First Attempts of the Use of 195Pt NMR of Phenylbenzothiazole Complexes as Spectroscopic Technique for the Cancer Diagnosis
Abstract
1. Introduction
2. Results and Discussion
2.1. Validation of 195Pt NMR Chemical Shifts Theoretical Methodology
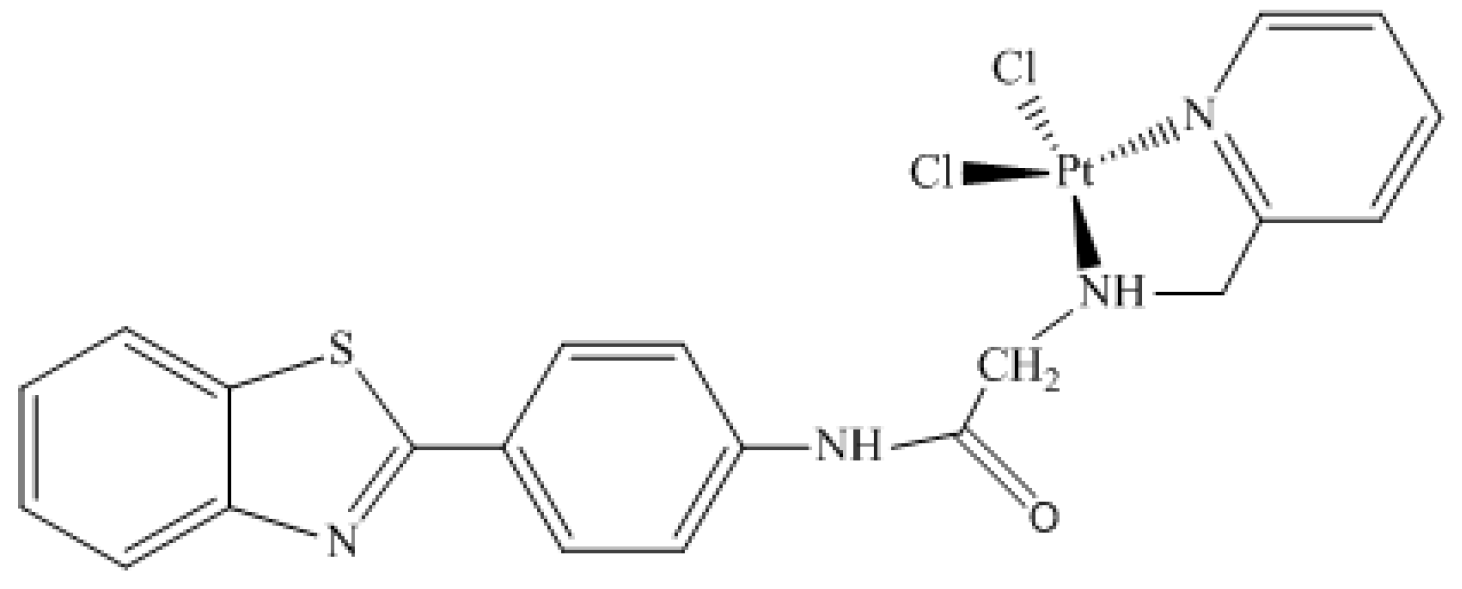
2.2. The 195Pt Nuclear Magnetic Resonance Chemical Shift
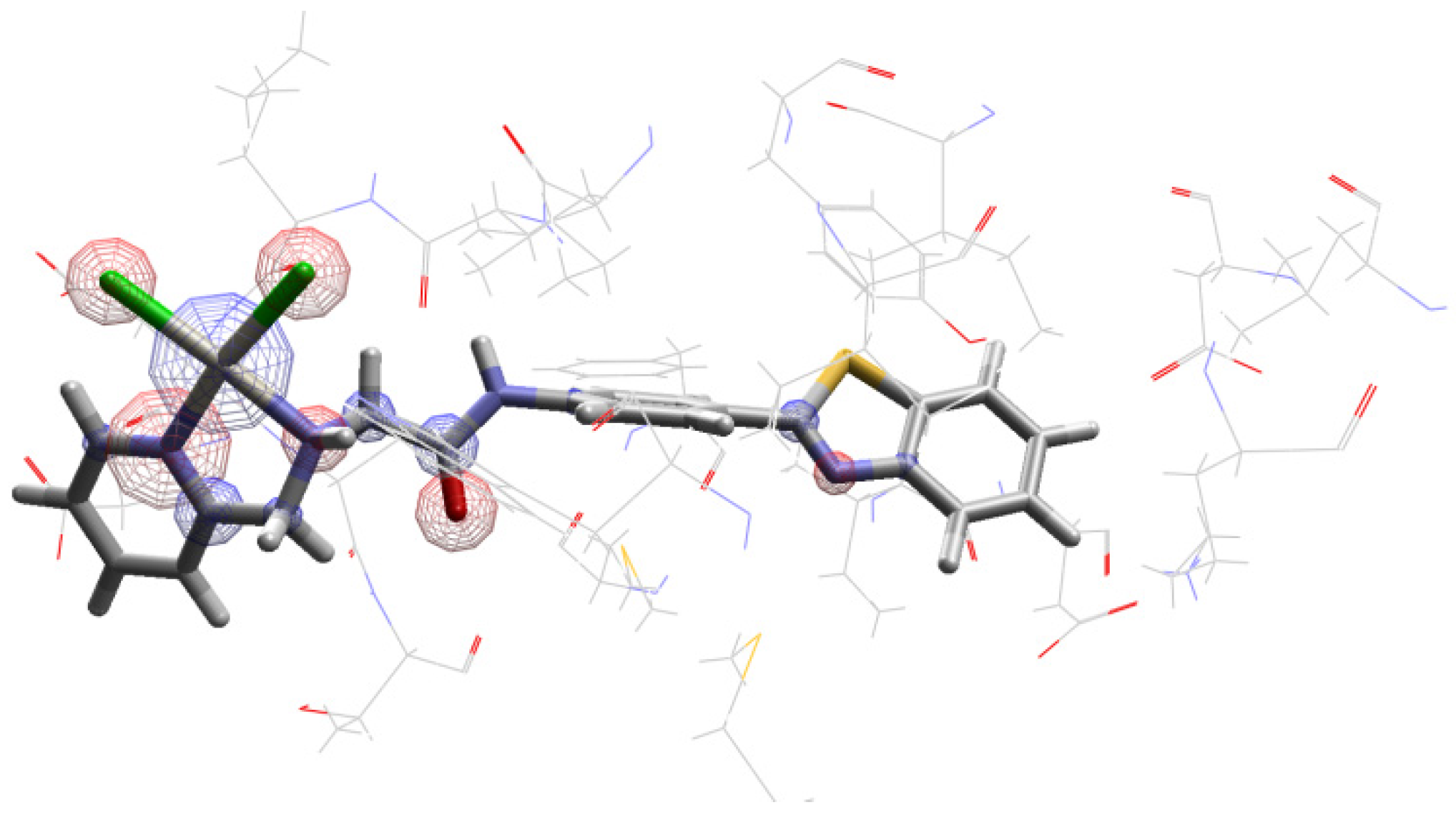
The 195Pt Chemical Shift in PI3K Enzyme Active Site
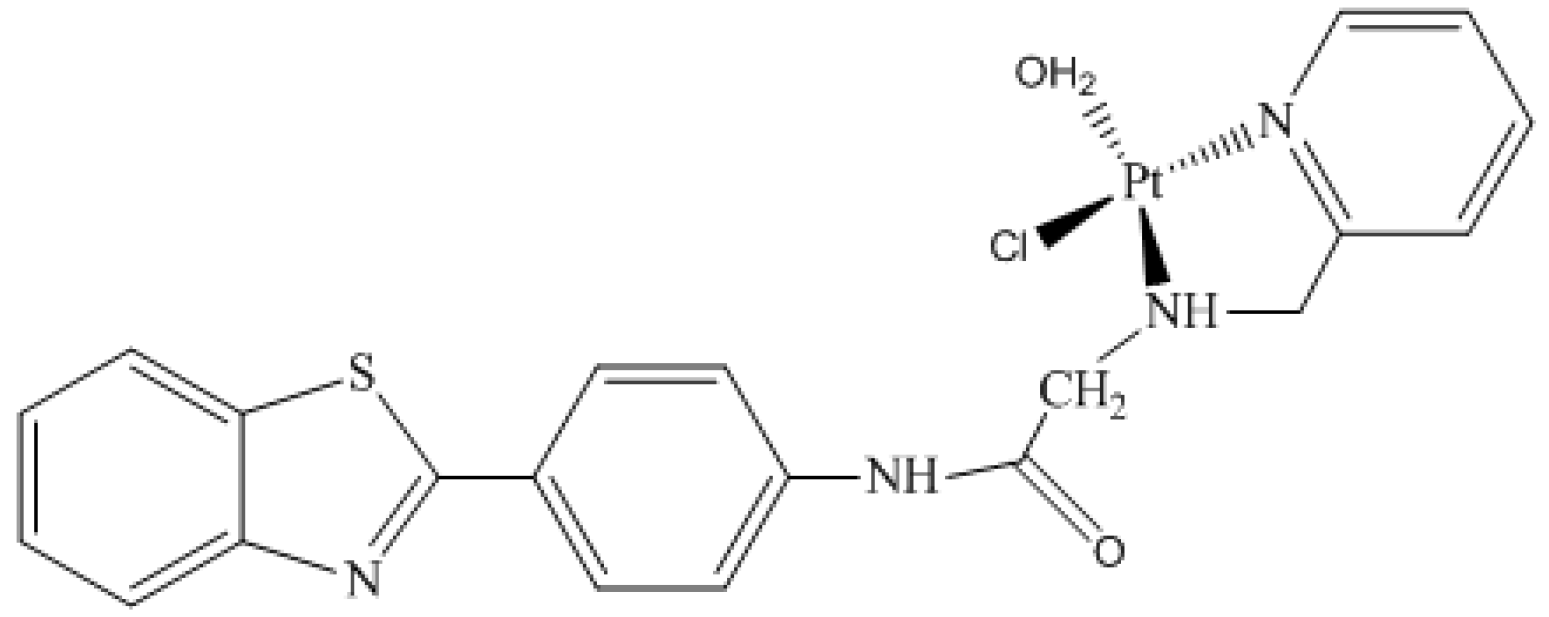
2.3. The 195Pt Chemical Shift for Monoaqua Platinum Complex
3. Methodology
3.1. Optimization and Molecular Dynamics (MD) Procedure
3.2. Nuclear Magnetic Resonance (NMR) Calculations
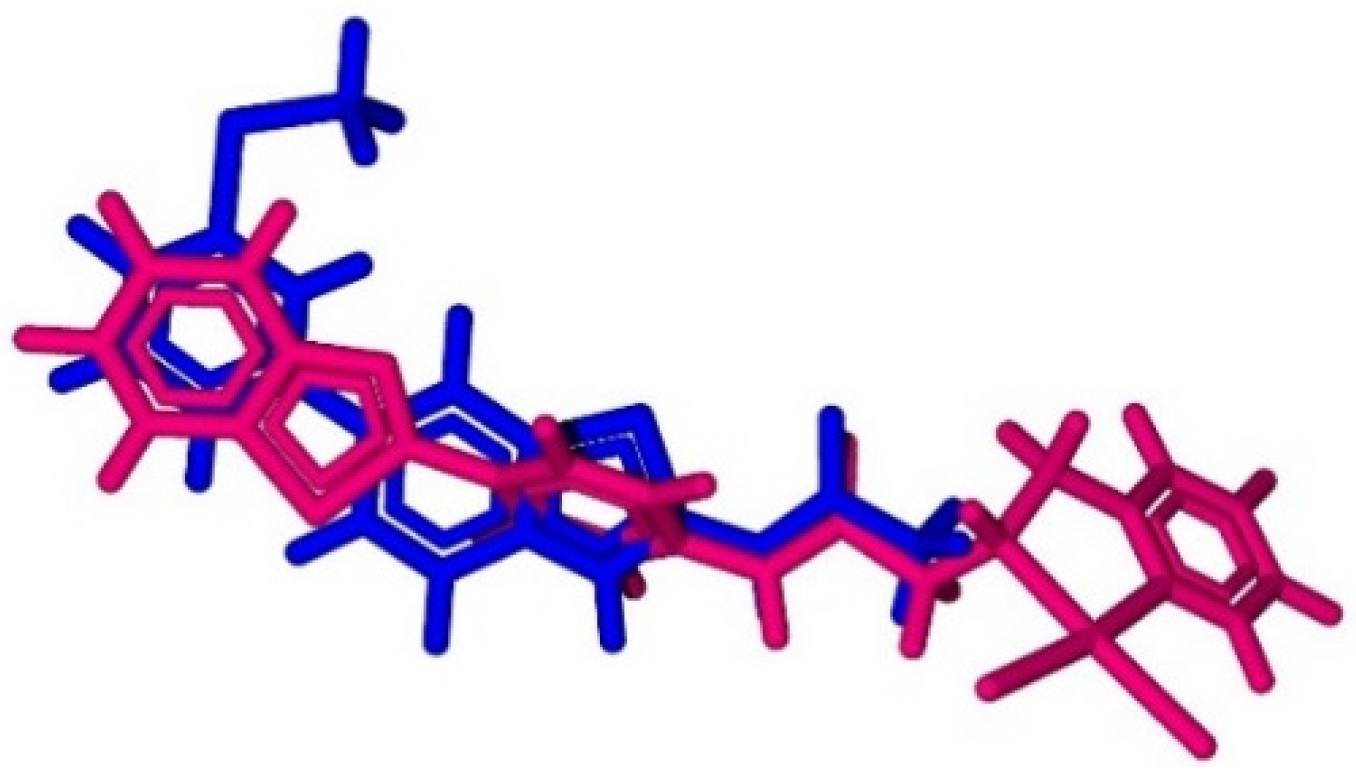
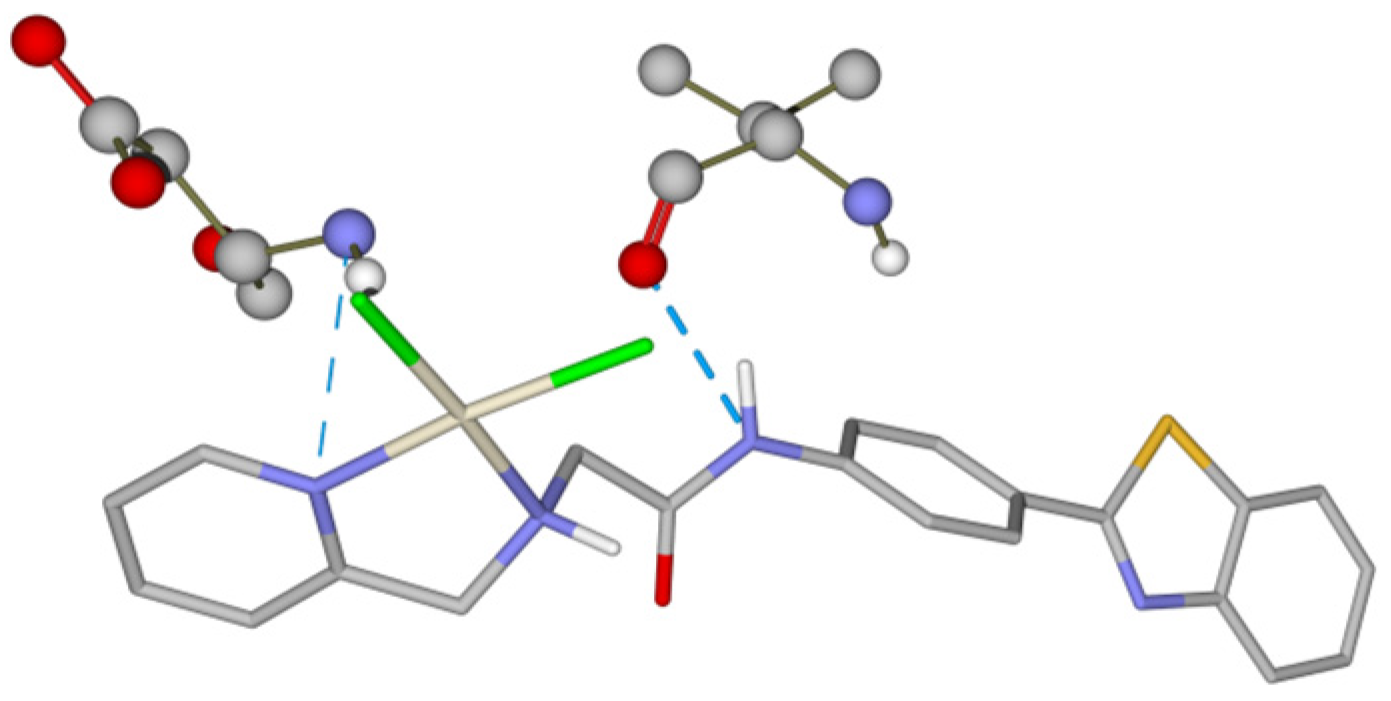
3.3. Molecular Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Jafari, S.; Saadatpour, Z.; Salmaninejad, A.; Momeni, F.; Mokhtari, M.; Nahand, J.S.; Rahmati, M.; Mirzaei, H.; Kianmehr, M. Breast cancer diagnosis: Imaging techniques and biochemical markers. J. Cell. Physiol. 2018, 233, 5200–5213. [Google Scholar] [CrossRef] [PubMed]
- Park, J.V.; Park, S.J.; Yoo, J.S. Finding characteristics of exceptional breast cancer subpopulations using subgroup mining and statistical test. Expert Syst. Appl. 2019, 118, 553–562. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, G.; Hao, X.; Ma, Y.; Ma, M.; Yan, X.; Jiang, X.; Bie, F.; Yuan, N. Potential biomarker for breast cancer screening: A systematic review and meta-analysis. Futur. Gener. Comput. Syst. 2019, 91, 518–526. [Google Scholar] [CrossRef]
