S.T.; He, Y. Silicon-based reproducible and active surface-enhanced Raman scattering substrates for sensitive, specific, and multiplex DNA detection. Appl. Phys. Lett. 2012, 100, 203104. [Google Scholar] [CrossRef]
- He, S.; Liu, K.-K.; Su, S.; Yan, J.; Mao, X.; Wang, D.; He, Y.; Li, L.-J.; Song, S.; Fan, C. Graphene-Based High-Efficiency Surface-Enhanced Raman Scattering-Active Platform for Sensitive and Multiplex DNA Detection. Anal. Chem. 2012, 84, 4622–4627. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Lazarides, A.A. Quantitative Amplification of Cy5 SERS in ‘Warm Spots’ Created by Plasmonic Coupling in Nanoparticle Assemblies of Controlled Structure. Available online: https://pubs.acs.org/doi/abs/10.1021/jp901355g (accessed on 15 June 2018).
- Li, M.; Cushing, S.K.; Liang, H.; Suri, S.; Ma, D.; Wu, N. Plasmonic nanorice antenna on triangle nanoarray for surface-enhanced Raman scattering detection of hepatitis B virus DNA. Anal. Chem. 2013, 85, 2072–2078. [Google Scholar] [CrossRef]
- Zhang, H.; Harpster, M.H.; Wilson, W.C.; Johnson, P.A. Surface-Enhanced Raman Scattering Detection of DNAs Derived from Virus Genomes Using Au-Coated Paramagnetic Nanoparticles. Langmuir 2012, 28, 4030–4037. [Google Scholar] [CrossRef]
- Kang, T.; Yoo, S.M.; Yoon, I.; Lee, S.Y.; Kim, B. Patterned Multiplex Pathogen DNA Detection by Au Particle-on-Wire SERS Sensor. Nano Lett. 2010, 10, 1189–1193. [Google Scholar] [CrossRef]
- Qin, L.; Banholzer, M.J.; Millstone, J.E.; Mirkin, C.A. Nanodisk Codes. Nano Lett. 2007, 7, 3849–3853. [Google Scholar] [CrossRef]
