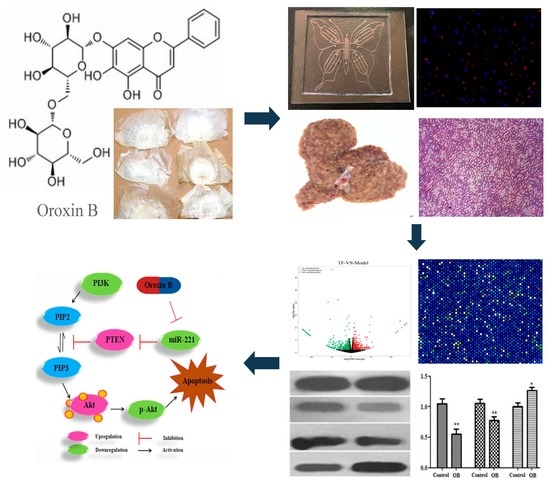
Oroxin B Induces Apoptosis by Down-Regulating MicroRNA-221 Resulting in the Inactivation of the PTEN/PI3K/AKT Pathway in Liver Cancer
Abstract
:1. Introduction
2. Results
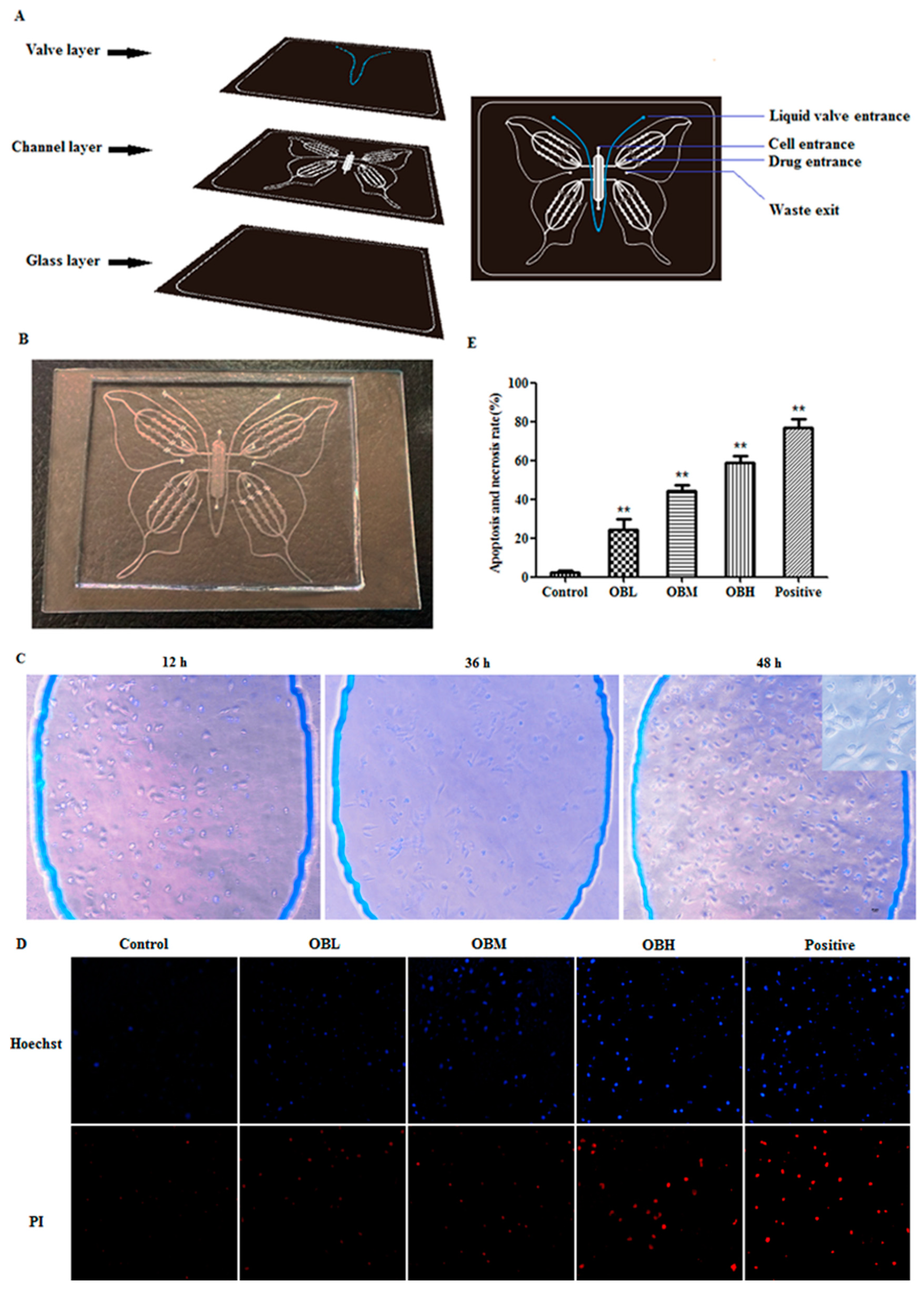
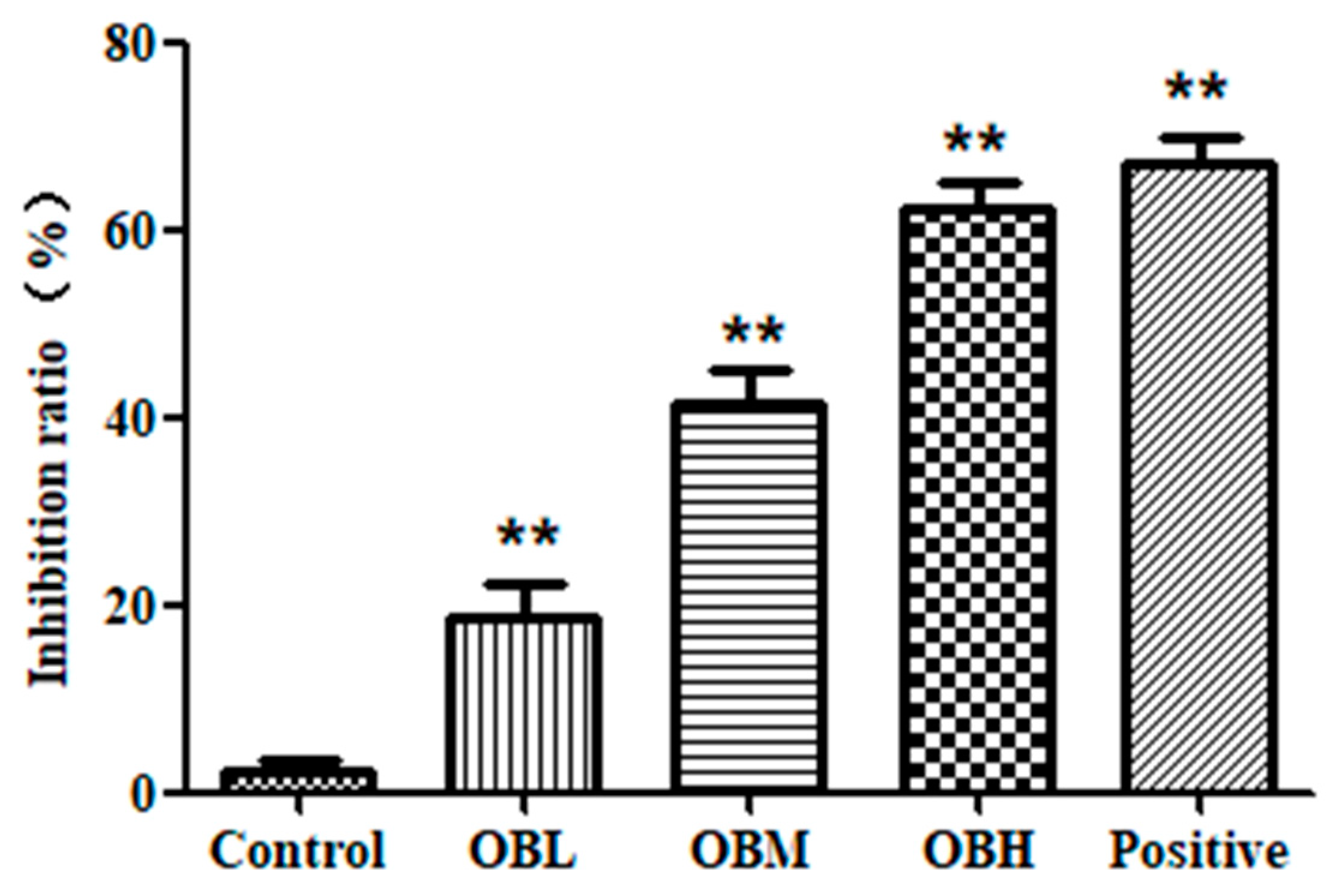
2.1. The Effect of Oroxin B on Proliferation of Human Hepatoma Cell Line HepG2
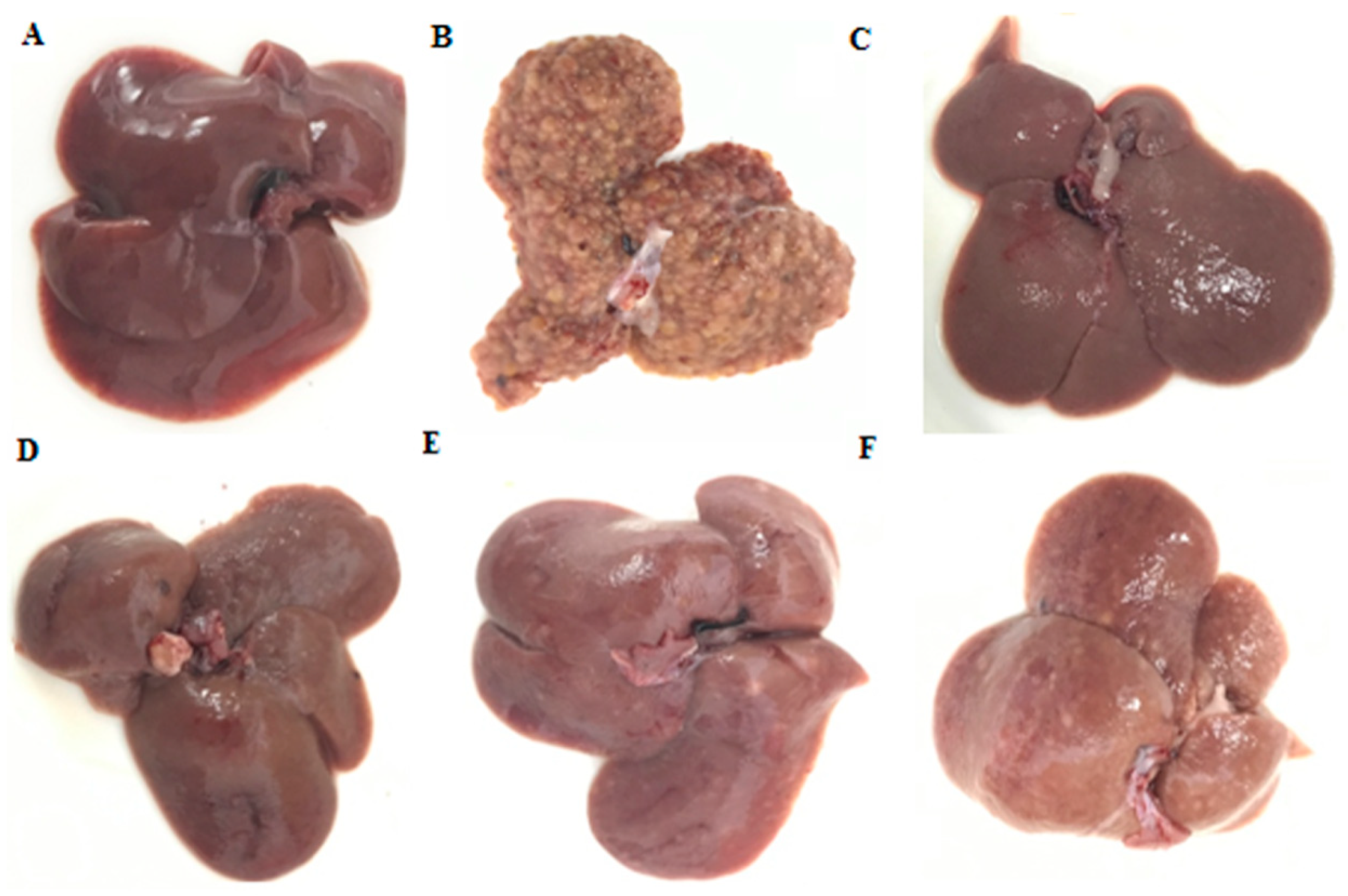
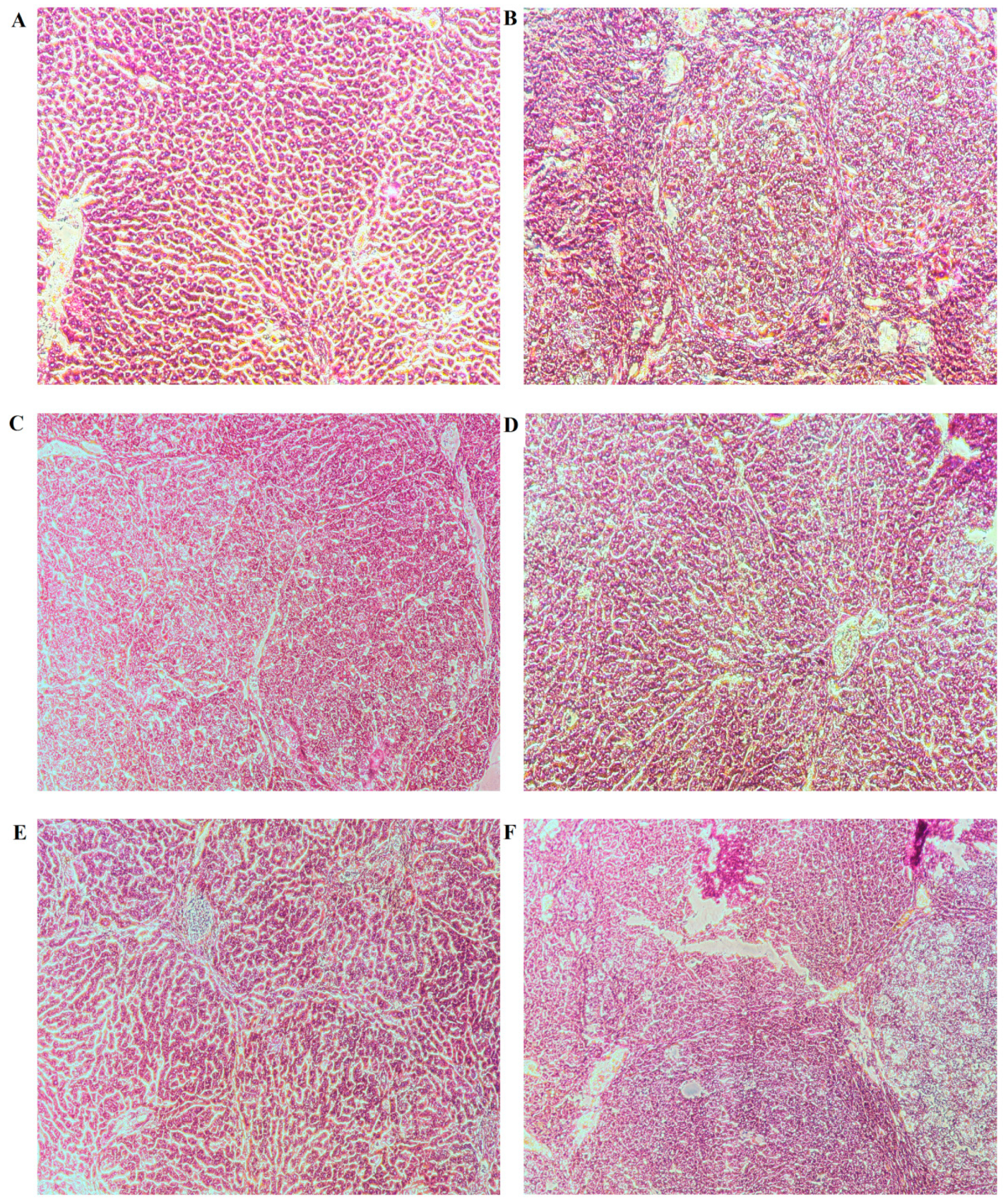
2.2. General Appearance of Liver and Histopathological Evaluation
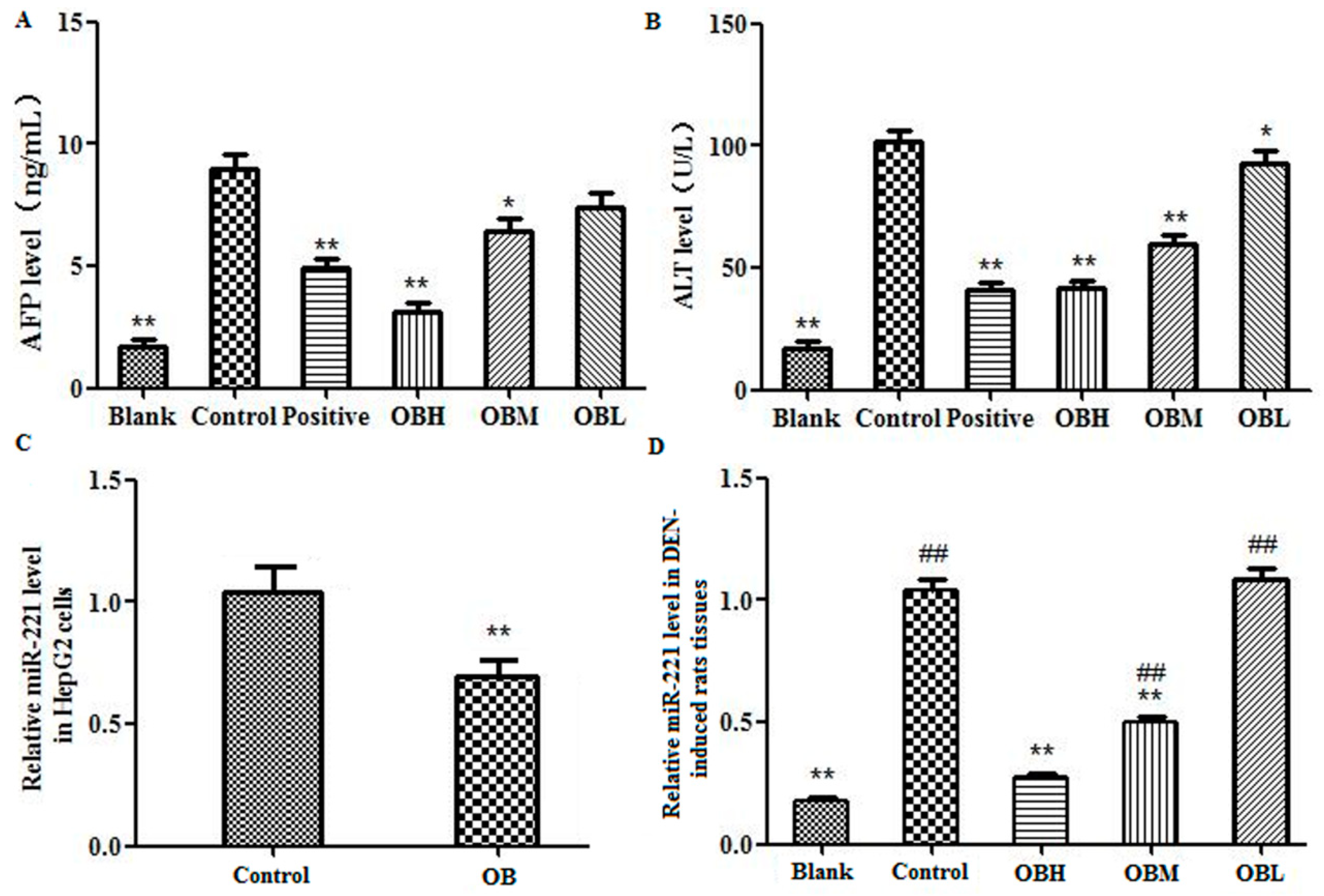
2.3. The Effect of OB on the Levels of AFP and ALT in Serum of DEN-Induced Rats
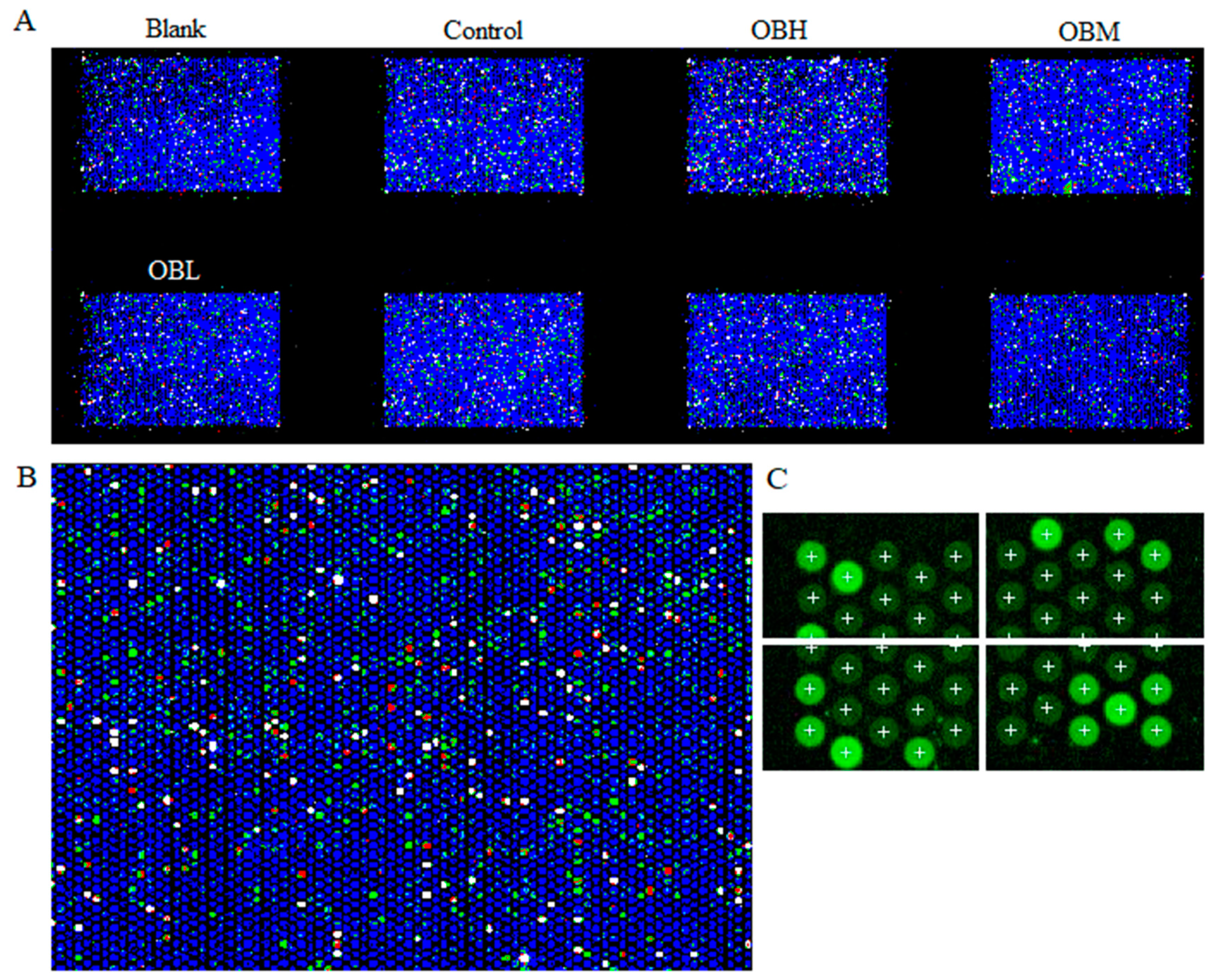
2.4. The Result of Microarrays
2.5. The Effect of OB on miR-221 Expression in HepG2 Cells and Liver Tissues
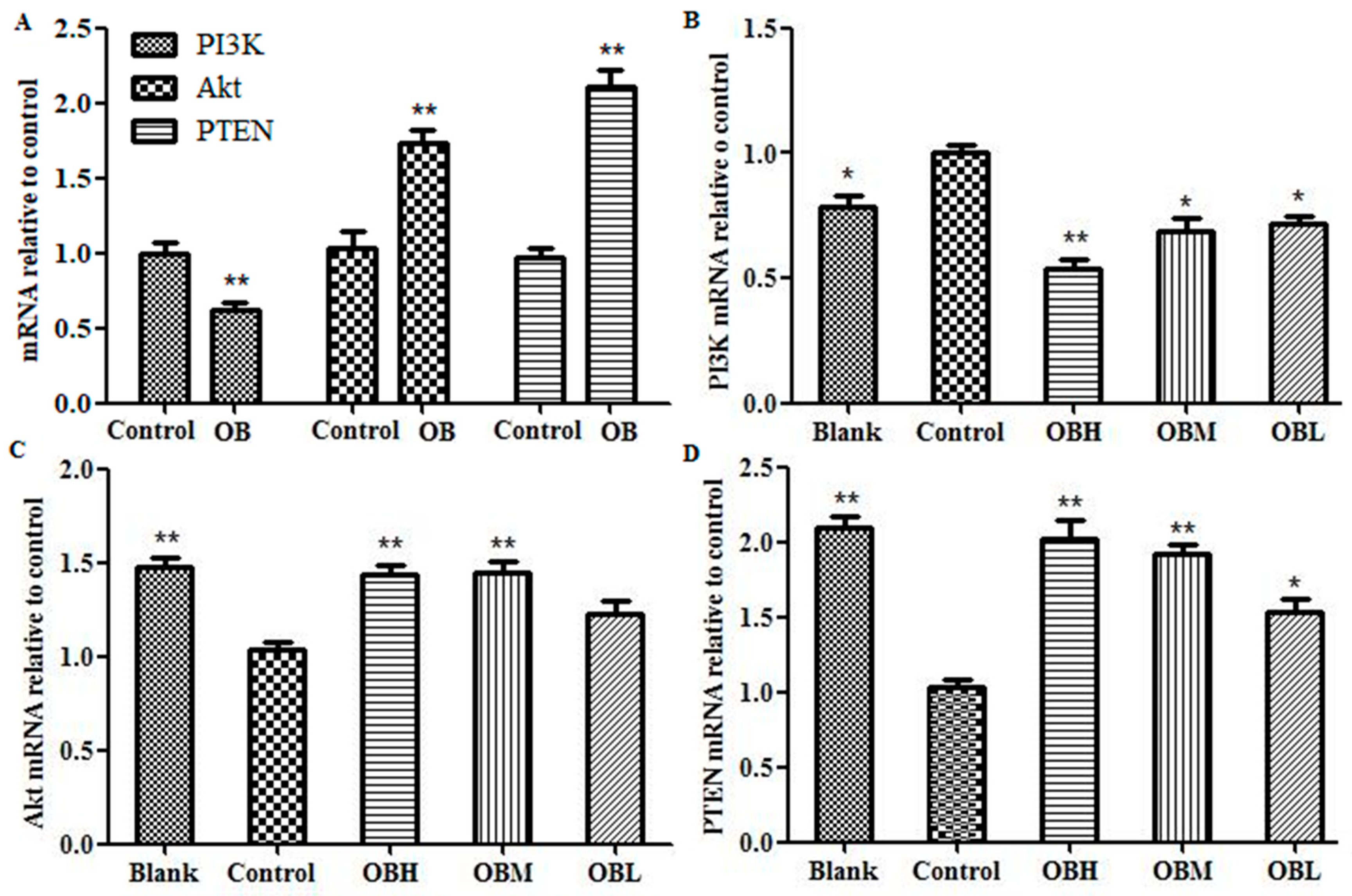
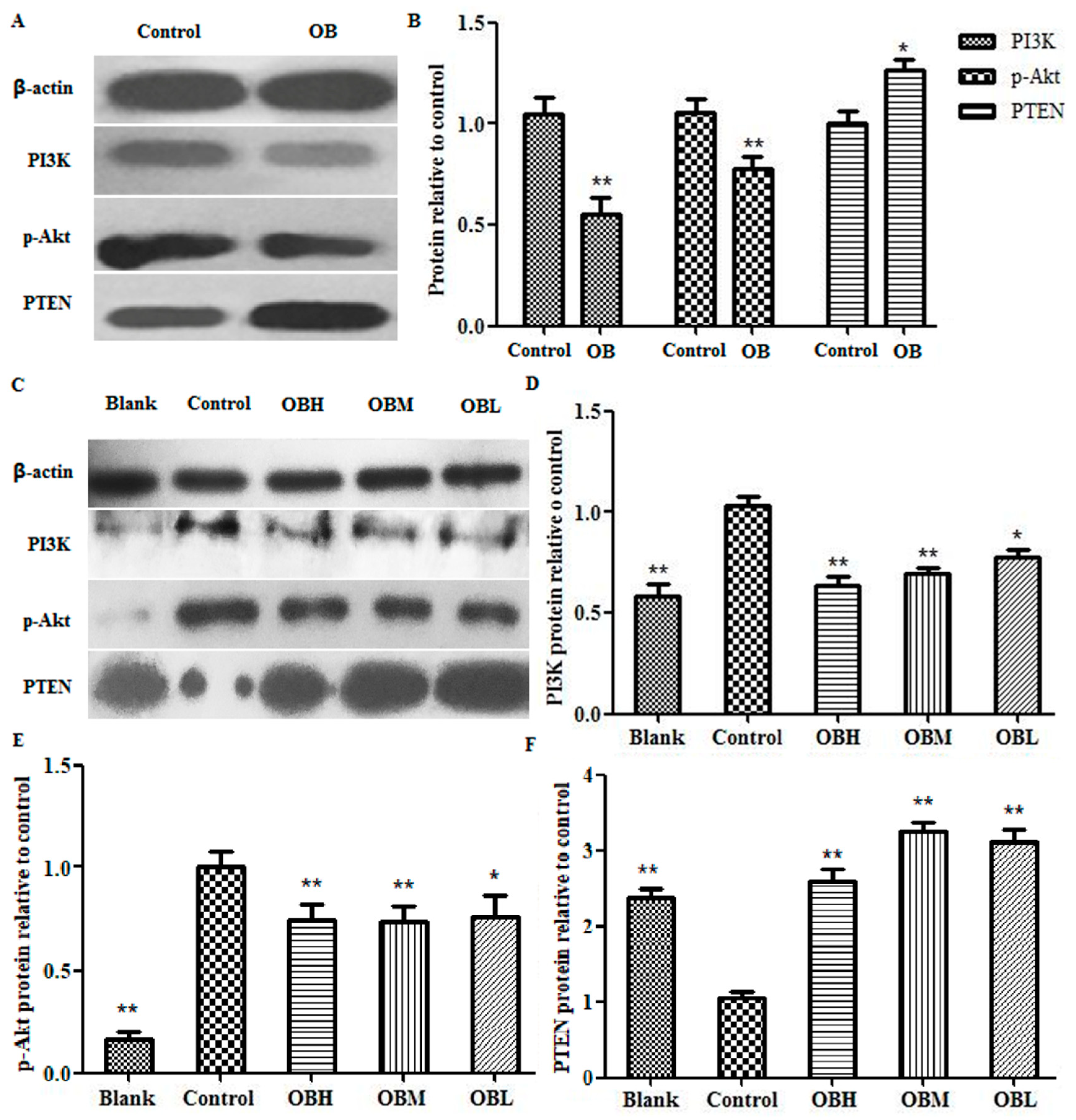
2.6. The Effect of OB on PI3K, PTEN, Akt mRNA and Protein Expression in HepG2 Cells and Liver Tissues
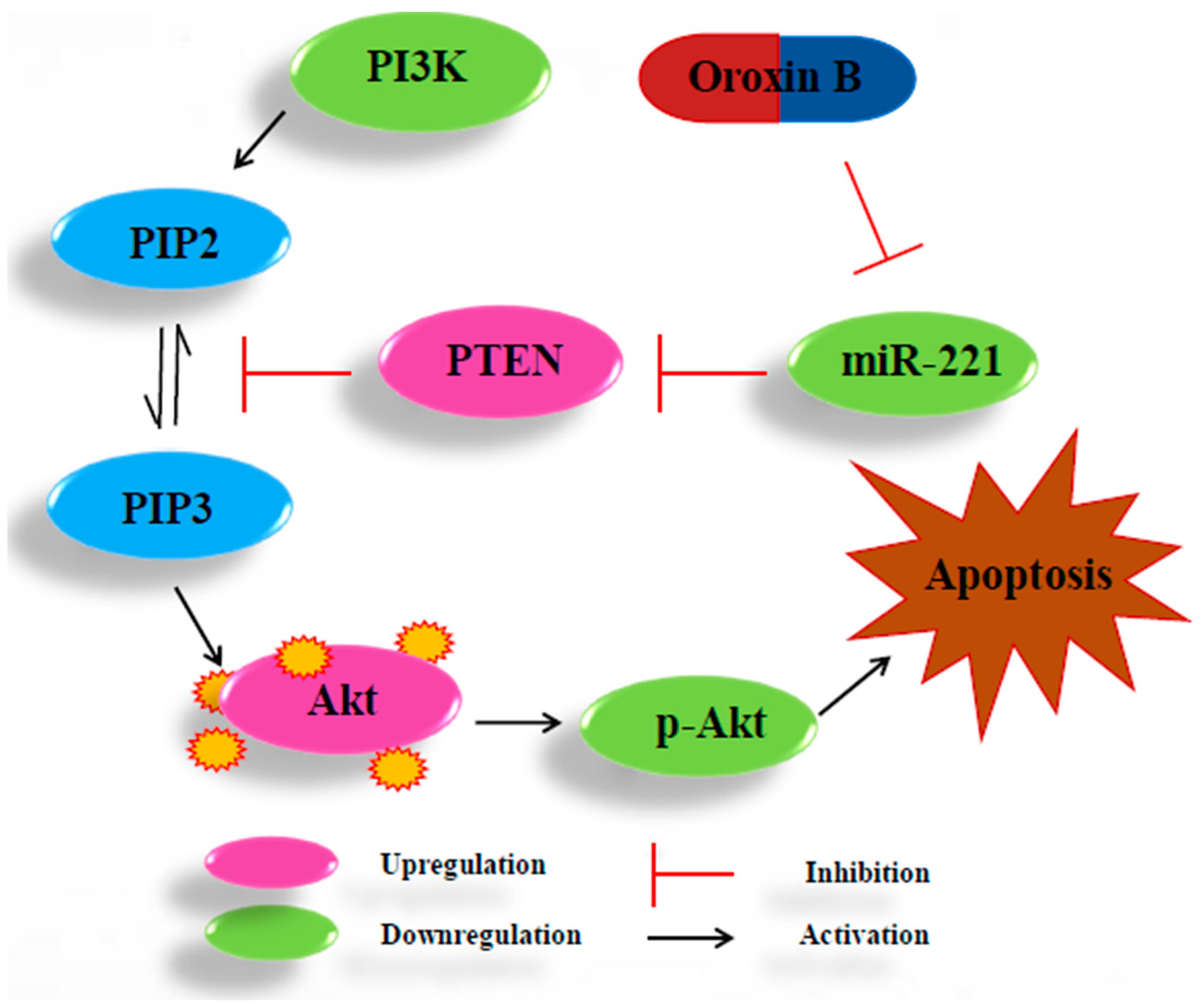
3. Discussion
4. Materials and Methods
4.1. Microfluidic Chip
4.2. Cell Culture
4.3. Proliferation Analysis
4.4. MTT Assay
4.5. Animals and Chemicals
4.6. Animals Experimental
4.7. Histopathology
4.8. ELISA Assays of ALT, and AFP
4.9. Microarray Analysis
4.10. RNA Isolation and Quantitative RT-PCR
4.11. Western Blotting Analysis
4.12. Statistics Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Xu, J.; Li, J.; Zheng, T.H.; Bai, L.; Liu, Z.J. MicroRNAs in the Occurrence and Developmentof Primary Hepatocellular Carcinoma. Adv. Clin. Exp. Med. 2016, 25, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Tang, Y.S.; Huang, S.F.; Ai, J.G.; Wang, H.X.; Zhang, L.P. HBx protein-induced upregulation of microRNA-221 promotes aberrant proliferation in HBV-related hepatocellular carcinoma by targeting estrogen receptor-α. Oncol. Rep. 2015, 33, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Nasser, M.W.; Wang, B.; Hsu, S.H.; Datta, J.; Kutay, J.H.; Yadav, A.; Nuovo, G.; Kumar, P.; Ghoshal, K. MicroRNA-122 Inhibits Tumorigenic Properties of Hepatocellular Carcinoma Cells and Sensitizes These Cells to Sorafenib. J. Biol. Chem. 2009, 284, 32015–32027. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Liao, Y.J.; Cai, M.Y.; Liu, T.H.; Chen, S.P.; Wu, P.H.; Wu, L.; Bian, X.W.; Guan, X.Y.; Zeng, Y.X. Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma In Vivo by Repressing Multiple Targets. PLoS Genet. 2015, 11, e1004873. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S.; Olaru, A.V.; An, F.; Luvsanjav, D.; Jin, Z.; Agarwal, R.; Tomuleasa, C.; Popescu, I.; Alexandrescu, S.; Dima, S. MicroRNA-21 inhibits Serpini1, a gene with novel tumour suppressive effects in gastric cancer. Dig. Liver Dis. 2012, 44, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Moshiri, F.; Callegari, E.; D’Abundo, L.; Corrà, F.; Lupini, L.; Sabbioni, S.; Negrini, M. Inhibiting the oncogenic mir-221 by microRNA sponge: Toward microRNA-based therapeutics for hepatocellular carcinoma. Gastroenterol. Hepatol. Bed Bench 2014, 7, 43–54. [Google Scholar]
