Enzymatic Cascades for Tailored 13C6 and 15N Enriched Human Milk Oligosaccharides
Abstract
1. Introduction
2. Results and Discussion
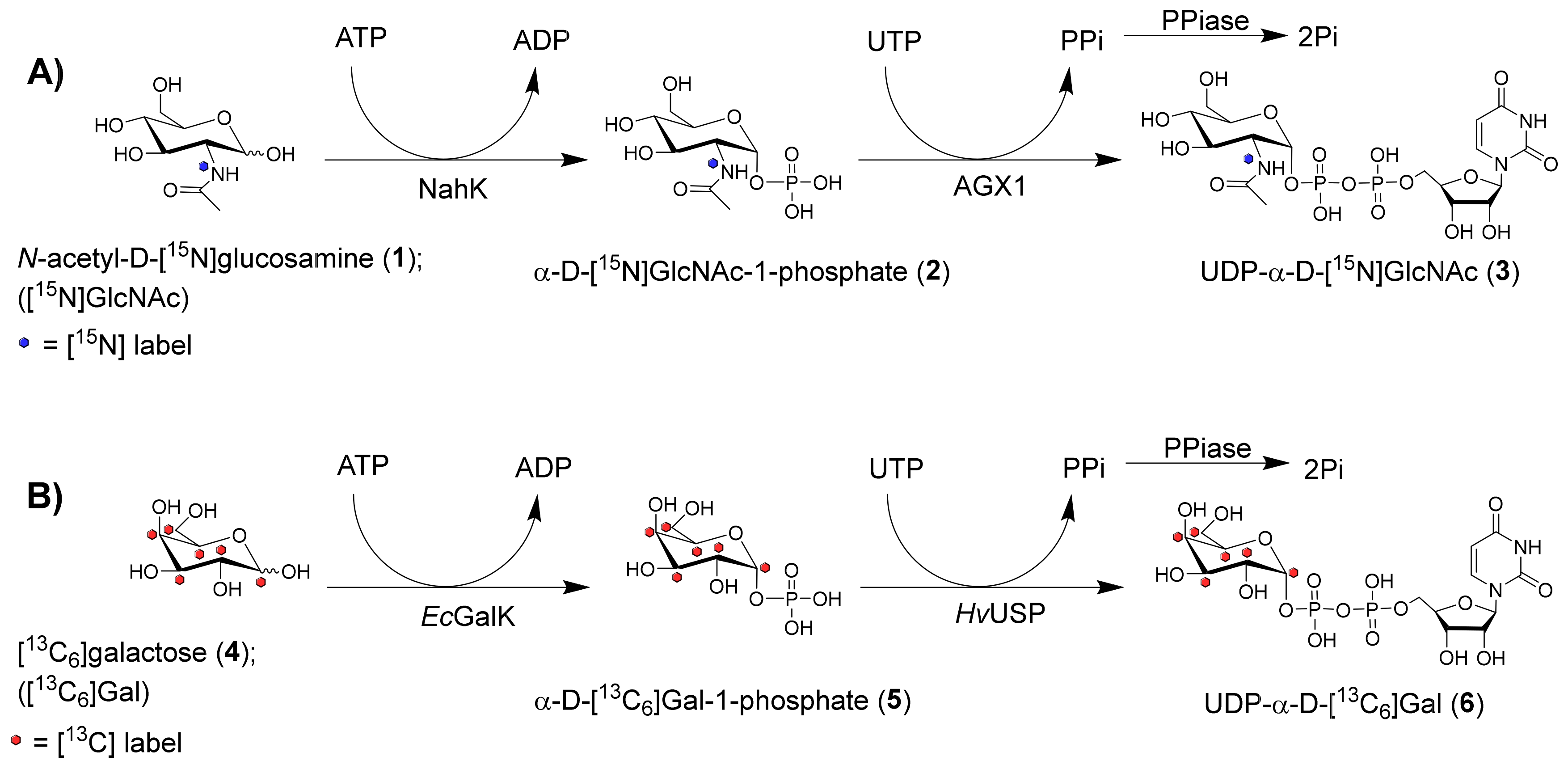
2.1. Nucleotide Sugar Synthesis
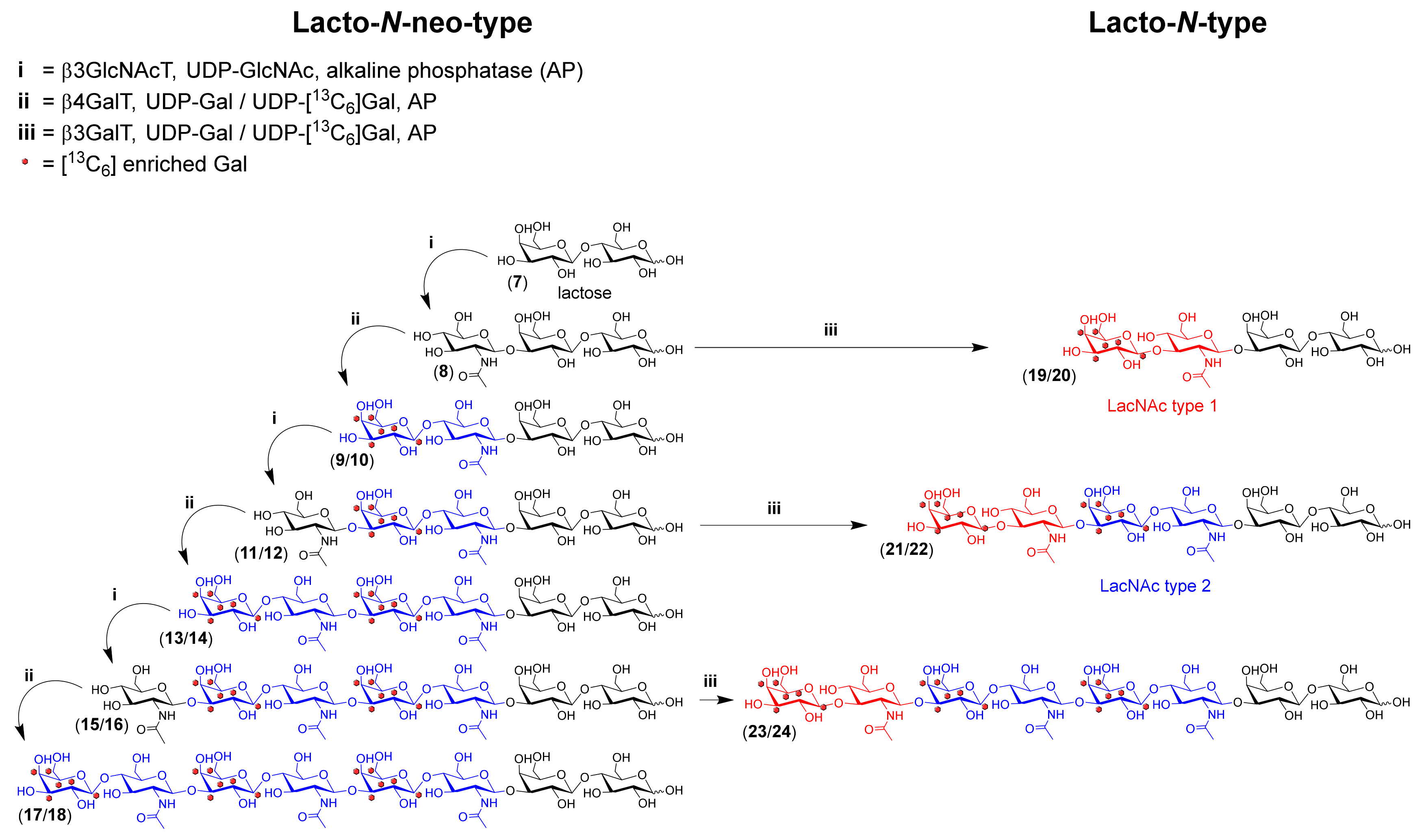
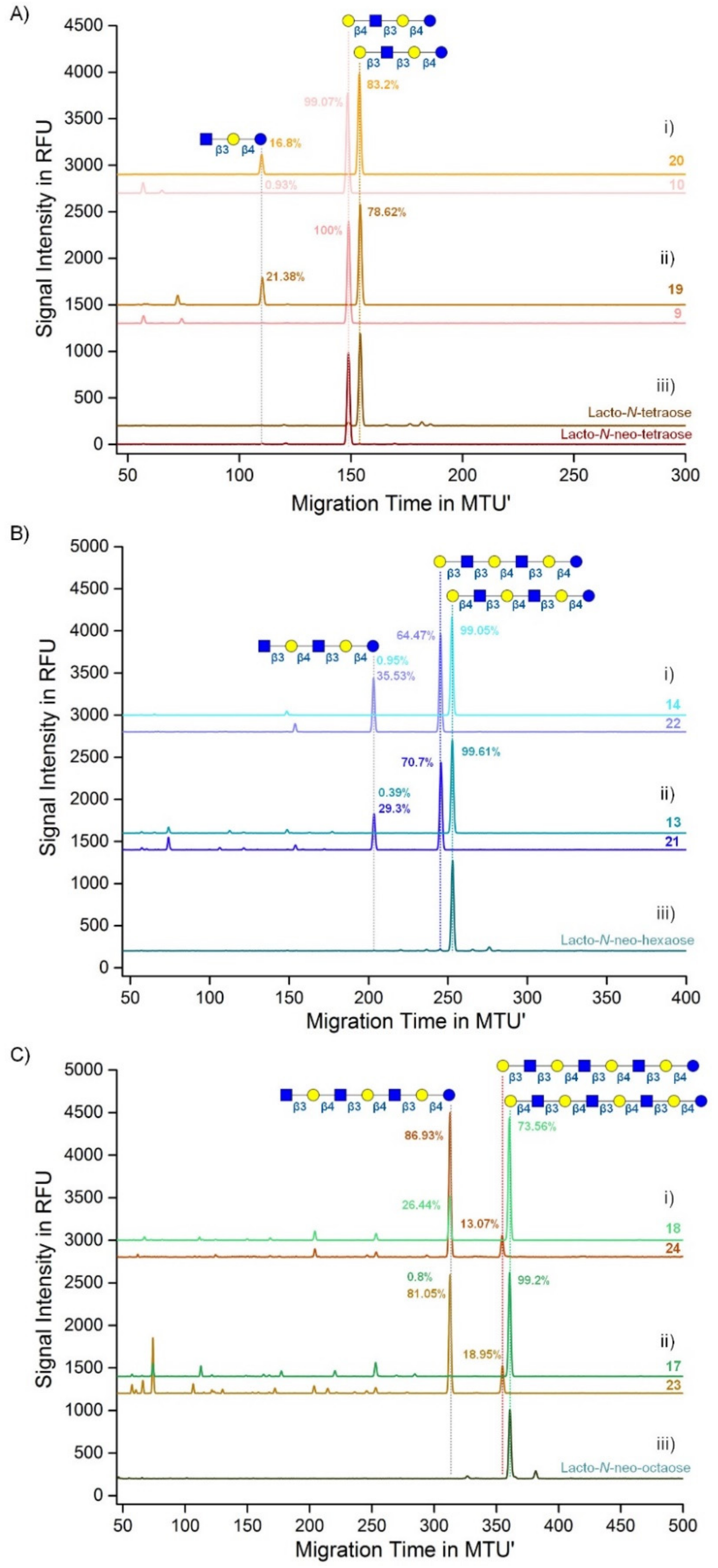
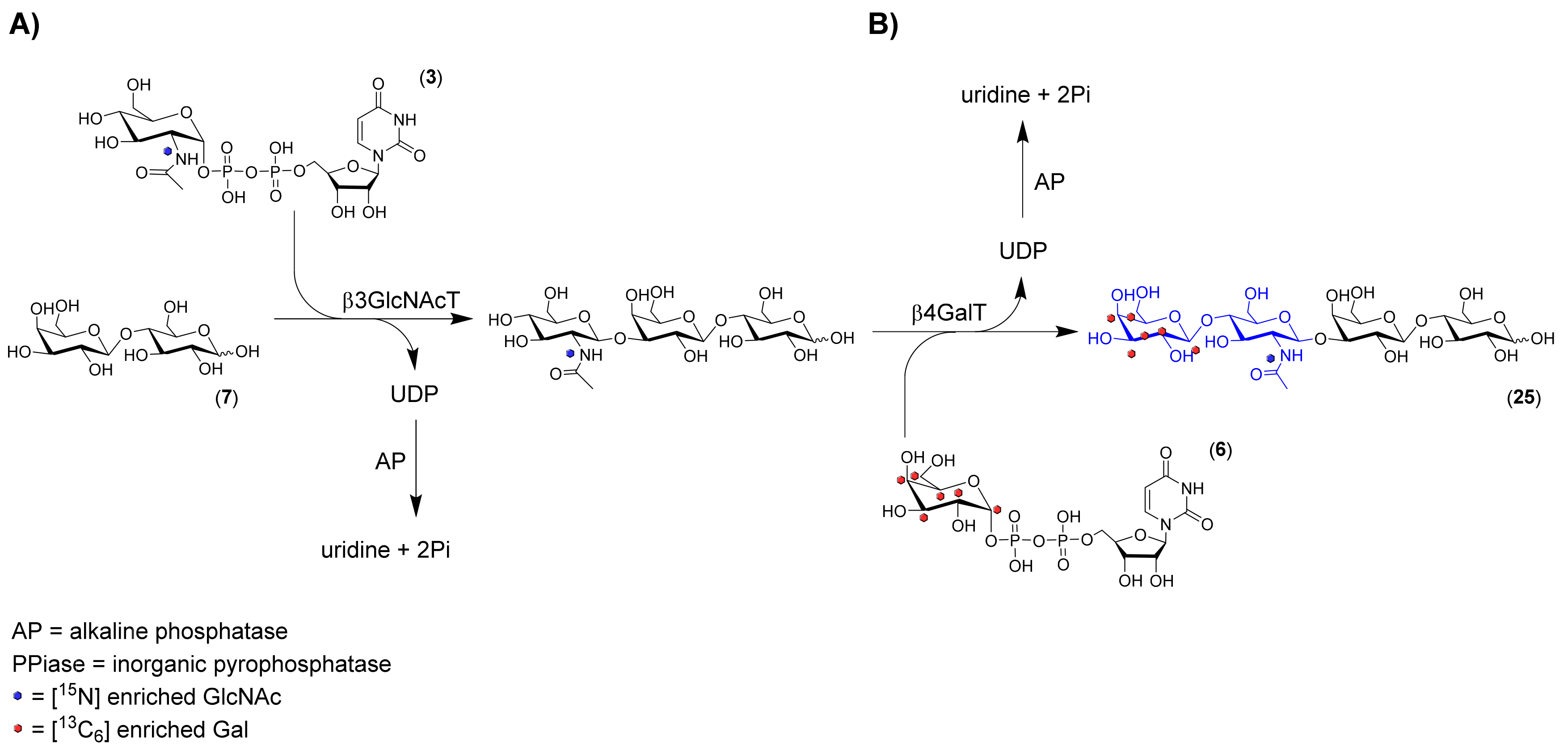
2.2. HMOS Synthesis
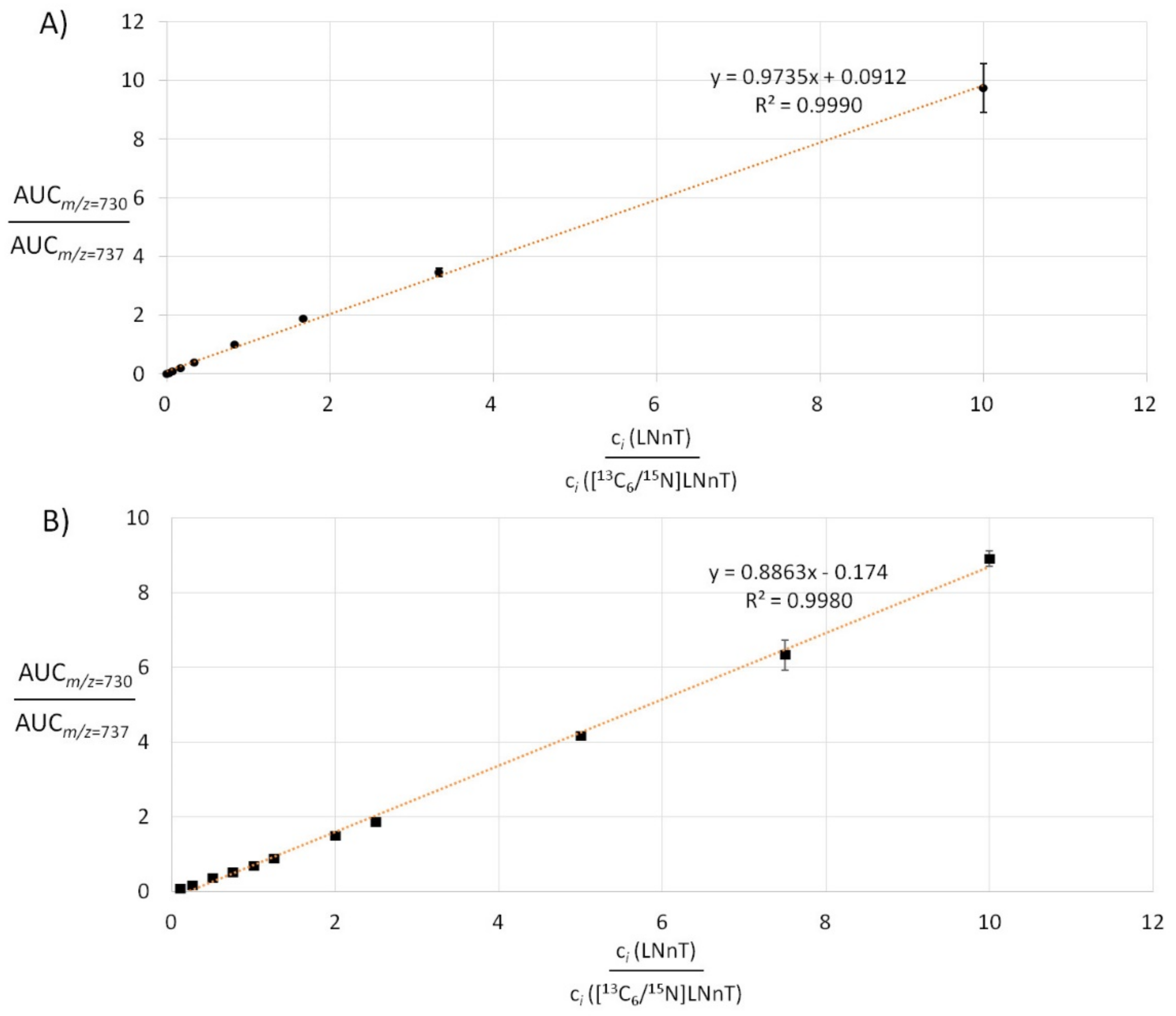
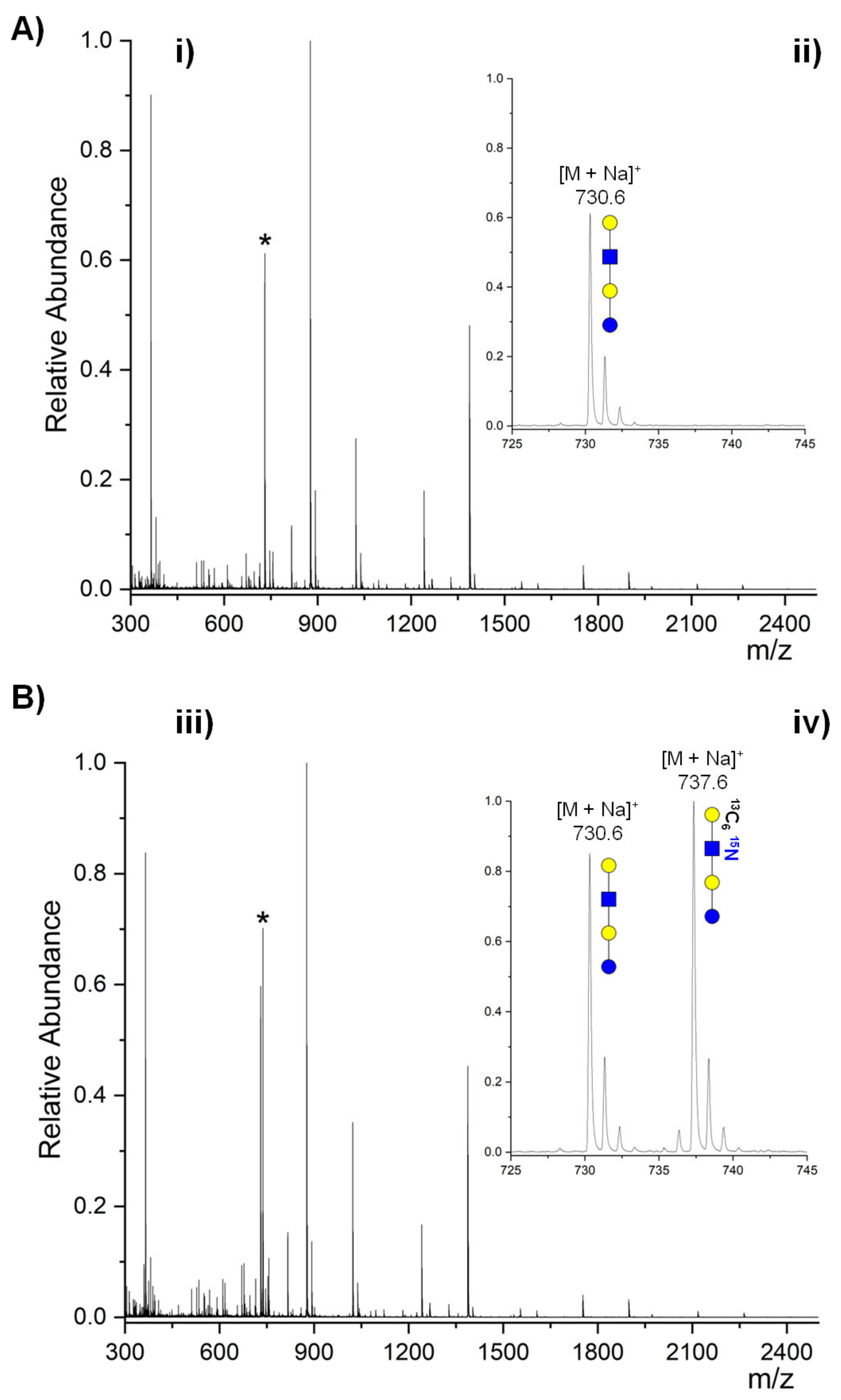
2.3. Absolute Compound Quantification in Complex Samples by MS
3. Materials and Methods
3.1. Recombinant Enzyme Preparation
3.2. Enzyme Activity Assay
3.3. HMOS Analysis via CE-LIF and xCGE-LIF
3.4. Nucleotide Sugar Synthesis
3.5. Sequential 13C6 and Native HMOS Synthesis
3.6. Sequential Synthesis of 13C6 and 15N-Labeled Lacto-N-Neo-Tetraose
3.7. TOC Analysis of the Purified [13C6/15N]Lacto-N-Neo-Tetraose
3.8. MS-Based Analysis
3.9. Nucleotide Sugar and HMOS Analysis via ESI-MS
3.10. Extraction of OS from Human Milk
3.11. MALDI-TOF-MS Analysis and Absolute Quantification of HMOS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. Human Milk Oligosaccharides (HMOS): Structure, Function, and Enzyme-Catalyzed Synthesis. Adv. Carbohydr. Chem. Biochem. 2015, 72, 113–190. [Google Scholar] [CrossRef] [PubMed]
- Kunz, C.; Rudloff, S.; Baier, W.; Klein, N.; Strobel, S. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annu. Rev. Nutr. 2000, 20, 699–722. [Google Scholar] [CrossRef] [PubMed]
- Coppa, G.V.; Pierani, P.; Zampini, L.; Carloni, I.; Carlucci, A.; Gabrielli, O. Oligosaccharides in human milk during different phases of lactation. Acta. Paediatr. Suppl. 1999, 88, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Newburg, D.S.; Neubauer, S.H. CHAPTER 4—Carbohydrates in Milks: Analysis, Quantities, and Significance. In Handbook of Milk Composition; Jensen, R.G., Ed.; Academic Press: San Diego, CA, USA, 1995; pp. 273–349. [Google Scholar] [CrossRef]
- Kobata, A. Structures and application of oligosaccharides in human milk. Proc. Jpn. Acad. Ser. B 2010, 86, 731–747. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, G.A.; Baumgärtner, F.; Albermann, C. Production of human milk oligosaccharides by enzymatic and whole-cell microbial biotransformations. J. Biotechnol. 2017, 258, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Thurl, S.; Munzert, M.; Boehm, G.; Matthews, C.; Stahl, B. Systematic review of the concentrations of oligosaccharides in human milk. Nutr. Rev. 2017, 75, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Bode, L.; Jantscher-Krenn, E. Structure-function relationships of human milk oligosaccharides. Adv. Nutr. 2012, 3, 383s–391s. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Davis, J.C.; Goonatilleke, E.; Smilowitz, J.T.; German, J.B.; Lebrilla, C.B. Absolute Quantitation of Human Milk Oligosaccharides Reveals Phenotypic Variations during Lactation. J. Nutr. 2017, 147, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Bych, K.; Mikš, M.H.; Johanson, T.; Hederos, M.J.; Vigsnæs, L.K.; Becker, P. Production of HMOs using microbial hosts—From cell engineering to large scale production. Curr. Opin. Biotechnol. 2019, 56, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Efsa Panel on Dietetic Products, Nutrition and Allergies. Statement on the safety of lacto-N-neotetraose and 2′-O-fucosyllactose as novel food ingredients in food supplements for children. EFSA J. 2015, 13, 4299. [Google Scholar] [CrossRef][Green Version]