- Banholzer, M.J.; Osberg, K.D.; Li, S.; Mangelson, B.F.; Schatz, G.C.; Mirkin, C.A. Silver-Based Nanodisk Codes. ACS Nano 2010, 4, 5446–5452. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zheng, P.-C.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q.; Liu, G.-K. Sub-attomolar HIV-1 DNA detection using surface-enhanced Raman spectroscopy. Analyst 2010, 135, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Kang, T.; Yoo, S.M.; Lee, S.Y.; Kim, B.; Choi, Y.-K. A well-ordered flower-like gold nanostructure for integrated sensors via surface-enhanced Raman scattering. Nanotechnology 2009, 20, 235302. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Richardson, J.; Brown, T.; Bartlett, P.N. SERS-melting: A new method for discriminating mutations in DNA sequences. J. Am. Chem. Soc. 2008, 130, 15589–15601. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Richardson, J.; Gaied, N.B.; Zhao, Z.; Brown, T.; Bartlett, P.N. The Use of an Electroactive Marker as a SERS Label in an E-melting Mutation Discrimination Assay. Electroanalysis 2009, 21, 2190–2197. [Google Scholar] [CrossRef]
- Hering, K.K.; Möller, R.; Fritzsche, W.; Popp, J. Microarray-based detection of dye-labeled DNA by SERRS using particles formed by enzymatic silver deposition. Chemphyschem 2008, 9, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.; Thompson, D.G.; Smith, W.E.; Faulds, K. Control of enhanced Raman scattering using a DNA-based assembly process of dye-coded nanoparticles. Nat. Nanotech. 2008, 3, 548–551. [Google Scholar] [CrossRef]
- Fu, X.; Cheng, Z.; Yu, J.; Choo, P.; Chen, L.; Choo, J. A SERS-based lateral flow assay biosensor for highly sensitive detection of HIV-1 DNA. Biosens. Bioelectron. 2016, 78, 530–537. [Google Scholar] [CrossRef]
- Wang, X.; Choi, N.; Cheng, Z.; Ko, J.; Chen, L.; Choo, J. Simultaneous Detection of Dual Nucleic Acids Using a SERS-Based Lateral Flow Assay Biosensor. Anal. Chem. 2017, 89, 1163–1169. [Google Scholar] [CrossRef]
