Effective Hydrogenation of 3-(2”-furyl)- and 3-(2”-thienyl)-1-(2’-hydroxyphenyl)-prop-2-en-1-one in Selected Yeast Cultures
Abstract
1. Introduction
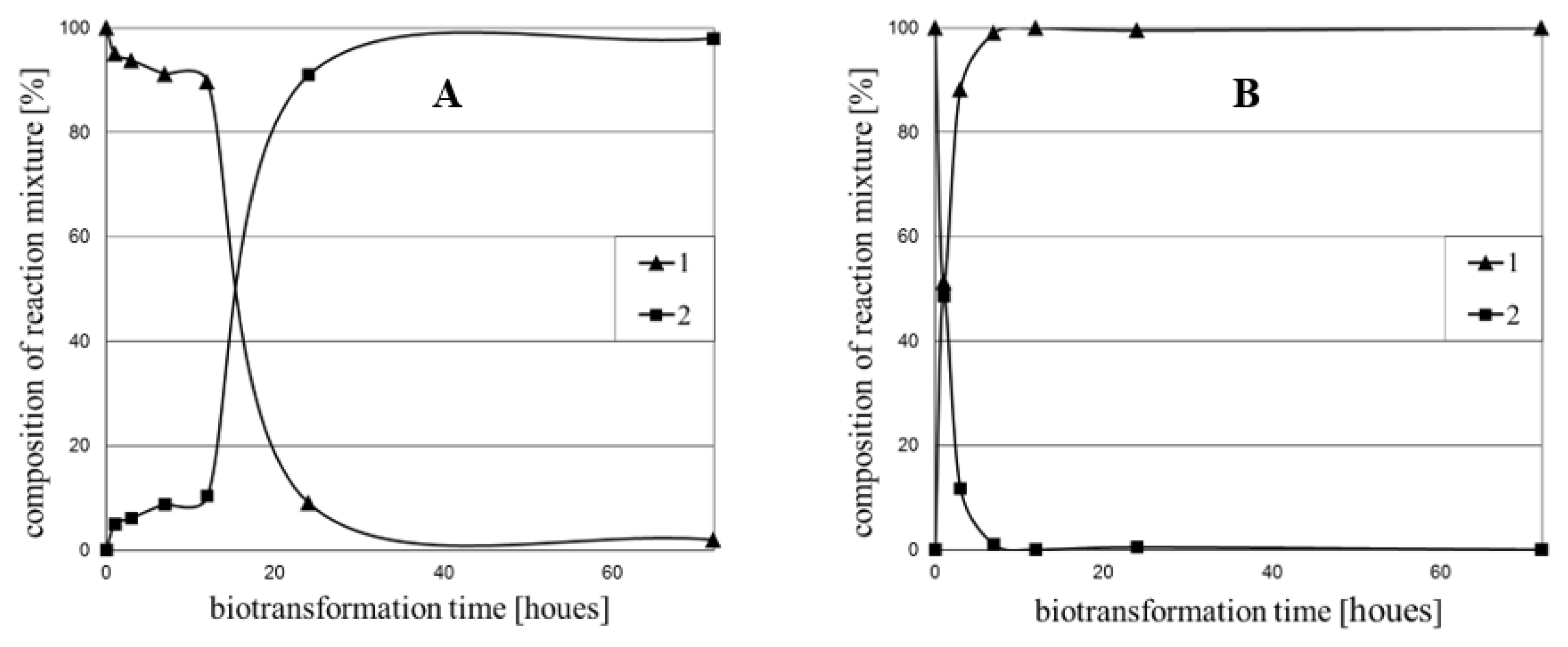