- Gramantieri, L.; Fornari, F.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Croce, C.M.; Bolondi, L.; Negrini, M. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin. Cancer Res. 2009, 15, 5073–5081. [Google Scholar] [CrossRef]
- Garofalo, M.; Leva, G.D.; Romano, G.; Nuovo, G.; Suh, S.S.; Ngankeu, A.; Taccioli, C.; Pichiorri, F.; Alder, H.; Secchiero, P. A miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009, 16, 498–509. [Google Scholar]
- Wong, Q.W.; Ching, A.K.; Chan, A.W.; Choy, K.W.; To, K.F.; Lai, P.B.; Wong, N. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin. Cancer Res. 2010, 16, 867–875. [Google Scholar] [CrossRef]
- Yang, P.; Fu, S.; Cao, Z.; Liao, H.; Huo, Z.; Pan, Y.; Zhang, G.; Gao, A.; Zhou, Q. Oroxin B selectively induces tumor-suppressive ER stress and concurrently inhibits tumor-adaptive ER stress in B-lymphoma cells for effective anti-lymphoma therapy. Toxicol. Appl. Pharmacol. 2015, 288, 269–279. [Google Scholar] [CrossRef]
- Qiu, J.Z.; Wang, D.C.; Zhang, Y.; Dong, J.; Wang, J.F.; Niu, X.D. Molecular Modeling Reveals the Novel Inhibition Mechanism and Binding Mode of Three Natural Compounds to Staphylococcal a-Hemolysin. PLoS ONE 2013, 8, e80197. [Google Scholar] [CrossRef] [PubMed]
- Li, N.N.; Meng, X.S.; Bao, Y.R.; Wang, S.; Li, T.J. Evidence for the Involvement of COX-2/VEGF and PTEN/Pl3K/AKT Pathway the Mechanism of Oroxin B Treated Liver Cancer. Pharmacogn. Mag. 2018, 14, 207–213. [Google Scholar] [PubMed]