- Zonouzy, V.T.; Niknami, S.; Ghofranipour, F.; Montazeri, A. An educational intervention based on the extended parallel process model to improve attitude, behavioral intention, and early breast cancer diagnosis: A randomized trial. Int. J. Women’s Health 2019, 11, 1–10. [Google Scholar] [CrossRef]
- Heneghan, H.M.; Miller, N.; Kelly, R.; Newell, J.; Kerin, M.J. Systemic miRNA-195 Differentiates Breast Cancer from Other Malignancies and Is a Potential Biomarker for Detecting Noninvasive and Early Stage Disease. Oncologist 2010, 15, 673–682. [Google Scholar] [CrossRef]
- Cui, X.; Li, Z.; Zhao, Y.; Song, A.; Shi, Y. Breast cancer identification via modeling of peripherally circulating miRNAs. PeerJ 2018, 6, e4551. [Google Scholar] [CrossRef]
- Khalkhali, I.; Mena, I.; Diggles, L. Review of imaging techniques for the diagnosis of breast cancer: A new role of prone scintimammography using technetium-99m sestamibi. Eur. J. Nucl. Med. 1994, 21, 357–362. [Google Scholar] [CrossRef]
- Pereira, B.T.L.; Silva, E.F.; Gonçalves, M.A.; Mancini, D.T.; Ramalho, T.C. Exploring EPR Parameters of99Tc Complexes for Designing New MRI Probes: Coordination Environment, Solvent, and Thermal Effects on the Spectroscopic Properties. J. Chem. 2017, 2017, 8102812. [Google Scholar] [CrossRef]