- Su, J.; Wang, D.; Nörbel, L.; Shen, J.; Zhao, Z.; Dou, Y.; Peng, T.; Shi, J.; Mathur, S.; Fan, C.; et al. Multicolor Gold-Silver Nano-Mushrooms as Ready-to-Use SERS Probes for Ultrasensitive and Multiplex DNA/miRNA Detection. Anal. Chem. 2017, 89, 2531–2538. [Google Scholar] [CrossRef]
- Shen, J.; Su, J.; Yan, J.; Zhao, B.; Wang, D.; Wang, S.; Li, K.; Liu, M.; He, Y.; Mathur, S.; et al. Bimetallic nano-mushrooms with DNA-mediated interior nanogaps for high-efficiency SERS signal amplification. Nano Res. 2015, 8, 731–742. [Google Scholar] [CrossRef]
- Sun, L.; Yu, C.; Irudayaraj, J. Raman Multiplexers for Alternative Gene Splicing. Anal. Chem. 2008, 80, 3342–3349. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yu, C.; Irudayaraj, J. Surface-Enhanced Raman Scattering Based Nonfluorescent Probe for Multiplex DNA Detection. Anal. Chem. 2007, 79, 3981–3988. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-M.; Wei, C.; Ma, W.-F.; An, Q.; Guo, J.; Hu, J.; Wang, C.-C. Multiplexed SERS detection of DNA targets in a sandwich-hybridization assay using SERS-encoded core–shell nanospheres. J. Mater. Chem. 2012, 22, 12100–12106. [Google Scholar] [CrossRef]
- Braun, G.; Lee, S.J.; Dante, M.; Nguyen, T.-Q.; Moskovits, M.; Reich, N. Surface-Enhanced Raman Spectroscopy for DNA Detection by Nanoparticle Assembly onto Smooth Metal Films. J. Am. Chem. Soc. 2007, 129, 6378–6379. [Google Scholar] [CrossRef] [PubMed]
- Khalil, I.; Yehye, W.A.; Julkapli, N.M.; Rahmati, S.; Sina, A.A.I.; Basirun, W.J.; Johan, M.R. Graphene oxide and gold nanoparticle based dual platform with short DNA probe for the PCR free DNA biosensing using surface-enhanced Raman scattering. Biosens. Bioelectron. 2019, 131, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Irudayaraj, J. Quantitative Surface-Enhanced Raman for Gene Expression Estimation. Biophys. J. 2009, 96, 4709–4716. [Google Scholar] [CrossRef]