- Vandenplas, Y.; Berger, B.; Carnielli, V.P.; Ksiazyk, J.; Lagstrom, H.; Sanchez Luna, M.; Migacheva, N.; Mosselmans, J.M.; Picaud, J.C.; Possner, M.; et al. Human Milk Oligosaccharides: 2′-Fucosyllactose (2′-FL) and Lacto-N-Neotetraose (LNnT) in Infant Formula. Nutrients 2018, 10, 1161. [Google Scholar] [CrossRef] [PubMed]
- Dotz, V.; Rudloff, S.; Meyer, C.; Lochnit, G.; Kunz, C. Metabolic fate of neutral human milk oligosaccharides in exclusively breast-fed infants. Mol. Nutr. Food Res. 2015, 59, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Galeotti, F.; Coppa, G.V.; Zampini, L.; Maccari, F.; Galeazzi, T.; Padella, L.; Santoro, L.; Gabrielli, O.; Volpi, N. On-line high-performance liquid chromatography-fluorescence detection-electrospray ionization-mass spectrometry profiling of human milk oligosaccharides derivatized with 2-aminoacridone. Anal. Biochem. 2012, 430, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Kottler, R.; Mank, M.; Hennig, R.; Muller-Werner, B.; Stahl, B.; Reichl, U.; Rapp, E. Development of a high-throughput glycoanalysis method for the characterization of oligosaccharides in human milk utilizing multiplexed capillary gel electrophoresis with laser-induced fluorescence detection. Electrophoresis 2013, 34, 2323–2336. [Google Scholar] [CrossRef]
- Echeverria, B.; Etxebarria, J.; Ruiz, N.; Hernandez, Á.; Calvo, J.; Haberger, M.; Reusch, D.; Reichardt, N.-C. Chemo-Enzymatic Synthesis of 13C Labeled Complex N-Glycans As Internal Standards for the Absolute Glycan Quantification by Mass Spectrometry. Anal. Chem. 2015, 87, 11460–11467. [Google Scholar] [CrossRef] [PubMed]
- Ninonuevo, M.R.; Ward, R.E.; LoCascio, R.G.; German, J.B.; Freeman, S.L.; Barboza, M.; Mills, D.A.; Lebrilla, C.B. Methods for the quantitation of human milk oligosaccharides in bacterial fermentation by mass spectrometry. Anal. Biochem. 2007, 361, 15–23. [Google Scholar] [CrossRef]
- Bartsch, H.; König, W.A.; Straβner, M.; Hintze, U. Quantitative determination of native and methylated cyclodextrins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Carbohydr. Res. 1996, 286, 41–53. [Google Scholar] [CrossRef]
- Baumgartner, F.; Jurzitza, L.; Conrad, J.; Beifuss, U.; Sprenger, G.A.; Albermann, C. Synthesis of fucosylated lacto-N-tetraose using whole-cell biotransformation. Bioorg. Med. Chem. 2015, 23, 6799–6806. [Google Scholar] [CrossRef]
- Han, N.S.; Kim, T.-J.; Park, Y.-C.; Kim, J.; Seo, J.-H. Biotechnological production of human milk oligosaccharides. Biotechnol. Adv. 2012, 30, 1268–1278. [Google Scholar] [CrossRef]
- Prudden, A.R.; Liu, L.; Capicciotti, C.J.; Wolfert, M.A.; Wang, S.; Gao, Z.; Meng, L.; Moremen, K.W.; Boons, G.-J. Synthesis of asymmetrical multiantennary human milk oligosaccharides. Proc. Natl. Acad. Sci. USA 2017, 114, 6954–6959. [Google Scholar] [CrossRef] [PubMed]
- Fischöder, T.; Cajic, S.; Reichl, U.; Rapp, E.; Elling, L. Enzymatic Cascade Synthesis Provides Novel Linear Human Milk Oligosaccharides as Reference Standards for xCGE-LIF Based High-Throughput Analysis. Biotechnol. J. 2019, 14, e1800305. [Google Scholar] [CrossRef] [PubMed]
- Zeuner, B.; Teze, D.; Muschiol, J.; Meyer, A.S. Synthesis of Human Milk Oligosaccharides: Protein Engineering Strategies for Improved Enzymatic Transglycosylation. Molecules 2019, 24, 2033. [Google Scholar] [CrossRef] [PubMed]