- Chin, C.D.; Laksanasopin, T.; Cheung, Y.K.; Steinmiller, D.; Linder, V.; Parsa, H. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat. Med. 2011, 17, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Yue, F.; Zhang, L.C.; Wang, J.R.; Wang, Y.Y.; Li, J.; Lin, B.C.; Qi, W. Application of microfluidic gradient chip in the analysis of lung cancer chemotherapy resistance. J. Pharm. Biomed. Anal. 2009, 49, 806–810. [Google Scholar]
- Wu, J.; Wheeldon, I.; Guo, Y.; Lu, T.; Du, Y.; Wang, B.; He, J.; Hu, Y.; Khademhosseini, A. A sandwiched microarray platform for benchtop cell-based high throughput screening. Biomaterials 2011, 32, 841–848. [Google Scholar] [CrossRef]
- Xu, Z.; Gao, Y.; Hao, Y.; Li, E.; Wang, Y.; Zhang, J.; Wang, W.; Gao, Z.; Wang, Q. Application of a microfluidic chip-based 3D co-culture to test drug sensitivity for individualized treatment of lung cancer. Biomaterials 2013, 34, 4109–4117. [Google Scholar] [CrossRef]
- Zhao, X.M.; Li, N.; Du, C.X. Chemical Constituents of Lowering LDL-C in Effective Part of Oroxylum indicum. Zhong Yao Cai 2016, 39, 555–558. [Google Scholar]
- Nagasaka, M.; Hashimoto, R.; Inoue, Y.; Ishiuchi, K.; Matsuno, M.; Itoh, Y. Anti-Tumorigenic Activity of Chrysin from Oroxylum indicum via Non-Genotoxic p53 Activation through the ATM-Chk2 Pathway. Molecules 2018, 23, 1394. [Google Scholar] [CrossRef]
- Lee, A.Y.; Kang, S.; Park, S.J.; Huang, J.; Im, D.S. Anti-Allergic Effect of Oroxylin A from Oroxylum indicum Using in vivo and in vitro Experiments. Biomol. Ther. 2016, 24, 283–290. [Google Scholar] [CrossRef]
- Hoshida, Y.; Fuchs, B.C.; Tanabe, K.K. Prevention of hepatocellular carcinoma: Potential targets, experimental models, and clinical challenges. Curr. Cancer Drug Targets 2012, 12, 1129–1159. [Google Scholar]