- Ye, S.; Yang, Y.; Xiao, J.; Zhang, S. Surface-enhanced Raman scattering assay combined with autonomous DNA machine for detection of specific DNA and cancer cells. Chem. Commun. 2012, 48, 8535–8537. [Google Scholar] [CrossRef]
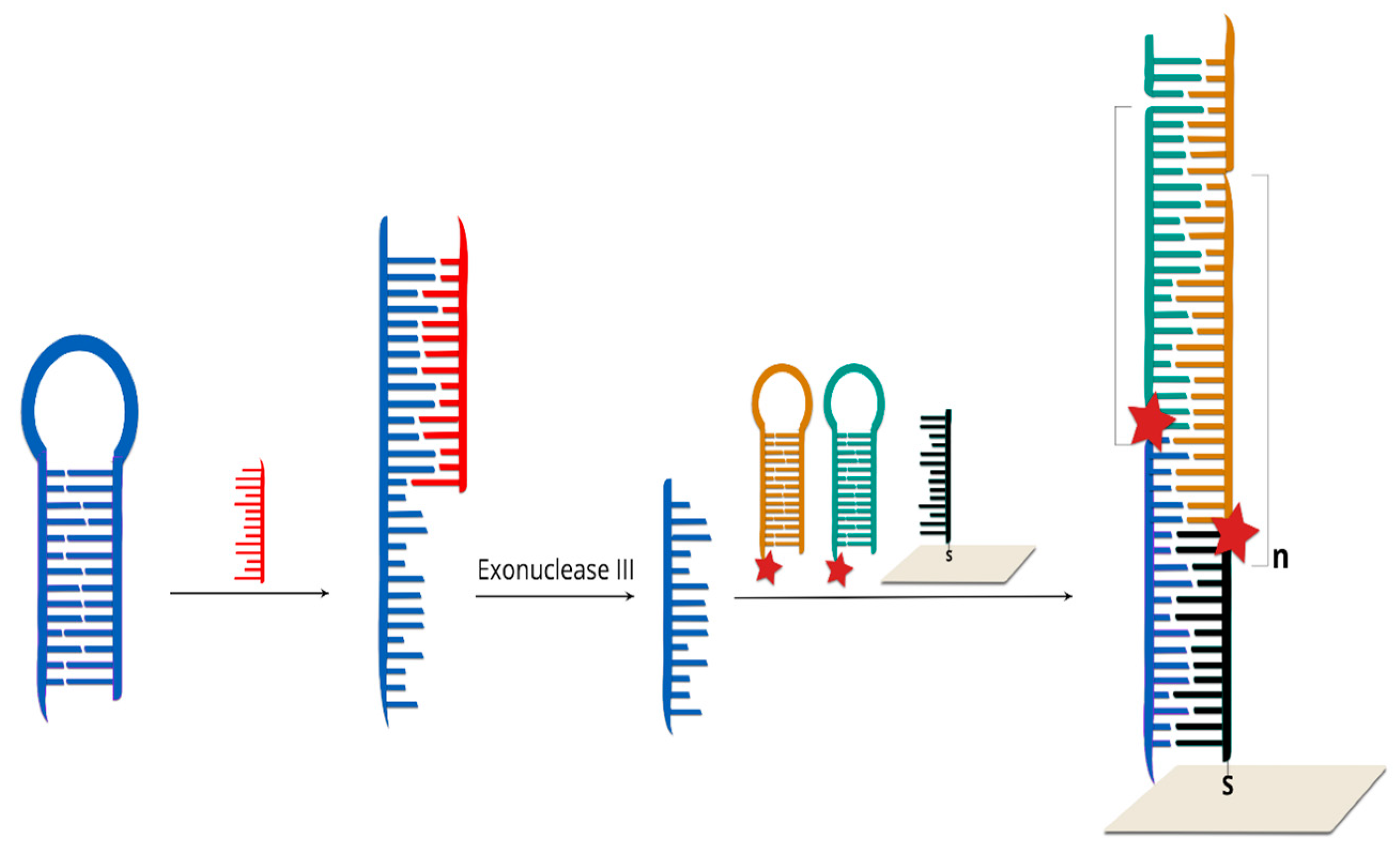
- Hu, J.; Zhang, C. Sensitive detection of nucleic acids with rolling circle amplification and surface-enhanced Raman scattering spectroscopy. Anal. Chem. 2010, 82, 8991–8997. [Google Scholar] [CrossRef]
- Sun, Y.; Peng, P.; Guo, R.; Wang, H.; Li, T. Exonuclease III-boosted cascade reactions for ultrasensitive SERS detection of nucleic acids. Biosens. Bioelectron. 2018, 104, 32–38. [Google Scholar] [CrossRef]
- Lowe, A.J.; Huh, Y.S.; Strickland, A.D.; Erickson, D.; Batt, C.A. Multiplex single nucleotide polymorphism genotyping utilizing ligase detection reaction coupled surface enhanced Raman spectroscopy. Anal. Chem. 2010, 82, 5810–5814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, Y.; Jiang, X.; Dong, C.; Song, C.; Han, C.; Wang, L. Ultrasensitive SERS detection of nucleic acids via simultaneous amplification of target-triggered enzyme-free recycling and multiple-reporter. Biosens. Bioelectron. 2019, 141, 111402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, H.; Lu, L.; Ai, K.; Zhang, G.; Cheng, X. Large-Area Silver-Coated Silicon Nanowire Arrays for Molecular Sensing Using Surface-Enhanced Raman Spectroscopy. Adv. Funct. Mater. 2008, 18, 2348–2355. [Google Scholar] [CrossRef]