- Nidetzky, B.; Gutmann, A.; Zhong, C. Leloir Glycosyltransferases as Biocatalysts for Chemical Production. ACS Catalysis 2018, 8, 6283–6300. [Google Scholar] [CrossRef]
- Schmaltz, R.M.; Hanson, S.R.; Wong, C.-H. Enzymes in the Synthesis of Glycoconjugates. Chem. Rev. 2011, 111, 4259–4307. [Google Scholar] [CrossRef]
- Sauerzapfe, B.; Namdjou, D.J.; Schumacher, T.; Linden, N.; Křenek, K.; Křen, V.; Elling, L. Characterization of recombinant fusion constructs of human β1,4-galactosyltransferase 1 and the lipase pre-propeptide from Staphylococcus hyicus. J. Mol. Catal. B Enzym. 2008, 50, 128–140. [Google Scholar] [CrossRef]
- Sauerzapfe, B.; Křenek, K.; Schmiedel, J.; Wakarchuk, W.W.; Pelantová, H.; Křen, V.; Elling, L. Chemo-enzymatic synthesis of poly-N-acetyllactosamine (poly-LacNAc) structures and their characterization for CGL2-galectin-mediated binding of ECM glycoproteins to biomaterial surfaces. Glycoconjugate J. 2009, 26, 141–159. [Google Scholar] [CrossRef]
- Logan, S.M.; Altman, E.; Mykytczuk, O.; Brisson, J.-R.; Chandan, V.; Michael, F.S.; Masson, A.; Leclerc, S.; Hiratsuka, K.; Smirnova, N.; et al. Novel biosynthetic functions of lipopolysaccharide rfaJ homologs from Helicobacter pylori. Glycobiology 2005, 15, 721–733. [Google Scholar] [CrossRef]
- Rech, C.; Rosencrantz, R.R.; Křenek, K.; Pelantová, H.; Bojarová, P.; Römer, C.E.; Hanisch, F.-G.; Křen, V.; Elling, L. Combinatorial One-Pot Synthesis of Poly-N-acetyllactosamine Oligosaccharides with Leloir-Glycosyltransferases. Adv. Synth. Catal. 2011, 353, 2492–2500. [Google Scholar] [CrossRef]
- Laaf, D.; Bojarová, P.; Pelantová, H.; Křen, V.; Elling, L. Tailored Multivalent Neo-Glycoproteins: Synthesis, Evaluation, and Application of a Library of Galectin-3-Binding Glycan Ligands. Bioconjugate Chem. 2017, 28, 2832–2840. [Google Scholar] [CrossRef]
- Fischöder, T.; Laaf, D.; Dey, C.; Elling, L. Enzymatic Synthesis of N-Acetyllactosamine (LacNAc) Type 1 Oligomers and Characterization as Multivalent Galectin Ligands. Molecules 2017, 22, 1320. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-w.; Xia, C.; Li, L.; Guan, W.-y.; Pettit, N.; Zhang, H.-c.; Chen, M.; Wang, P.G. Characterization and synthetic application of a novel β1,3-galactosyltransferase from Escherichia coli O55:H7. Bioorg. Med. Chem. 2009, 17, 4910–4915. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.P.; Maxwell, E.S.; Burton, R.M. Enzymatic Syntheses of C14-Labeled Uridine Diphosphoglucose, Galactose 1-Phosphate, and Uridine Diphosphogalactose1. J. Am. Chem. Soc. 1959, 81, 6514–6517. [Google Scholar] [CrossRef]
- Virgilio, S.D.; Glushka, J.; Moremen, K.; Pierce, M. Enzymatic synthesis of natural and 13C enriched linear poly-N-acetyllactosamines as ligands for galectin-1. Glycobiology 1999, 9, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.F.; Thellend, A.; Piffeteau, A.; Vidal-Cros, A. Chemoenzymatic synthesis of stable isotope labeled UDP-N-[2H]-acetyl-glucosamine and [2H]-acetyl-chitooligosaccharides. Glycoconjugate J. 2006, 23, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Wahl, C.; Hirtz, D.; Elling, L. Multiplexed Capillary Electrophoresis as Analytical Tool for Fast Optimization of Multi-Enzyme Cascade Reactions—Synthesis of Nucleotide Sugars. Biotechnol. J. 2016, 11, 1298–1308. [Google Scholar] [CrossRef] [PubMed]