- Mann, C.D.; Neal, C.P.; Garcea, G.; Manson, M.M.; Dennison, A.R.; Berry, D.P. Phytochemicals as potential chemopreventive and chemotherapeutic agents in hepatocarcinogenesis. Eur. J. Cancer Prev. 2009, 18, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Nakae, D.; Yoshiji, H.; Mizumoto, Y.; Horiguchi, K.; Shiraiwa, K.; Tamura, K.; Denda, A.; Konishi, Y. High incidence of hepatocellular carcinomas induced by a choline deficient L-amino acid defined diet in rats. Cancer Res. 1992, 52, 5042–5045. [Google Scholar] [PubMed]
- Liu, H.; Chang, J.K.; Hou, J.Q.; Zhao, Z.H.; Zhang, L.D. Inhibition of miR-221 influences bladder cancer cell proliferation and apoptosis. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3193–3199. [Google Scholar] [PubMed]
- Francesca, F.; Maddalena, M.; Marzia, G.; Elisa, C.; Angelo, V.; Hisamitsu, M. p53/mdm2 Feedback Loop Sustains miR-221 Expression and Dictates the Response to Anticancer Treatments in Hepatocellular Carcinoma. Mol. Cancer Res. 2014, 12, 203–216. [Google Scholar]
- Rong, M.; Chen, G.; Dang, Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer 2013, 13, 21. [Google Scholar] [CrossRef]
- Visone, R.; Russo, L.; Pallante, P.; De, M.I.; Ferraro, A.; Leone, V. MicroRNAs miR-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip 1 protein levels and cell cycle. Endocr. Relat. Cancer 2007, 14, 791–798. [Google Scholar] [CrossRef]
- Zhao, G.; Cai, C.; Yang, T.; Qiu, X.; Liao, B.; Li, W.; Ji, Z.; Zhao, J.; Zhao, H.; Guo, M. MicroRNA-221 Induces Cell Survival and Cisplatin Resistance through PI3K/Akt Pathway in Human Osteosarcoma. PLoS ONE 2013, 8, e53906. [Google Scholar]