- Wabuyele, M.B.; Vo-Dinh, T. Detection of Human Immunodeficiency Virus Type 1 DNA Sequence Using Plasmonics Nanoprobes. Anal. Chem. 2005, 77, 7810–7815. [Google Scholar] [CrossRef]
- Ngo, H.T.; Wang, H.-N.; Burke, T.; Ginsburg, G.S.; Vo-Dinh, T. Multiplex detection of disease biomarkers using SERS molecular sentinel-on-chip. Anal. Bioanal. Chem. 2014, 406, 3335–3344. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Zeng, J.; Zhao, F.; Lin, S.H.; Raja, B.; Strych, U.; Willson, R.C.; Shih, W.-C. Label-free, in situ SERS monitoring of individual DNA hybridization in microfluidics. Nanoscale 2014, 6, 8521–8526. [Google Scholar] [CrossRef]
- Kim, S.; Tran Ngoc, H.; Kim, J.; Yoo, S.Y.; Chung, H. Toehold-mediated DNA displacement-based surface-enhanced Raman scattering DNA sensor utilizing an Au-Ag bimetallic nanodendrite substrate. Anal. Chim. Acta 2015, 885, 132–139. [Google Scholar] [CrossRef]
- Wang, H.-N.; Dinh, T.V. Multiplex detection of breast cancer biomarkers using plasmonic molecular sentinel nanoprobes. Nanotechnology 2009, 20, 065101. [Google Scholar] [CrossRef]
- Wang, H.-N.; Dhawan, A.; Du, Y.; Batchelor, D.; Leonard, D.N.; Misra, V.; Vo-Dinh, T. Molecular sentinel-on-chip for SERS-based biosensing. Phys. Chem. Chem. Phys. 2013, 15, 6008–6015. [Google Scholar] [CrossRef][Green Version]
- Wu, L.; Garrido-Maestu, A.; Guerreiro, J.R.L.; Carvalho, S.; Abalde-Cela, S.; Prado, M.; Diéguez, L. Amplification-free SERS analysis of DNA mutation in cancer cells with single-base sensitivity. Nanoscale 2019, 11, 7781–7789. [Google Scholar] [CrossRef]
- Guo, J.; Chen, Y.; Jiang, Y.; Ju, H. Polyadenine-Modulated DNA Conformation Monitored by Surface-Enhanced Raman Scattering (SERS) on Multibranched Gold Nanoparticles and Its Sensing Application. Chem. Eur. J. 2017, 23, 9332–9337. [Google Scholar] [CrossRef] [PubMed]