- Wahl, C.; Spiertz, M.; Elling, L. Characterization of a new UDP-sugar pyrophosphorylase from Hordeum vulgare (barley). J. Biotechnol. 2017, 258, 51–55. [Google Scholar] [CrossRef]
- Fischöder, T.; Wahl, C.; Zerhusen, C.; Elling, L. Repetitive Batch Mode Facilitates Enzymatic Synthesis of the Nucleotide Sugars UDP-Gal, UDP-GlcNAc, and UDP-GalNAc on a Multi-Gram Scale. Biotechnol. J. 2019, 14, 1800386. [Google Scholar] [CrossRef]
- Zhang, X.; Han, X.; Xia, K.; Xu, Y.; Yang, Y.; Oshima, K.; Haeger, S.M.; Perez, M.J.; McMurtry, S.A.; Hippensteel, J.A.; et al. Circulating heparin oligosaccharides rapidly target the hippocampus in sepsis, potentially impacting cognitive functions. Proc. Natl. Acad. Sci. USA 2019, 116, 9208–9213. [Google Scholar] [CrossRef]
- Abeln, M.; Borst, K.M.; Cajic, S.; Thiesler, H.; Kats, E.; Albers, I.; Kuhn, M.; Kaever, V.; Rapp, E.; Münster-Kühnel, A.; et al. Sialylation Is Dispensable for Early Murine Embryonic Development in Vitro. ChemBioChem 2017, 18, 1305–1316. [Google Scholar] [CrossRef]
- Hennig, R.; Cajic, S.; Borowiak, M.; Hoffmann, M.; Kottler, R.; Reichl, U.; Rapp, E. Towards personalized diagnostics via longitudinal study of the human plasma N-glycome. Biochim. Biophys. Acta 2016, 1860, 1728–1738. [Google Scholar] [CrossRef] [PubMed]
- Hennig, R.; Rapp, E.; Kottler, R.; Cajic, S.; Borowiak, M.; Reichl, U. N-Glycosylation Fingerprinting of Viral Glycoproteins by xCGE-LIF. Methods Mol. Biol. 2015, 1331, 123–143. [Google Scholar] [CrossRef] [PubMed]
- Konze, S.A.; Cajic, S.; Oberbeck, A.; Hennig, R.; Pich, A.; Rapp, E.; Buettner, F.F.R. Quantitative Assessment of Sialo-Glycoproteins and N-Glycans during Cardiomyogenic Differentiation of Human Induced Pluripotent Stem Cells. ChemBioChem 2017, 18, 1317–1331. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, J.; Rapp, E.; Reichl, U. N-glycan analysis by CGE-LIF: Profiling influenza A virus hemagglutinin N-glycosylation during vaccine production. Electrophoresis 2008, 29, 4203–4214. [Google Scholar] [CrossRef] [PubMed]
- Thiesler, C.T.; Cajic, S.; Hoffmann, D.; Thiel, C.; van Diepen, L.; Hennig, R.; Sgodda, M.; Weibetamann, R.; Reichl, U.; Steinemann, D.; et al. Glycomic Characterization of Induced Pluripotent Stem Cells Derived from a Patient Suffering from Phosphomannomutase 2 Congenital Disorder of Glycosylation (PMM2-CDG). Mol. Cell. Proteom. 2016, 15, 1435–1452. [Google Scholar] [CrossRef] [PubMed]
- Mittermayr, S.; Bones, J.; Guttman, A. Unraveling the Glyco-Puzzle: Glycan Structure Identification by Capillary Electrophoresis. Anal. Chem. 2013, 85, 4228–4238. [Google Scholar] [CrossRef]
- Ceroni, A.; Maass, K.; Geyer, H.; Geyer, R.; Dell, A.; Haslam, S.M. GlycoWorkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans. J. Proteome Res. 2008, 7, 1650–1659. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Marth, J.D.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. Symbol nomenclature for glycan representation. Proteomics 2009, 9, 5398–5399. [Google Scholar] [CrossRef]
- Li, Y.; Xue, M.; Sheng, X.; Yu, H.; Zeng, J.; Thon, V.; Chen, Y.; Muthana, M.M.; Wang, P.G.; Chen, X. Donor substrate promiscuity of bacterial beta1-3-N-acetylglucosaminyltransferases and acceptor substrate flexibility of beta1-4-galactosyltransferases. Bioorg. Med. Chem. 2016, 24, 1696–1705. [Google Scholar] [CrossRef]