- Duffy, M.J.; Harbeck, N.; Nap, M.; Molina, R.; Nicolini, A.; Senkus, E.; Cardoso, F. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur. J. Cancer 2017, 75, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Kurozumi, S.; Yamaguchi, Y.; Kurosumi, M.; Ohira, M.; Matsumoto, H.; Horiguchi, J. Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J. Hum. Genet. 2017, 62, 15–24. [Google Scholar] [CrossRef]
- Mavroidi, B.; Sagnou, M.; Stamatakis, K.; Paravatou-petsotas, M.; Pelecanou, M.; Methenitis, C. Palladium (II) and platinum (II) complexes of derivatives of 2-(40-aminophenyl) benzothiazole as potential anticancer agents. Inorg. Chim. Acta 2016, 444, 63–75. [Google Scholar] [CrossRef]
- Mancini, D.T.; Souza, E.F.; Caetano, M.S.; Ramalho, T.C. 99Tc NMR as a promising technique for structural investigation of biomolecules: Theoretical studies on the solvent and thermal effects of phenylbenzothiazole complex. Magn. Reson. Chem. 2014, 53, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Rosernberg, B.; Vancamp, L.; Krigas, T. Inhibition of Cell Division in Escherichia coli by Electrolysis Products from a Platinum Electrode. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef]
- Weiss, R.B.; Christian, M.C. New Cisplatin Analogues in Development. A review. Drugs 1993, 46, 360–377. [Google Scholar] [CrossRef]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef]