- Di, C.A.; Pandolfi, P.P. The Multiple Roles of PTEN in Tumor Suppression. Cell 2000, 100, 387–390. [Google Scholar]
- Xue, Q.; Sun, K.; Deng, H.J.; Lei, S.T.; Dong, J.Q.; Li, G.X. Anti-miRNA-221 sensitizes human colorectal carcinoma cells to radiation by upregulating PTEN. World J. Gastroenterol. 2013, 19, 9307–9317. [Google Scholar] [CrossRef]
- Zhang, C.; Kang, C.; Wang, P.; Cao, Y.; Yu, S. MicroRNA-221 and -222 Regulate Radiation Sensitivity by Targeting the PTEN Pathway. Int. J. Radiat. Oncol. Biol. Phys. 2011, 80, 240–248. [Google Scholar] [CrossRef]
- Xie, Q.; Yan, Y.R.; Huang, Z.P.; Zhong, X.Y.; Huang, L. MicroRNA-221 targeting PI3-K/Akt signaling axis induces cell proliferation and BCNU resistance in human glioblastoma. Neuropathology 2014, 34, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.X.; Wang, X.; Sun, C.; Zheng, X.; Wei, H.; Tian, Z.; Sun, R. Chronic Alcohol Consumption Promotes Diethylnitrosamine-Induced Hepatocarcinogenesis via Immune Disturbances. Sci. Rep. 2017, 7, 2567–2574. [Google Scholar] [CrossRef] [PubMed]
- Bruna, P.M.R.; Camilla, P.S.; Chun, L.; Andrew, S.G.; Lynne, M.A.; Wang, X.D. Inhibition of diethylnitrosamine-initiated alcohol-promoted hepatic inflammation and precancerous lesions by flavonoid luteolin is associated with increased sirtuin 1 activity in mice. HepatoBiliary Surg. Nutr. 2015, 4, 124–134. [Google Scholar]
- Femke, H.; Isabelle, C.; Hans, V.V. Experimental mouse models for hepatocellular carcinoma Research. Int. J. Exp. Path. 2009, 90, 367–386. [Google Scholar]
