Multi-Armed 1,2,3-Selenadiazole and 1,2,3-Thiadiazole Benzene Derivatives as Novel Glyoxalase-I Inhibitors
Abstract
:1. Introduction
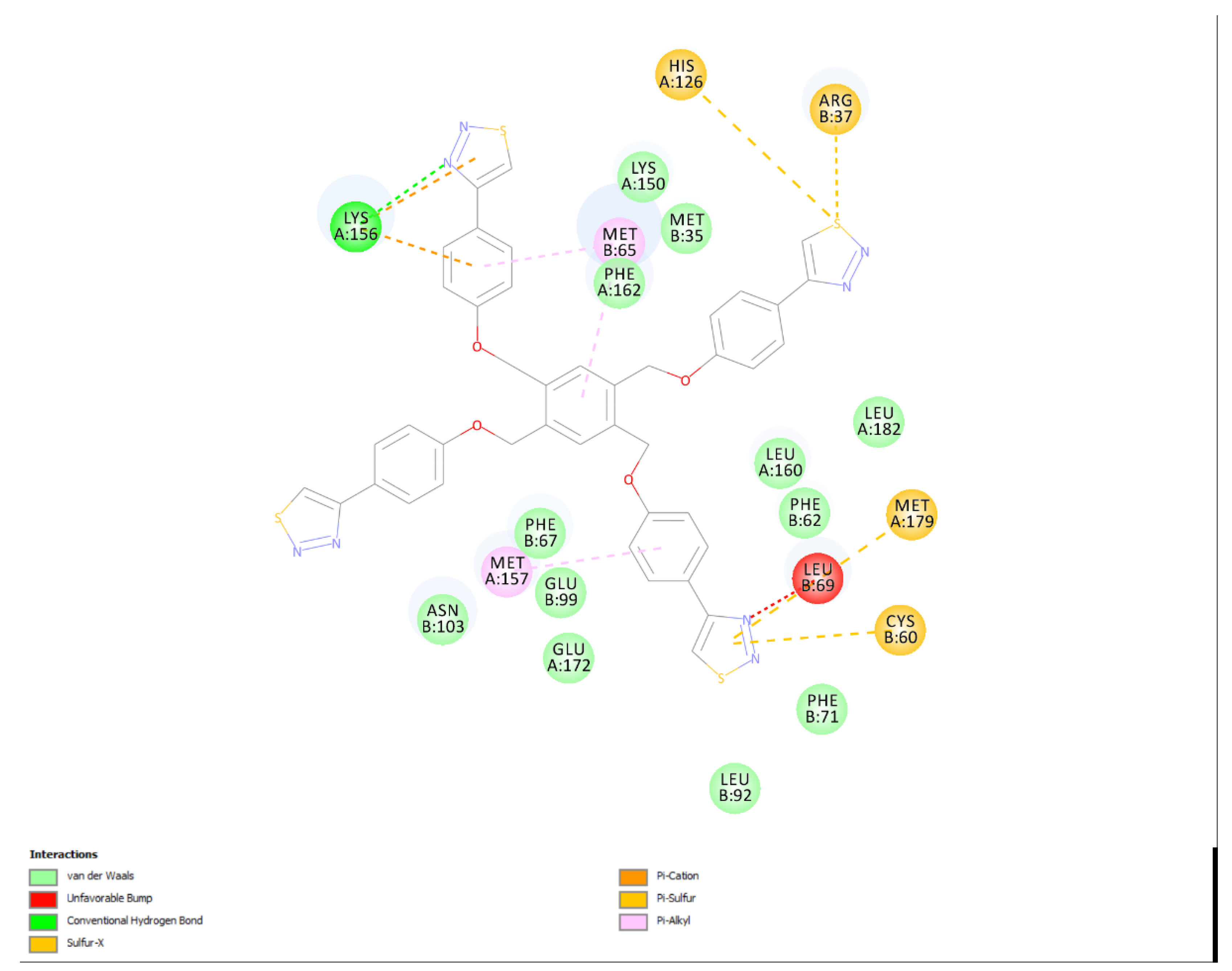
2. Results and Discussion
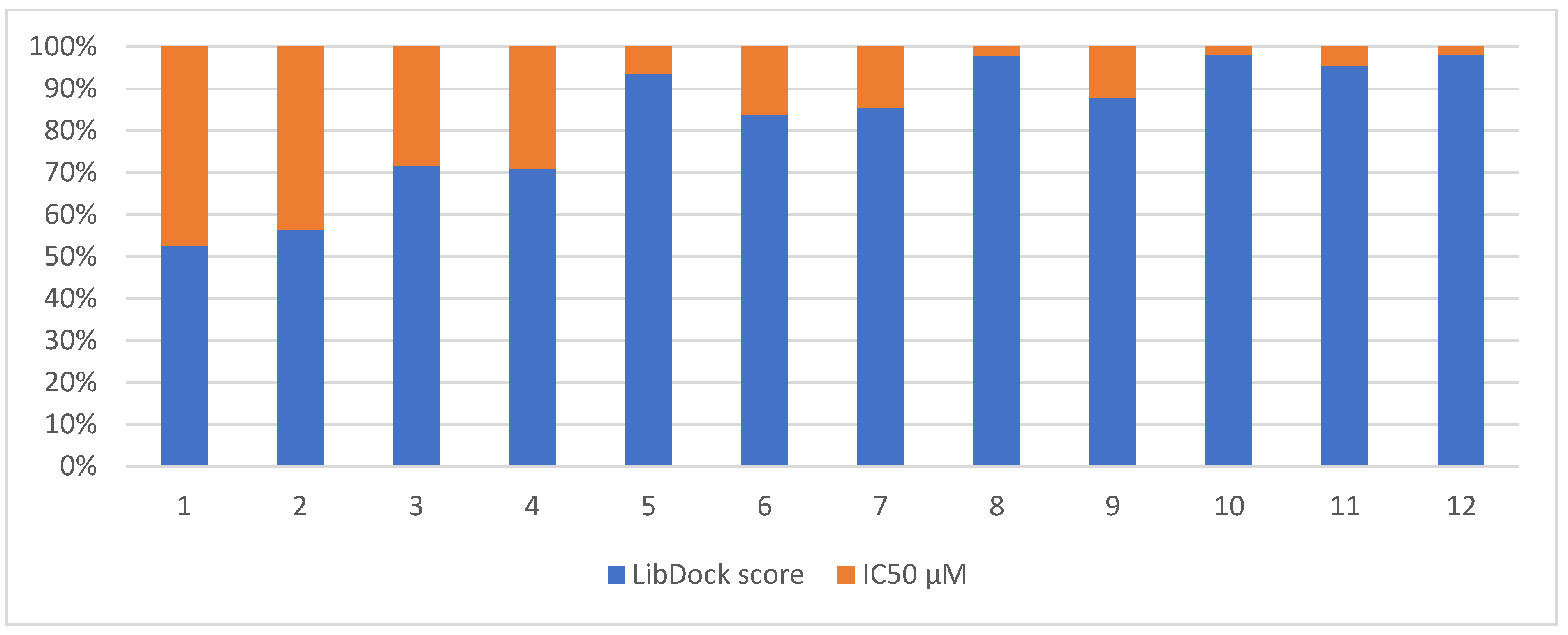
Correlation between the in silico and the in vitro results
3. Materials and Methods
3.1. Computational Materials
3.2. Experimental Materials
3.3. Computational Methods
3.3.1. Ligand preparation
3.3.2. Preparation of Glo-I Enzyme
3.3.3. Molecular Docking
3.3.4. In vitro Enzyme Assay
3.3.5. Correlation between the Docking Scores and in vitro IC50 Values
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mhaidat, N.M.; Al-Smadi, M.; Al-Momani, F.; Alzoubi, K.H.; Mansi, I.; Al-Balas, Q. Synthesis, antimicrobial and in vitro antitumor activities of a series of 1,2,3-thiadiazole and 1,2,3-selenadiazole derivatives. Drug Des. Devel. Ther. 2015, 9, 3645–3652. [Google Scholar] [CrossRef] [PubMed]
- Al-Smadi, M.; Ratrout, S. New 1,2,3-Selenadiazole and 1,2,3-Thiadiazole Derivatives. Molecules 2004, 9, 957. [Google Scholar] [CrossRef] [PubMed]
- Aziz, N.M.; Rowland, J.H. Trends and advances in cancer survivorship research: challenge and opportunity. Semin. Radiat. Oncol. 2003, 13, 248–266. [Google Scholar] [CrossRef]
- Xue, M.; Rabbani, N.; Thornalley, P.J. Glyoxalase in ageing. Semin. Cell. Dev. Biol. 2011, 22, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 1990, 269, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.D.; Ridderström, M.; Olin, B.; Kavarana, M.J.; Creighton, D.J.; Mannervik, B. Reaction Mechanism of Glyoxalase I Explored by an X-ray Crystallographic Analysis of the Human Enzyme in Complex with a Transition State Analogue. Biochemistry 1999, 38, 13480–13490. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.A.; Thornalley, P.J. The formation of methylglyoxal from triose phosphates. Investigation using a specific assay for methylglyoxal. Eur. J. Biochem. 1993, 212, 101–105. [Google Scholar] [CrossRef]