- Deo, K.M.; Ang, D.L.; Mcghie, B.; Rajamanickam, A.; Dhiman, A.; Khoury, A.; Holland, J.; Bjelosevic, A.; Pages, B.; Gordon, C.; et al. platinum coordination compounds with potent anticancer activity. Coord. Chem. Rev. 2018, 375, 148–163. [Google Scholar] [CrossRef]
- Brabec, V.; Hrabina, O.; Kasparkova, J. Cytotoxic platinum coordination compounds. DNA binding agents. Coord. Chem. Rev. J. 2017, 351, 2–31. [Google Scholar] [CrossRef]
- Solomon, V.R.; Hu, C.; Lee, H. Hybrid pharmacophore design and synthesis of isatin-benzothiazole analogs for their anti-breast cancer activity. Bioorg. Med. Chem. 2009, 17, 7585–7592. [Google Scholar] [CrossRef]
- Abdelgawad, M.A.; Belal, A.; Omar, H.A.; Hegazy, L.; Rateb, M.E. Synthesis, Anti-Breast Cancer Activity, and Molecular Modeling of Some Benzothiazole and Benzoxazole Derivatives. Arch. Pharm. Chem. Life Sci. 2013, 346, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S. Kinetic aspects of platinum anticancer agents. Polyhedron 2017, 138, 109–124. [Google Scholar] [CrossRef]
- Benedetti, M.; Girelli, C.R.; Antonucci, D.; Fanizzi, F.P. [PtCl(η1-CH2-CH2OR)(NN). and [PtCl(η2-CH2CH2)(NN).+, NN = dinitrogen ligand, complexes. Sterical and electronic effects evidenced by NMR analysis. J. Organomet. Chem. 2014, 771, 40–46. [Google Scholar] [CrossRef]