- 9337Faulds, K.; Fruk, L.; Robson, D.C.; Thompson, D.G.; Enright, A.; Smith, W.E.; Graham, D. A new approach for DNA detection by SERRS. Faraday Discuss. 2006, 132, 261–268. [Google Scholar]
- Jung, J.; Chen, L.; Lee, S.; Kim, S.; Seong, G.H.; Choo, J.; Lee, E.K.; Oh, C.-H.; Lee, S. Fast and sensitive DNA analysis using changes in the FRET signals of molecular beacons in a PDMS microfluidic channel. Anal. Bioanal. Chem. 2007, 387, 2609–2615. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.T.; Wang, H.-N.; Fales, A.M.; Nicholson, B.P.; Woods, C.W.; Vo-Dinh, T. DNA bioassay-on-chip using SERS detection for dengue diagnosis. Analyst 2014, 139, 5655–5659. [Google Scholar] [CrossRef] [PubMed]
- Fabris, L.; Dante, M.; Braun, G.; Lee, S.J.; Reich, N.O.; Moskovits, M.; Nguyen, T.-Q.; Bazan, G.C. A Heterogeneous PNA-Based SERS Method for DNA Detection. J. Am. Chem. Soc. 2007, 129, 6086–6087. [Google Scholar] [CrossRef]
- Fang, C.; Agarwal, A.; Buddharaju, K.D.; Khalid, N.M.; Salim, S.M.; Widjaja, E.; Garland, M.V.; Balasubramanian, N.; Kwong, D.-L. DNA detection using nanostructured SERS substrates with Rhodamine B as Raman label. Biosens. Bioelectron. 2008, 24, 216–221. [Google Scholar] [CrossRef]
- Lee, S.-W.; Chen, Y.-W.; Kuan, E.C.; Lan, M.-Y. Dual-function nanostructured platform for isolation of nasopharyngeal carcinoma circulating tumor cells and EBV DNA detection. Biosens. Bioelectron. 2019, 142, 111509. [Google Scholar] [CrossRef]
- Lin, T.-W.; Wu, H.-Y.; Tasi, T.-T.; Lai, Y.-H.; Shen, H.-H. Surface-enhanced Raman spectroscopy for DNA detection by the self-assembly of Ag nanoparticles onto Ag nanoparticle–graphene oxide nanocomposites. Phys. Chem. Chem. Phys. 2015, 17, 18443–18448. [Google Scholar] [CrossRef]