- Rulli, A.; Carli, L.; Romani, R.; Baroni, T.; Giovannini, E.; Rosi, G.; Talesa, V. Expression of glyoxalase I and II in normal and breast cancer tissues. Breast Cancer Res. Treat 2001, 66, 67–72. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Tennant, D.A.; Duran, R.V.; Gottlieb, E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 2010, 10, 267–277. [Google Scholar] [CrossRef]
- Geng, X.; Ma, J.; Zhang, F.; Xu, C. Glyoxalase I in tumor cell proliferation and survival and as a potential target for anticancer therapy. Oncol. Res. Treat 2014, 37, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J.; Rabbani, N. Glyoxalase in tumourigenesis and multidrug resistance. Semin. Cell Dev. Biol. 2011, 22, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Mearini, E.; Romani, R.; Mearini, L.; Antognelli, C.; Zucchi, A.; Baroni, T.; Porena, M.; Talesa, V.N. Differing expression of enzymes of the glyoxalase system in superficial and invasive bladder carcinomas. Eur. J. Cancer 2002, 38, 1946–1950. [Google Scholar] [CrossRef]
- Hosoda, F.; Arai, Y.; Okada, N.; Shimizu, H.; Miyamoto, M.; Kitagawa, N.; Katai, H.; Taniguchi, H.; Yanagihara, K.; Imoto, I. Integrated genomic and functional analyses reveal glyoxalase I as a novel metabolic oncogene in human gastric cancer. Oncogene 2015, 34, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-L.; Tsai, M.-M.; Tsai, C.-Y.; Huang, Y.-H.; Chen, C.-Y.; Chi, H.-C.; Tseng, Y.-H.; Chao, I.-W.; Lin, W.-C.; Wu, S.-M. Glyoxalase-I is a novel prognosis factor associated with gastric cancer progression. PLoS ONE 2012, 7, e34352. [Google Scholar] [CrossRef]
- Wang, Y.; Kuramitsu, Y.; Tokuda, K.; Okada, F.; Baron, B.; Akada, J.; Kitagawa, T.; Nakamura, K. Proteomic analysis indicates that overexpression and nuclear translocation of lactoylglutathionelyase (GLO1) is associated with tumor progression in murine fibrosarcoma. Electrophoresis 2014, 35, 2195–2202. [Google Scholar] [PubMed]
- Wang, Y.; Kuramitsu, Y.; Ueno, T.; Suzuki, N.; Yoshino, S.; Iizuka, N.; Akada, J.; Kitagawa, T.; Oka, M.; Nakamura, K. Glyoxalase I (GLO1) is up-regulated in pancreatic cancerous tissues compared with related non-cancerous tissues. Anticancer Res. 2012, 32, 3219–3222. [Google Scholar] [PubMed]