- Benedetti, M.; Antonucci, D.; Fanizzi, F.P.; Papadia, P.; de Castro, F. General cooperative effects of single atom ligands on a metal: A 195 Pt NMR chemical shift as a function of coordinated halido ligands’ ionic radii overall sum. Dalton Trans. 2015, 44, 15377–15381. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Papadia, P.; Girelli, C.R.; De Castro, F.; Capitelli, F.; Fanizzi, F.P. X-ray structures versus NMR signals in pentacoordinate [PtX2(η2-CH2=CH2)(Me2phen). (X = Cl, Br, I) complexes. Inorganica Chim. Acta 2015, 428, 8–13. [Google Scholar] [CrossRef]
- Benedetti, M.; Antonucci, D.; Girelli, C.R.; Fanizzi, F.P. Hindrance, Donor Ability of Men_NN Chelates and Overall Stability of Pentacoordinate [PtCl2(η2-CH2=CH2) (Men_NN). Complexes as Observed by η2-Olefin 1JPt,C Modulation: An NMR Study. Eur. J. Inorg. Chem. 2015, 2308–2316. [Google Scholar] [CrossRef]
- Paschoal, D.; Guerra, C.F.; de Oliveira, M.A.; Ramalho, T.C.; Dos Santos, H.F. Predicting Pt-195 NMR Chemical Shift Using New Relativistic All-Electron Basis Set. J. Comput. Chem. 2016, 37, 2360–2373. [Google Scholar] [CrossRef]
- Still, B.M.; Kumar, P.G.A.; Aldrich-Wright, J.R.; Price, W.S. 195Pt NMR—Theory and application. Chem. Soc. Rev. 2007, 36, 665–686. [Google Scholar] [CrossRef]