- Ujita, M.; Misra, A.K.; McAuliffe, J.; Hindsgaul, O.; Fukuda, M. Poly-N-acetyllactosamine Extension inN-Glycans and Core 2- and Core 4-branchedO-Glycans Is Differentially Controlled by i-Extension Enzyme and Different Members of the β1,4-Galactosyltransferase Gene Family. J. Biol. Chem. 2000, 275, 15868–15875. [Google Scholar] [CrossRef]
- Bode, L.; Kuhn, L.; Kim, H.Y.; Hsiao, L.; Nissan, C.; Sinkala, M.; Kankasa, C.; Mwiya, M.; Thea, D.M.; Aldrovandi, G.M. Human milk oligosaccharide concentration and risk of postnatal transmission of HIV through breastfeeding. Am. J. Clin. Nutr. 2012, 96, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Courtade, L.; Han, S.; Lee, S.; Mian, F.M.; Buck, R.; Forsythe, P. Attenuation of food allergy symptoms following treatment with human milk oligosaccharides in a mouse model. Allergy 2015, 70, 1091–1102. [Google Scholar] [CrossRef] [PubMed]
- Jantscher-Krenn, E.; Zherebtsov, M.; Nissan, C.; Goth, K.; Guner, Y.S.; Naidu, N.; Choudhury, B.; Grishin, A.V.; Ford, H.R.; Bode, L. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut 2012, 61, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Morrow, A.L.; Ruiz-Palacios, G.M.; Altaye, M.; Jiang, X.; Guerrero, M.L.; Meinzen-Derr, J.K.; Farkas, T.; Chaturvedi, P.; Pickering, L.K.; Newburg, D.S. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J. Pediatr. 2004, 145, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, N.; Odenwald, H.; Kukkonen, A.K.; Kuitunen, M.; Savilahti, E.; Kunz, C. FUT2-dependent breast milk oligosaccharides and allergy at 2 and 5 years of age in infants with high hereditary allergy risk. Eur. J. Nutr. 2017, 56, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Asakuma, S.; Urashima, T.; Akahori, M.; Obayashi, H.; Nakamura, T.; Kimura, K.; Watanabe, Y.; Arai, I.; Sanai, Y. Variation of major neutral oligosaccharides levels in human colostrum. Eur. J. Clin. Nutr. 2008, 62, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Austin, S.; Cuany, D.; Michaud, J.; Diehl, B.; Casado, B. Determination of 2′-Fucosyllactose and Lacto-N-neotetraose in Infant Formula. Molecules 2018, 23, 2650. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Warren, C.D.; Ruiz-Palacios, G.M.; Pickering, L.K.; Newburg, D.S. Milk oligosaccharide profiles by reversed-phase HPLC of their perbenzoylated derivatives. Anal. Biochem. 1997, 251, 89–97. [Google Scholar] [CrossRef]
- Gabrielli, O.; Zampini, L.; Galeazzi, T.; Padella, L.; Santoro, L.; Peila, C.; Giuliani, F.; Bertino, E.; Fabris, C.; Coppa, G.V. Preterm milk oligosaccharides during the first month of lactation. Pediatrics 2011, 128, e1520–e1531. [Google Scholar] [CrossRef]
- Kunz, C.; Rudloff, S.; Hintelmann, A.; Pohlentz, G.; Egge, H. High-pH anion-exchange chromatography with pulsed amperometric detection and molar response factors of human milk oligosaccharides. J. Chromatogr. B Biomed. Sci. Appl. 1996, 685, 211–221. [Google Scholar] [CrossRef]
- McGuire, M.K.; Meehan, C.L.; McGuire, M.A.; Williams, J.E.; Foster, J.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; Mbugua, S.; Moore, S.E.; et al. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am. J. Clin. Nutr. 2017, 105, 1086–1100. [Google Scholar] [CrossRef] [PubMed]