- Chen, Y.; Fang, L.; Li, G.; Zhang, J.; Li, C.; Ma, M.; Guan, C.; Bai, F.; Lyu, J.; Meng, Q.H. Synergistic inhibition of colon cancer growth by the combination of methylglyoxal and silencing of glyoxalase I mediated by the STAT1 pathway. Oncotarget 2017, 8, 54838–54857. [Google Scholar] [CrossRef] [PubMed]
- Burdelski, C.; Shihada, R.; Hinsch, A.; Angerer, A.; Gobel, C.; Friedrich, E.; Hube-Magg, C.; Burdak-Rothkamm, S.; Kluth, M.; Simon, R.; et al. High-Level Glyoxalase 1 (GLO1) expression is linked to poor prognosis in prostate cancer. Prostate 2017, 77, 1528–1538. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Zhou, Q.; Zhang, Z.; Zhu, Y.; Duan, T.; Feng, Y. Metformin sensitizes endometrial cancer cells to chemotherapy by repressing glyoxalase I expression. J. Obstet. Gynaecol. Res. 2012, 38, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Al-Shar’i, N.; Hassan, M.; Al-Balas, Q.; Almaaytah, A. Identification of Possible Glyoxalase II Inhibitors as Anticancer Agents by a Customized 3D Structure-Based Pharmacophore Model. Jordan J. Pharm. Sci. 2015, 108, 1–21. [Google Scholar]
- Belanger, M.; Yang, J.; Petit, J.M.; Laroche, T.; Magistretti, P.J.; Allaman, I. Role of the glyoxalase system in astrocyte-mediated neuroprotection. J. Neurosci. 2011, 31, 18338–18352. [Google Scholar] [CrossRef] [PubMed]
- Davidson, S.D.; Milanesa, D.M.; Mallouh, C.; Choudhury, M.S.; Tazaki, H.; Konno, S. A possible regulatory role of glyoxalase I in cell viability of human prostate cancer. Urol. Res. 2002, 30, 116–121. [Google Scholar] [CrossRef] [PubMed]
- More, S.S.; Vince, R. Inhibition of glyoxalase I: the first low-nanomolar tight-binding inhibitors. J. Med. Chem. 2009, 52, 4650–4656. [Google Scholar] [CrossRef] [PubMed]
- More, S.S.; Vince, R. Design, synthesis, and binding studies of bidentate Zn-chelating peptidic inhibitors of glyoxalase-I. Bioorg. Med. Chem. Lett. 2007, 17, 3793–3797. [Google Scholar] [CrossRef]
- More, S.S.; Vince, R. A metabolically stable tight-binding transition-state inhibitor of glyoxalase-I. Bioorg. Med. Chem. Lett. 2006, 16, 6039–6042. [Google Scholar] [CrossRef]