- Thomsen, R.; Christensen, M.H. MolDock: A New Technique for High-Accuracy Molecular Docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg. J. Chem. Phys. 1985, 82, 270–283. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced Basis Sets of Split Valence, Triple Zeta Valence and Quadruple Zeta Valence. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Gaussian, Inc.: Wallingford, CT, USA, 2009.
- Barone, V.; Cossi, M.; Tomasi, J. Geometry optimization of molecular structures in solution by the polarizable continuum model. J. Comput. Chem. 1998, 19, 404–417. [Google Scholar] [CrossRef]
- Cancès, E.; Mennucci, B.; Tomasi, J. A new integral equation formalism for the polarizable continuum model: Theoretical background and applications to isotropic and anisotropic dielectrics. J. Chem. Phys. 1997, 107, 3032–3041. [Google Scholar] [CrossRef]
- Gonçalves, M.A.; Santos, L.S.; Prata, D.M.; Peixoto, F.C.; da Cunha, E.F.F.; Ramalho, T.C. Optimal wavelet signal compression as an efficient alternative to investigate molecular dynamics simulations: Application to thermal and solvent effects of MRI probes. Theor. Chem. Acc. 2017, 136, 15. [Google Scholar] [CrossRef]
- Iyengar, S.S.; Frisch, M.J. Effect of time-dependent basis functions and their superposition error on atom- centered density matrix propagation (ADMP): Connections to wavelet theory of multiresolution analysis. J. Chem. Phys. 2004, 121, 5061–5070. [Google Scholar] [CrossRef]
- SciLab v 2.7. 1989–2003 INRIA/ENPC. Available online: www.scilab.org (accessed on 30 October 2019).