- Nijman, R.M.; Liu, Y.; Bunyatratchata, A.; Smilowitz, J.T.; Stahl, B.; Barile, D. Characterization and Quantification of Oligosaccharides in Human Milk and Infant Formula. J. Agric. Food Chem. 2018, 66, 6851–6859. [Google Scholar] [CrossRef] [PubMed]
- Nissan, C.; Naidu, N.; Choudhury, B.; Bode, L. A new HPLC-based method to profile and quantify Human Milk Oligosaccharides from as little as 1 uL milk. FASEB J. 2010, 24, 556–620. [Google Scholar] [CrossRef]
- Sprenger, N.; Lee, L.Y.; De Castro, C.A.; Steenhout, P.; Thakkar, S.K. Longitudinal change of selected human milk oligosaccharides and association to infants’ growth, an observatory, single center, longitudinal cohort study. PLoS ONE 2017, 12, e0171814. [Google Scholar] [CrossRef] [PubMed]
- Thurl, S.; Munzert, M.; Henker, J.; Boehm, G.; Müller-Werner, B.; Jelinek, J.; Stahl, B. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br. J. Nutr. 2010, 104, 1261–1271. [Google Scholar] [CrossRef]
- Bao, Y.; Zhu, L.; Newburg, D.S. Simultaneous quantification of sialyloligosaccharides from human milk by capillary electrophoresis. Anal. Biochem. 2007, 370, 206–214. [Google Scholar] [CrossRef]
- Newburg, D.S.; Shen, Z.; Warren, C.D. Quantitative Analysis of Human Milk Oligosaccharides by Capillary Electrophoresis. In Short and Long Term Effects of Breast Feeding on Child Health; Koletzko, B., Michaelsen, K.F., Hernell, O., Eds.; Springer US: Boston, MA, USA, 2002; pp. 381–382. [Google Scholar] [CrossRef]
- Shen, Z.; Warren, C.D.; Newburg, D.S. High-performance capillary electrophoresis of sialylated oligosaccharides of human milk. Anal. Biochem. 2000, 279, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Song, J.F.; Weng, M.Q.; Wu, S.M.; Xia, Q.C. Analysis of neutral saccharides in human milk derivatized with 2-aminoacridone by capillary electrophoresis with laser-induced fluorescence detection. Anal. Biochem. 2002, 304, 126–129. [Google Scholar] [CrossRef]
- Niñonuevo, M.R.; Perkins, P.D.; Francis, J.; Lamotte, L.M.; LoCascio, R.G.; Freeman, S.L.; Mills, D.A.; German, J.B.; Grimm, R.; Lebrilla, C.B. Daily Variations in Oligosaccharides of Human Milk Determined by Microfluidic Chips and Mass Spectrometry. J. Agric. Food Chem. 2008, 56, 618–626. [Google Scholar] [CrossRef]
- Nwosu, C.C.; Aldredge, D.L.; Lee, H.; Lerno, L.A.; Zivkovic, A.M.; German, J.B.; Lebrilla, C.B. Comparison of the Human and Bovine Milk N-Glycome via High-Performance Microfluidic Chip Liquid Chromatography and Tandem Mass Spectrometry. J. Proteome Res. 2012, 11, 2912–2924. [Google Scholar] [CrossRef]
- Pfenninger, A.; Karas, M.; Finke, B.; Stahl, B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MS(n) (part 2: Application to isomeric mixtures). J. Am. Soc. Mass Spectrom. 2002, 13, 1341–1348. [Google Scholar] [CrossRef]
- Pfenninger, A.; Karas, M.; Finke, B.; Stahl, B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MSn (Part 1: Methodology). J. Am. Soc. Mass Spectrom. 2002, 13, 1331–1340. [Google Scholar] [CrossRef]
- Blank, D.; Gebhardt, S.; Maass, K.; Lochnit, G.; Dotz, V.; Blank, J.; Geyer, R.; Kunz, C. High-throughput mass finger printing and Lewis blood group assignment of human milk oligosaccharides. Anal. Bioanal. Chem. 2011, 401, 2495–2510. [Google Scholar] [CrossRef] [PubMed]