- Santel, T.; Pflug, G.; Hemdan, N.Y.; Schafer, A.; Hollenbach, M.; Buchold, M.; Hintersdorf, A.; Lindner, I.; Otto, A.; Bigl, M.; et al. Curcumin inhibits glyoxalase 1: A possible link to its anti-inflammatory and anti-tumor activity. PLoS ONE 2008, 3, e3508. [Google Scholar] [CrossRef]
- Takasawa, R.; Takahashi, S.; Saeki, K.; Sunaga, S.; Yoshimori, A.; Tanuma, S. Structure–activity relationship of human GLO I inhibitory natural flavonoids and their growth inhibitory effects. Bioorg. Med. Chem. 2008, 16, 3969–3975. [Google Scholar] [CrossRef]
- Takasawa, R.; Saeki, K.; Tao, A.; Yoshimori, A.; Uchiro, H.; Fujiwara, M.; Tanuma, S. Delphinidin, a dietary anthocyanidin in berry fruits, inhibits human glyoxalase I. Bioorg. Med. Chem. 2010, 18, 7029–7033. [Google Scholar] [CrossRef]
- Al-Balas, Q.; Hassan, M.; Al-Oudat, B.; Alzoubi, H.; Mhaidat, N.; Almaaytah, A. Generation of the First Structure-Based Pharmacophore Model Containing a Selective “Zinc Binding Group” Feature to Identify Potential Glyoxalase-1 Inhibitors. Molecules 2012, 17, 13740. [Google Scholar] [CrossRef]
- Al-Balas, Q.A.; Hassan, M.A.; Al-Shar’i, N.A.; El-Elimat, T.; Almaaytah, A.M. Computational and experimental exploration of the structure–activity relationships of flavonoids as potent glyoxalase-I inhibitors. Drug Dev. Res. 2017, 79, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Qosay, A.A.-B.; Mohammad, A.H.; Ghazi, A.A.J.; Nizar, A.A.-S.; Ammar, M.A.; Tamam, E.-E. Novel Thiazole Carboxylic Acid Derivatives Possessing a “Zinc Binding Feature” as Potential Human Glyoxalase-I Inhibitors. Lett. Drug Des. Dis. 2017, 14, 1324–1334. [Google Scholar] [CrossRef]
- Al-Smadi, M.; Hanold, N.; Meier, H. Multiple 1,2,3-Thiadiazoles. J. Heterocycl. Chem. A 1997, 34, 605–611. [Google Scholar] [CrossRef]
- Al-Smadi, M.; Meier, H. Multi-Arm 1,2,3-Thiadiazole Systems. Liebigs Ann. Chem. 1997, 11, 2357–2361. [Google Scholar] [CrossRef]
- Al-Smadi, M.; Ratrout, S. New Multi-1,2,3-Selenadiazole Aromatic Derivatives. Molecules 2005, 10, 1126–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Not available. |