- Lim, D.-K.; Jeon, K.-S.; Kim, H.M.; Nam, J.-M.; Suh, Y.D. Nanogap-engineerable Raman-active nanodumbbells for single-molecule detection. Nat. Mater. 2010, 9, 60–67. [Google Scholar] [CrossRef]
- Li, J.F.; Huang, Y.F.; Ding, Y.; Yang, Z.L.; Li, S.B.; Zhou, X.S.; Fan, F.R.; Zhang, W.; Zhou, Z.Y.; Wu, D.Y.; et al. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature 2010, 464, 392–395. [Google Scholar] [CrossRef]
- Wee, E.J.H.; Wang, Y.; Tsao, S.C.-H.; Trau, M. Simple, Sensitive and Accurate Multiplex Detection of Clinically Important Melanoma DNA Mutations in Circulating Tumour DNA with SERS Nanotags. Theranostics 2016, 6, 1506–1513. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, T.; Li, C.S.; Song, Y.; Lou, H.; Guan, D.; Jin, L. Surface Enhanced Raman Spectroscopy (SERS) for the Multiplex Detection of Braf, Kras, and Pik3ca Mutations in Plasma of Colorectal Cancer Patients. Theranostics 2018, 8, 1678–1689. [Google Scholar] [CrossRef] [PubMed]
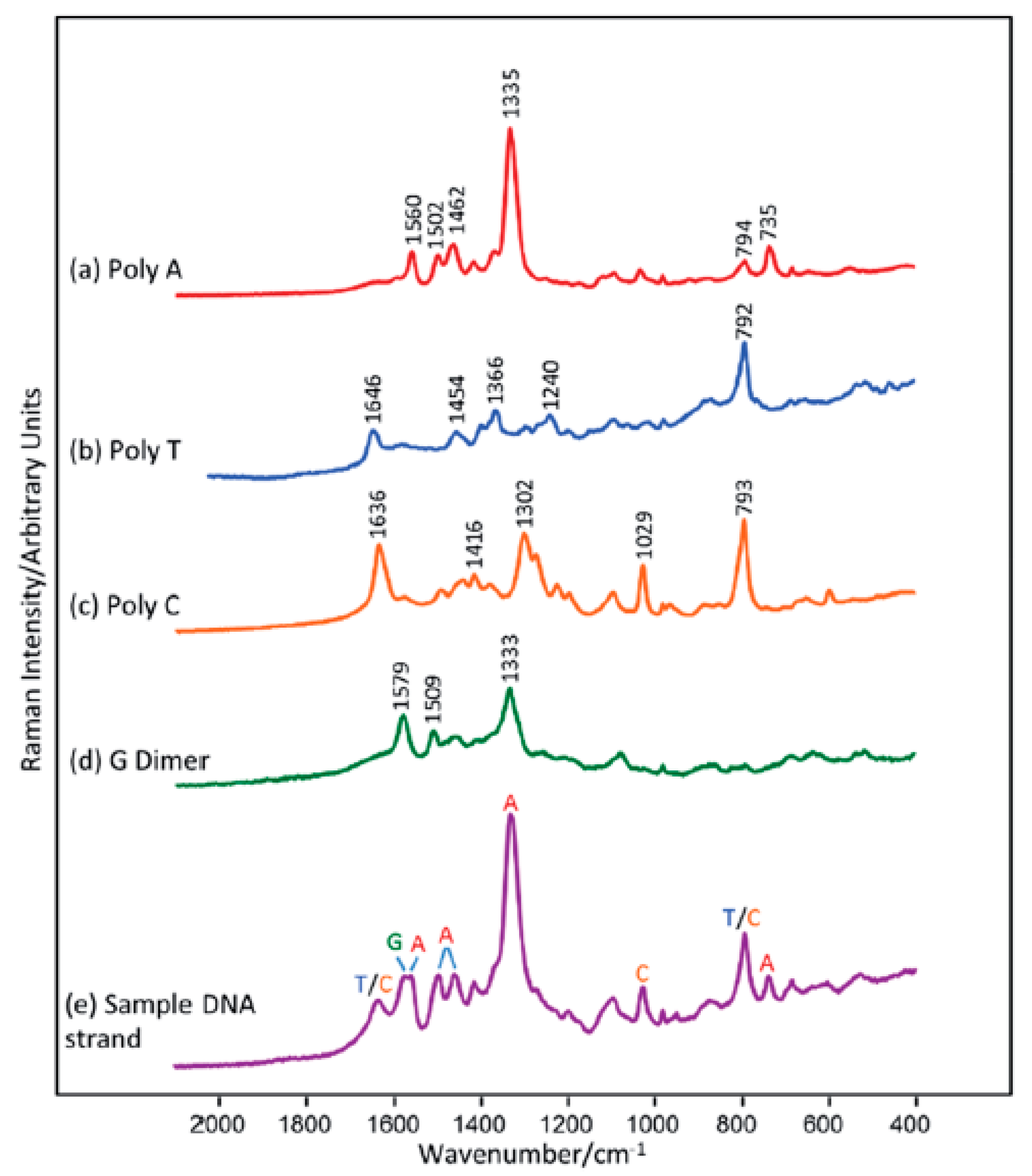
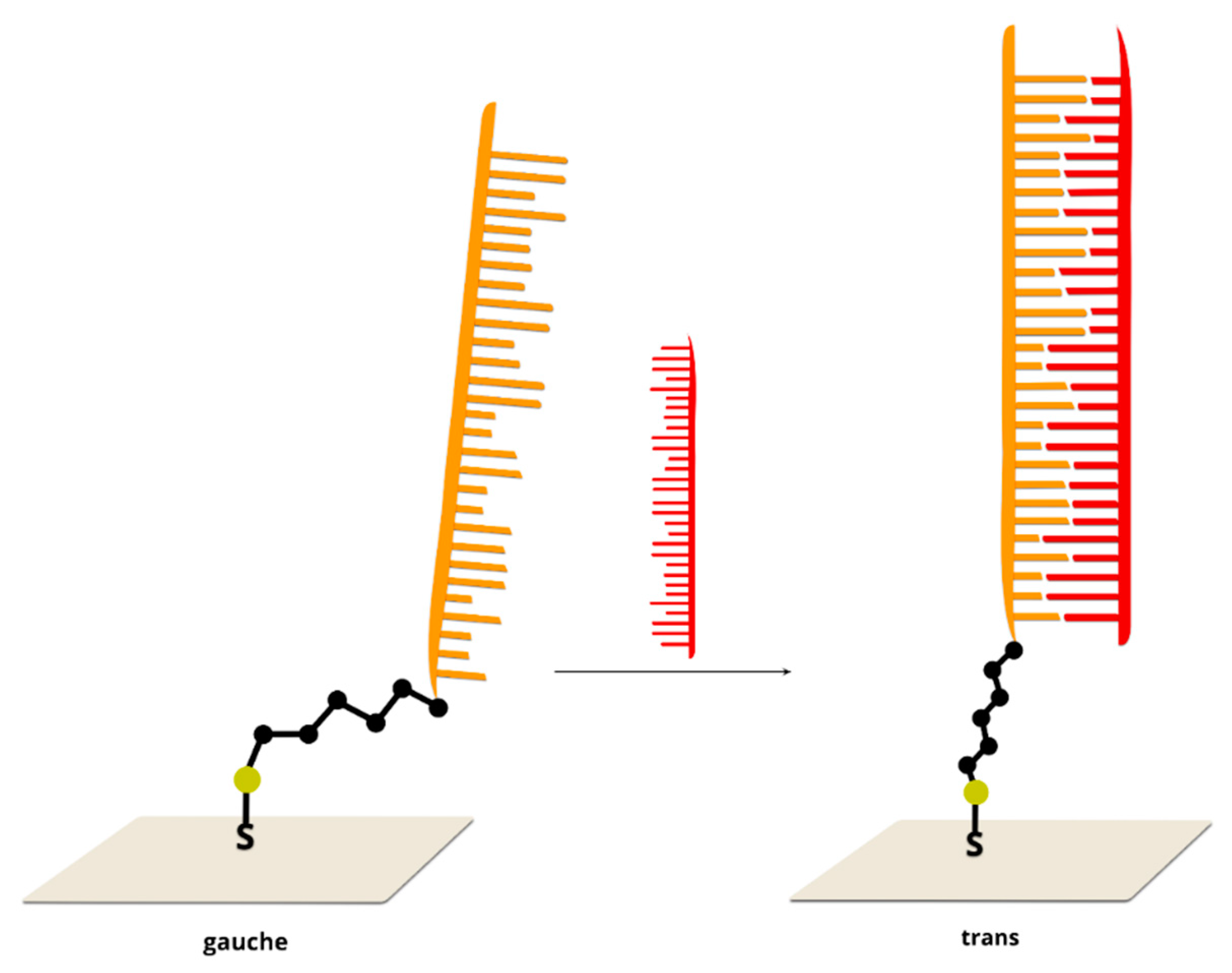
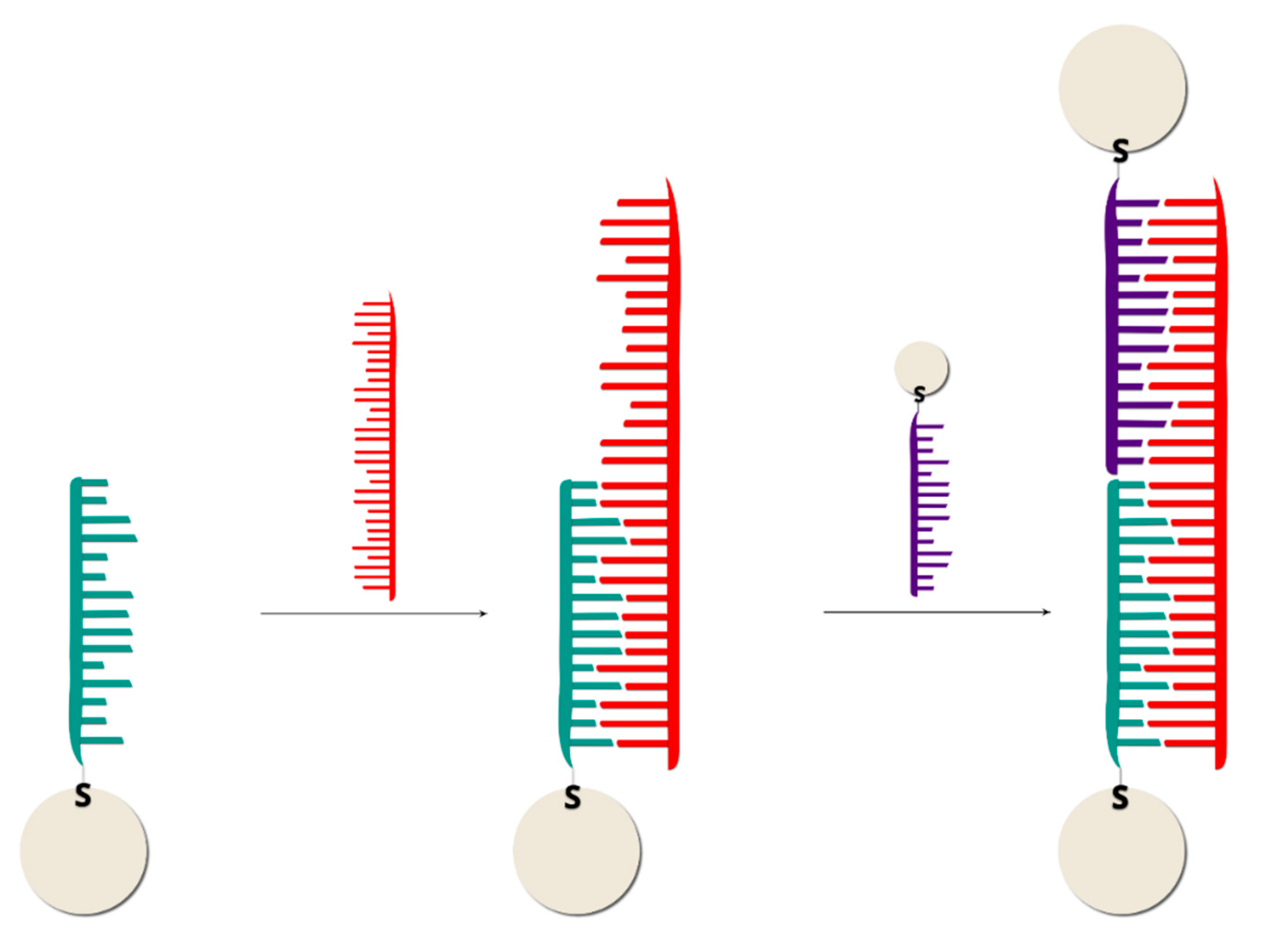
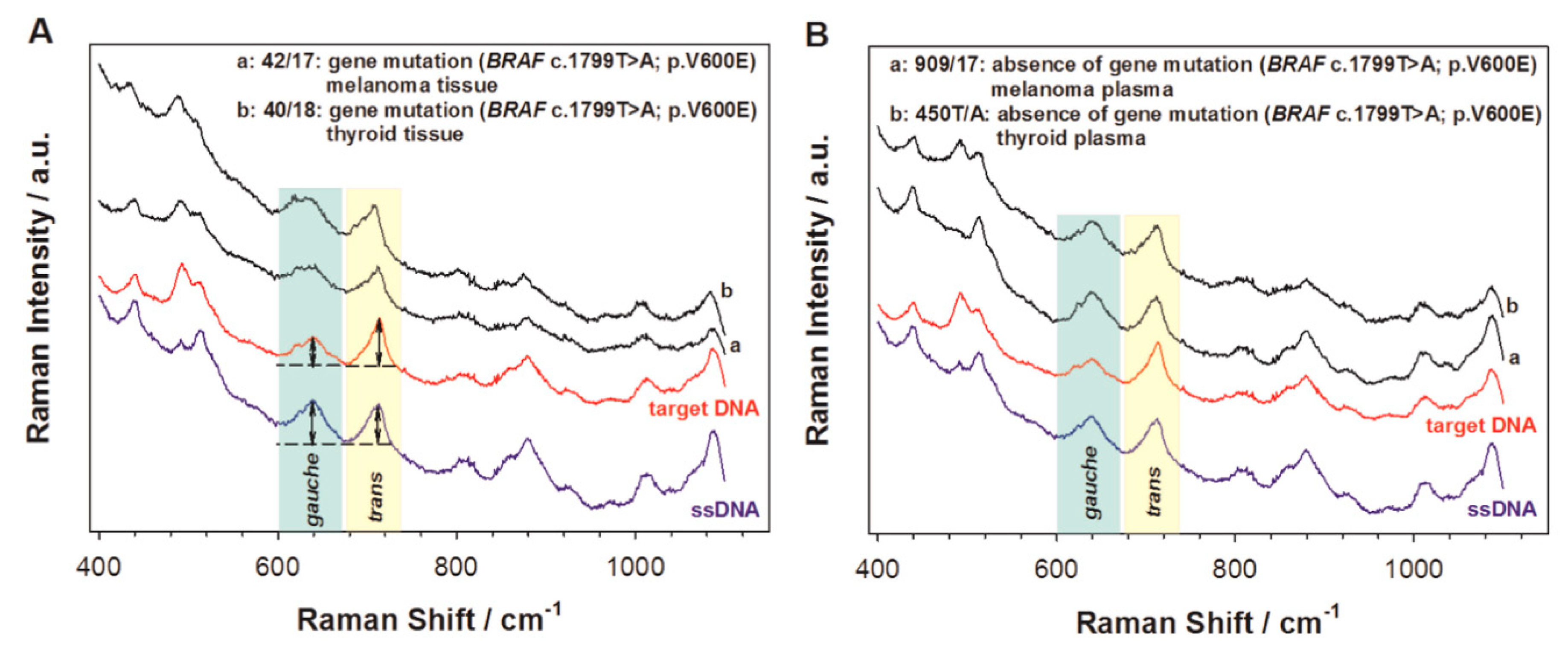
| LP | Method | Principle of the Method | Sensitivity | Specificity * | Cost Per Reaction (USD) | Sample Preparation | Method Complexity | Main Application |
|---|---|---|---|---|---|---|---|---|
| 1 | allele-specific PCR | amplification refractory mutation system | 1%–10% | + | 1 | hours | low | germline and somatic mutation genotyping |
| 2 | qPCR | fluorescently labeled probes | 1%–5% | +++ | 2 | hours | very low | germline and somatic mutation genotyping, gene expression, copy number |
| 3 | PCR–HRM | detecting small differences in PCR amplicon melting (dissociation) curves | 1%–5% | ++ | 2 | hours | very low | germline and somatic mutation genotyping |