- Stahl, B.; Steup, M.; Karas, M.; Hillenkamp, F. Analysis of neutral oligosaccharides by matrix-assisted laser desorption ionization mass spectrometry. Anal. Chem. 1991, 63, 1463–1466. [Google Scholar] [CrossRef]
- Stahl, B.; Thurl, S.; Zeng, J.; Karas, M.; Hillenkamp, F.; Steup, M.; Sawatzki, G. Oligosaccharides from human milk as revealed by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Biochem. 1994, 223, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Addona, T.A.; Abbatiello, S.E.; Schilling, B.; Skates, S.J.; Mani, D.R.; Bunk, D.M.; Spiegelman, C.H.; Zimmerman, L.J.; Ham, A.-J.L.; Keshishian, H.; et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring–based measurements of proteins in plasma. Nat. Biotechnol. 2009, 27, 633. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, E.; Wu, J.; Karl, J.; Liao, H.; Zolg, W.; Guild, B. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics 2004, 4, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Mirgorodskaya, O.A.; Kozmin, Y.P.; Titov, M.I.; Körner, R.; Sönksen, C.P.; Roepstorff, P. Quantitation of peptides and proteins by matrix-assisted laser desorption/ionization mass spectrometry using 18O-labeled internal standards. Rapid Commun. Mass Spectrom. 2000, 14, 1226–1232. [Google Scholar] [CrossRef]
- Van Erven, G.; de Visser, R.; Merkx, D.W.H.; Strolenberg, W.; de Gijsel, P.; Gruppen, H.; Kabel, M.A. Quantification of Lignin and Its Structural Features in Plant Biomass Using (13)C Lignin as Internal Standard for Pyrolysis-GC-SIM-MS. Anal. Chem. 2017, 89, 10907–10916. [Google Scholar] [CrossRef]
- Wilkinson, W.R.; Gusev, A.I.; Proctor, A.; Houalla, M.; Hercules, D.M. Selection of internal standards for quantitative analysis by matrix-assisted laser desorption-ionization (MALDI) time-of-flight mass spectrometry. Fresenius’ J. Anal. Chem. 1997, 357, 241–248. [Google Scholar] [CrossRef]
- Tao, N.; DePeters, E.J.; Freeman, S.; German, J.B.; Grimm, R.; Lebrilla, C.B. Bovine milk glycome. J. Dairy Sci. 2008, 91, 3768–3778. [Google Scholar] [CrossRef] [PubMed]
- Urashima, T.; Taufik, E.; Fukuda, K.; Asakuma, S. Recent advances in studies on milk oligosaccharides of cows and other domestic farm animals. Biosci. Biotechnol. Biochem. 2013, 77, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Nicolardi, S.; Palmblad, M.; Dalebout, H.; Bladergroen, M.; Tollenaar, R.A.E.M.; Deelder, A.M.; van der Burgt, Y.E.M. Quality control based on isotopic distributions for high-throughput MALDI-TOF and MALDI-FTICR serum peptide profiling. J. Am. Soc. Mass Spectrom. 2010, 21, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Leo, F.; Asakuma, S.; Fukuda, K.; Senda, A.; Urashima, T. Determination of sialyl and neutral oligosaccharide levels in transition and mature milks of Samoan women, using anthranilic derivatization followed by reverse phase high performance liquid chromatography. Biosci. Biotechnol. Biochem. 2010, 74, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Andersson, B.; Porras, O.; Hanson, L.A.; Lagergard, T.; Svanborg-Eden, C. Inhibition of attachment of Streptococcus pneumoniae and Haemophilus influenzae by human milk and receptor oligosaccharides. J. Infect. Dis. 1986, 153, 232–237. [Google Scholar] [CrossRef]