| Index | Chemical Structure | LibDock Score | IC50 µM |
|---|---|---|---|
| 1 | 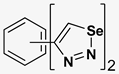 | 71.218 | 64.00 |
| 2 | 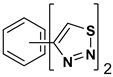 | 76.639 | 61.10 ± 9.1 |
| 3 |  | 77.4038 | 32.10 ± 5.5 |
| 4 |  | 83.3944 | 34.01 ± 1.9 |
| 5 | 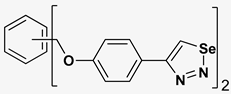 | 109.94 | 7.90 ± 1.8 |
| 6 | 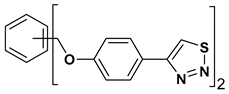 | 117.882 | 23.60 ± 1.7 |
| 7 | 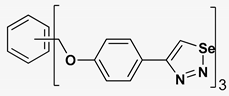 | 135.237 | 23.06 ± 1.6 |
| 8 | 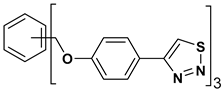 | 143.245 | 3.00 ± 0.15 |
| 9 | 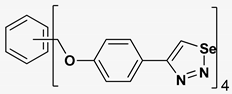 | 139.985 | 19.53 ± 2.0 |
| 10 | 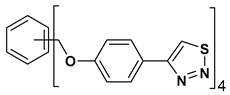 | 119.431 | 2.40 ± 0.13 |
| 11 | 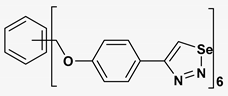 | 173.358 | 8.30 ± 0.56 |
| 12 | 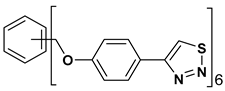 | 133.129 | 2.67 ± 0.31 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Balas, Q.A.; Al-Smadi, M.L.; Hassan, M.A.; Al Jabal, G.A.; Almaaytah, A.M.; Alzoubi, K.H. Multi-Armed 1,2,3-Selenadiazole and 1,2,3-Thiadiazole Benzene Derivatives as Novel Glyoxalase-I Inhibitors. Molecules 2019, 24, 3210. https://doi.org/10.3390/molecules24183210
Al-Balas QA, Al-Smadi ML, Hassan MA, Al Jabal GA, Almaaytah AM, Alzoubi KH. Multi-Armed 1,2,3-Selenadiazole and 1,2,3-Thiadiazole Benzene Derivatives as Novel Glyoxalase-I Inhibitors. Molecules. 2019; 24(18):3210. https://doi.org/10.3390/molecules24183210
Chicago/Turabian StyleAl-Balas, Qosay A., Mousa L. Al-Smadi, Mohammad A. Hassan, Ghazi A. Al Jabal, Ammar M. Almaaytah, and Karem H. Alzoubi. 2019. "Multi-Armed 1,2,3-Selenadiazole and 1,2,3-Thiadiazole Benzene Derivatives as Novel Glyoxalase-I Inhibitors" Molecules 24, no. 18: 3210. https://doi.org/10.3390/molecules24183210
APA StyleAl-Balas, Q. A., Al-Smadi, M. L., Hassan, M. A., Al Jabal, G. A., Almaaytah, A. M., & Alzoubi, K. H. (2019). Multi-Armed 1,2,3-Selenadiazole and 1,2,3-Thiadiazole Benzene Derivatives as Novel Glyoxalase-I Inhibitors. Molecules, 24(18), 3210. https://doi.org/10.3390/molecules24183210