| 4 | Sanger Sequencing | fluorescently labeled nucleotides | 10%–20% | ++++ | 5 | day | high | low frequency somatic mutation detection (liquid biopsy), very precise copy number detection |
| 5 | ddPCR | fluorescently labeled probes and emultion PCR | 0.01%–1% | ++++ | 3 | hours | medium | low frequency somatic mutation detection (liquid biopsy), very precise copy number detection |
| 6 | NGS | massively paralelled sequencing | 0.1%–5% | +++++ | 250 | 1–2 weeks | very high | low frequency somatic mutations detection (liquid biopsy), panel sequencing (10–400), whole genome sequencing |
| 7 | SERS | measurement of the Raman spectrum of Raman reporter | 0.001%–1% | ++++ | n.a | hours | very low | germline and somatic mutation genotyping |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pyrak, E.; Krajczewski, J.; Kowalik, A.; Kudelski, A.; Jaworska, A. Surface Enhanced Raman Spectroscopy for DNA Biosensors—How Far Are We? Molecules 2019, 24, 4423. https://doi.org/10.3390/molecules24244423
Pyrak E, Krajczewski J, Kowalik A, Kudelski A, Jaworska A. Surface Enhanced Raman Spectroscopy for DNA Biosensors—How Far Are We? Molecules. 2019; 24(24):4423. https://doi.org/10.3390/molecules24244423
Chicago/Turabian StylePyrak, Edyta, Jan Krajczewski, Artur Kowalik, Andrzej Kudelski, and Aleksandra Jaworska. 2019. "Surface Enhanced Raman Spectroscopy for DNA Biosensors—How Far Are We?" Molecules 24, no. 24: 4423. https://doi.org/10.3390/molecules24244423
APA StylePyrak, E., Krajczewski, J., Kowalik, A., Kudelski, A., & Jaworska, A. (2019). Surface Enhanced Raman Spectroscopy for DNA Biosensors—How Far Are We? Molecules, 24(24), 4423. https://doi.org/10.3390/molecules24244423