- Duan, T.; Sun, W.; Zhang, M.; Ge, J.; He, Y.; Zhang, J.; Zheng, Y.; Yang, W.; Shen, H.M.; Yang, J. Dietary restriction protects against diethylnitrosamine-induced hepatocellular tumorigenesis by restoring the disturbed gene expression profile. Sci. Rep. 2017, 7, 43745–43758. [Google Scholar] [CrossRef]
- Zhang, X.L.; Yu, H.; Xiong, Y.Y.; Ma, S.T.; Zhao, L.; She, S.F. Resveratrol Down-Regulates Myosin Light Chain Kinase, Induces Apoptosis and Inhibits Diethylnitrosamine-Induced Liver Tumorigenesis in Rats. Int. J. Mol. Sci. 2013, 14, 1940–1951. [Google Scholar] [CrossRef]
- Chou, W.W.; Wang, Y.T.; Liao, Y.C.; Wang, S.N.; Juo, S.H.H. Decreased MicroRNA-221 is Associated with High Levels of TNF-α in Human Adipose Tissue-Derived Mesenchymal Stem Cells from Obese Woman. Cell Physiol. Biochem. 2013, 32, 127–137. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Gao, Y.; Cai, L.; Li, F.; Lou, Y.; Xu, N.; Kang, Y.; Yang, H. MicroRNA-221 is involved in the regulation of osteoporosis through regulates RUNX2 protein expression and osteoblast differentiation. Am. J. Transl. Res. 2017, 9, 126–135. [Google Scholar]
- Yusuke, G.; Satoko, K.; Rika, N.; Akira, K.; Mayuko, K.; Hideki, E. MicroRNA expression signature of castration-resistant prostate cancer: The microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br. J. Cancer 2015, 113, 1055–1065. [Google Scholar]
Sample Availability: Not available. |
| Name | Log FC ([OB] vs. [Control]) | Regulation | Active_Sequence |
|---|---|---|---|
| miR-128-3p | −2.63 | Down | AAAGAGACCGGTTCACTGTG |