- Idanpaan-Heikkila, I.; Simon, P.M.; Zopf, D.; Vullo, T.; Cahill, P.; Sokol, K.; Tuomanen, E. Oligosaccharides interfere with the establishment and progression of experimental pneumococcal pneumonia. J. Infect. Dis. 1997, 176, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Puccio, G.; Alliet, P.; Cajozzo, C.; Janssens, E.; Corsello, G.; Sprenger, N.; Wernimont, S.; Egli, D.; Gosoniu, L.; Steenhout, P. Effects of Infant Formula With Human Milk Oligosaccharides on Growth and Morbidity: A Randomized Multicenter Trial. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Coppa, G.V.; Zampini, L.; Galeazzi, T.; Gabrielli, O. Prebiotics in human milk: A review. Dig. Liver Dis. 2006, 38 (Suppl. 2), S291–S294. [Google Scholar] [CrossRef]
- Gyorgy, P.; Norris, R.F.; Rose, C.S. Bifidus factor. I. A variant of Lactobacillus bifidus requiring a special growth factor. Arch. Biochem. Biophys. 1954, 48, 193–201. [Google Scholar] [CrossRef]
- Kitaoka, M.; Tian, J.; Nishimoto, M. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl. Environ. Microbiol. 2005, 71, 3158–3162. [Google Scholar] [CrossRef]
- Steenhout, P.; Sperisen, P.; Martin, F.-P.; Sprenger, N.; Wernimont, S.; Pecquet, S.; Berger, B. Term Infant Formula Supplemented with Human Milk Oligosaccharides (2′Fucosyllactose and Lacto-N-neotetraose) Shifts Stool Microbiota and Metabolic Signatures Closer to that of Breastfed Infants. FASEB J. 2016, 30, 275–277. [Google Scholar] [CrossRef]
- De Leoz, M.L.; Kalanetra, K.M.; Bokulich, N.A.; Strum, J.S.; Underwood, M.A.; German, J.B.; Mills, D.A.; Lebrilla, C.B. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: A proof-of-concept study. J. Proteome Res. 2015, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- De Leoz, M.L.; Wu, S.; Strum, J.S.; Ninonuevo, M.R.; Gaerlan, S.C.; Mirmiran, M.; German, J.B.; Mills, D.A.; Lebrilla, C.B.; Underwood, M.A. A quantitative and comprehensive method to analyze human milk oligosaccharide structures in the urine and feces of infants. Anal. Bioanal. Chem. 2013, 405, 4089–4105. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
| Compound | Amount [µmol] | Average Yield [%] | Calculated m/z [M − H]− | Observed m/z [M − H]− |
|---|---|---|---|---|
| 3 (UDP-[15N]GlcNAc) | 200 | 100% | 607.1 (a) | 607.2 |
| 6 (UDP-[13C6]Gal) | 400 | 100% | 571.3 (a) | 570.9 |
| UDP-GlcNAc (a) | 606.1 | 606.2 | ||
| UDP-Gal (a) | 565.3 (a) | 564.9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fischöder, T.; Cajic, S.; Grote, V.; Heinzler, R.; Reichl, U.; Franzreb, M.; Rapp, E.; Elling, L. Enzymatic Cascades for Tailored 13C6 and 15N Enriched Human Milk Oligosaccharides. Molecules 2019, 24, 3482. https://doi.org/10.3390/molecules24193482
Fischöder T, Cajic S, Grote V, Heinzler R, Reichl U, Franzreb M, Rapp E, Elling L. Enzymatic Cascades for Tailored 13C6 and 15N Enriched Human Milk Oligosaccharides. Molecules. 2019; 24(19):3482. https://doi.org/10.3390/molecules24193482
Chicago/Turabian StyleFischöder, Thomas, Samanta Cajic, Valerian Grote, Raphael Heinzler, Udo Reichl, Matthias Franzreb, Erdmann Rapp, and Lothar Elling. 2019. "Enzymatic Cascades for Tailored 13C6 and 15N Enriched Human Milk Oligosaccharides" Molecules 24, no. 19: 3482. https://doi.org/10.3390/molecules24193482
APA StyleFischöder, T., Cajic, S., Grote, V., Heinzler, R., Reichl, U., Franzreb, M., Rapp, E., & Elling, L. (2019). Enzymatic Cascades for Tailored 13C6 and 15N Enriched Human Milk Oligosaccharides. Molecules, 24(19), 3482. https://doi.org/10.3390/molecules24193482