- Wolinski, K.; Hinton, J.F.; Pulay, P. Efficient Implementation of the Gauge-Independent Atomic Orbital Method for NMR Chemical Shift Calculations. J. Am. Chem. Soc. 1990, 112, 8251–8260. [Google Scholar] [CrossRef]
- Cheeseman, J.R.; Trucks, G.W.; Keith, T.A.; Frisch, M.J. A comparison of models for calculating nuclear magnetic resonance shielding tensors. J. Chem. Phys. 1996, 104, 5497–5509. [Google Scholar] [CrossRef]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef] [PubMed]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1997, 78, 1396. [Google Scholar] [CrossRef]
- Ramalho, T.C.; Pereira, C.M.P.; Martins, T.L.C.; Figueroa-Villar, J.D.; Flores, A.F.C. Theoretical and experimental 13C and 15N NMR investigation of guanylhydrazones in solution. Magn. Reson. Chem. 2003, 41, 983–988. [Google Scholar] [CrossRef]
- D’Angelo, N.D.; Kim, T.S.; Andrews, K.; Booker, S.K.; Caenepeel, S.; Chen, K.; Amico, D.D.; Freeman, D.; Jiang, J.; Liu, L.; et al. Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/ Mammalian Target of Rapamycin (mTOR) Dual Inhibitors. J. Med. Chem. 2011, 54, 1789–1811. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, T.C.; Taft, C. Thermal and solvent effects on the NMR and UV parameters of some bioreductive drugs. J. Chem. Phy. 2005, 123, 54319. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, A.P.; França, T.C.; Oliveira, A.A.; Da Cunha, E.F.F.; Ramalho, T.C. Design of New Chemotherapeutics Against the Deadly Anthrax Disease. Docking and Molecular Dynamics Studies of Inhibitors Containing Pyrrolidine and Riboamidrazone Rings on Nucleoside Hydrolase from Bacillus anthracis. J. Biomol. Struct. Dyn. 2011, 28, 455–470. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
| Level of Approximation | 195Pt (ppm) |
|---|---|
| δe(PBEPBE//B3LYP) | −1960.49 |
| δe(PBEPBE//B3LYP/PCM(H2O)_ | −2296.13 |
| δe(PBEPBE/PCM(H2O)//B3LYP/PCM(H2O)) | −1783.80 |
| δ310K(PBEPBE//ADMP) | −2256.00 |
| δ310K (PBEPBE/PCM(H2O)//ADMP/PCM(H2O)) | −4069.56 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, B.T.L.; Gonçalves, M.A.; Mancini, D.T.; Kuca, K.; Ramalho, T.C. First Attempts of the Use of 195Pt NMR of Phenylbenzothiazole Complexes as Spectroscopic Technique for the Cancer Diagnosis. Molecules 2019, 24, 3970. https://doi.org/10.3390/molecules24213970
Pereira BTL, Gonçalves MA, Mancini DT, Kuca K, Ramalho TC. First Attempts of the Use of 195Pt NMR of Phenylbenzothiazole Complexes as Spectroscopic Technique for the Cancer Diagnosis. Molecules. 2019; 24(21):3970. https://doi.org/10.3390/molecules24213970
Chicago/Turabian StylePereira, Bruna T. L., Mateus A. Gonçalves, Daiana T. Mancini, Kamil Kuca, and Teodorico C. Ramalho. 2019. "First Attempts of the Use of 195Pt NMR of Phenylbenzothiazole Complexes as Spectroscopic Technique for the Cancer Diagnosis" Molecules 24, no. 21: 3970. https://doi.org/10.3390/molecules24213970
APA StylePereira, B. T. L., Gonçalves, M. A., Mancini, D. T., Kuca, K., & Ramalho, T. C. (2019). First Attempts of the Use of 195Pt NMR of Phenylbenzothiazole Complexes as Spectroscopic Technique for the Cancer Diagnosis. Molecules, 24(21), 3970. https://doi.org/10.3390/molecules24213970