| miR-129-1-3p | −4.21 | Down | CTTTTTGGGGTAAGGG |
| miR-221-3p | −2.93 | Down | GAAACCCAGCAGACAATGTA |
| miR-300-3p | −2.98 | Down | GAAGAGAGCTTGCCCTTG |
| miR-324-5p | −2.81 | Down | ACACCAATGCCCTAGGG |
| miR-328a-3p | −2.93 | Down | ACGGAAGGGCAGAGAGGGC |
| miR-335 | −4.32 | Down | ACATTTTTCGTTATTGCTCT |
| miR-127-3p | −3.57 | Up | AGCCAAGCTCAGACGGAT |
| miR-212-3p | 5.44 | Up | TGGCCGTGACTGGA |
| miR-3099 | 2.69 | Up | TCCCCAACCTCTTTCT |
| miR-30b-3p | 3.47 | Up | GACGTAAACATCCACATCCCA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, N.; Men, W.; Zheng, Y.; Wang, H.; Meng, X. Oroxin B Induces Apoptosis by Down-Regulating MicroRNA-221 Resulting in the Inactivation of the PTEN/PI3K/AKT Pathway in Liver Cancer. Molecules 2019, 24, 4384. https://doi.org/10.3390/molecules24234384
Li N, Men W, Zheng Y, Wang H, Meng X. Oroxin B Induces Apoptosis by Down-Regulating MicroRNA-221 Resulting in the Inactivation of the PTEN/PI3K/AKT Pathway in Liver Cancer. Molecules. 2019; 24(23):4384. https://doi.org/10.3390/molecules24234384
Chicago/Turabian StyleLi, Nannan, Wenxiao Men, Yibo Zheng, Hechen Wang, and Xiansheng Meng. 2019. "Oroxin B Induces Apoptosis by Down-Regulating MicroRNA-221 Resulting in the Inactivation of the PTEN/PI3K/AKT Pathway in Liver Cancer" Molecules 24, no. 23: 4384. https://doi.org/10.3390/molecules24234384
APA StyleLi, N., Men, W., Zheng, Y., Wang, H., & Meng, X. (2019). Oroxin B Induces Apoptosis by Down-Regulating MicroRNA-221 Resulting in the Inactivation of the PTEN/PI3K/AKT Pathway in Liver Cancer. Molecules, 24(23), 4384. https://doi.org/10.3390/molecules24234384