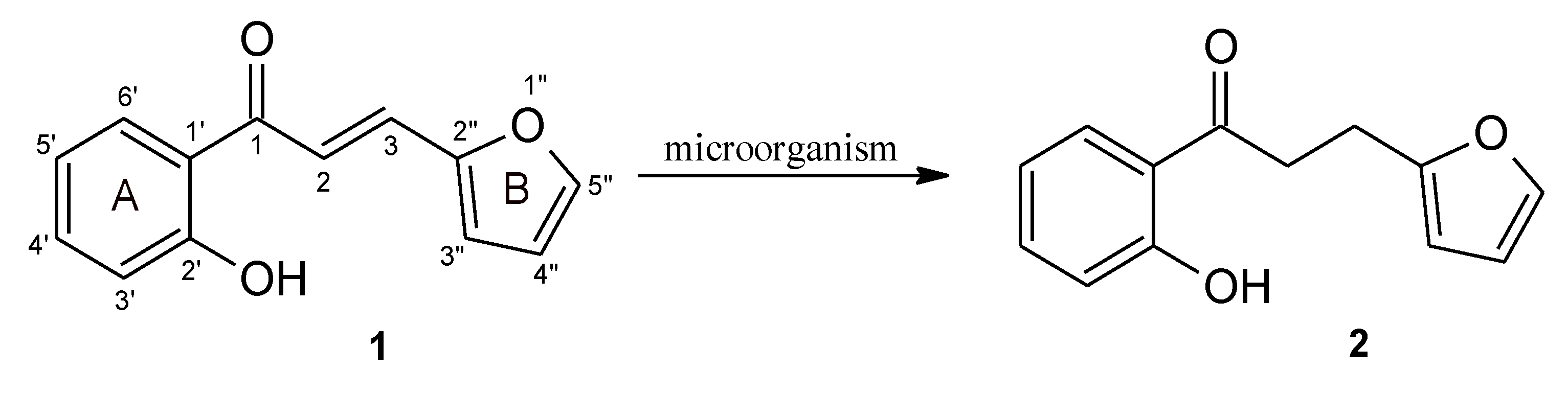
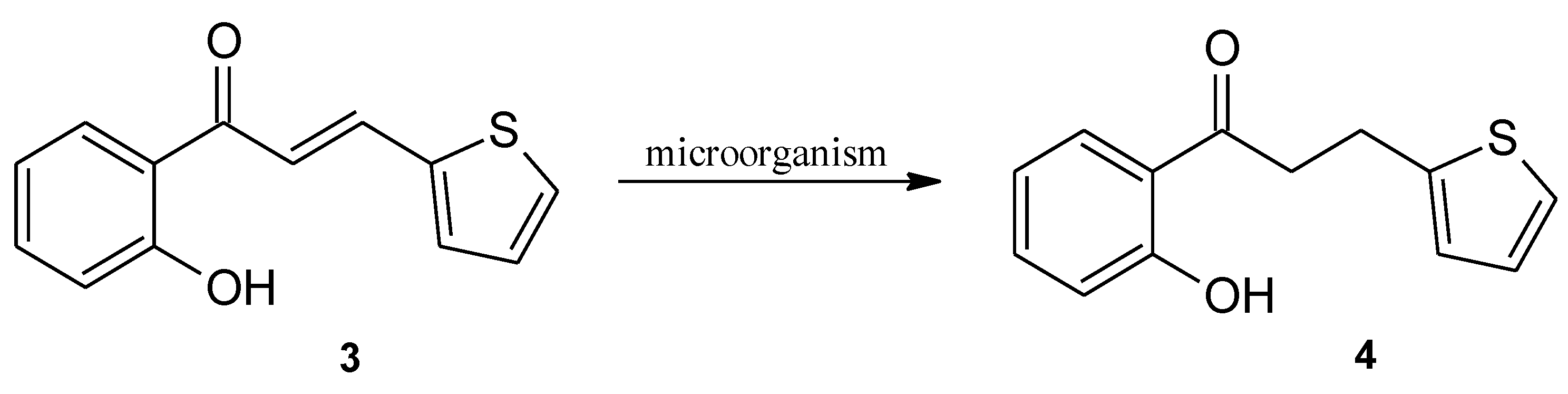
2. Results and Discussion
Biotransformation
3. Materials and Methods
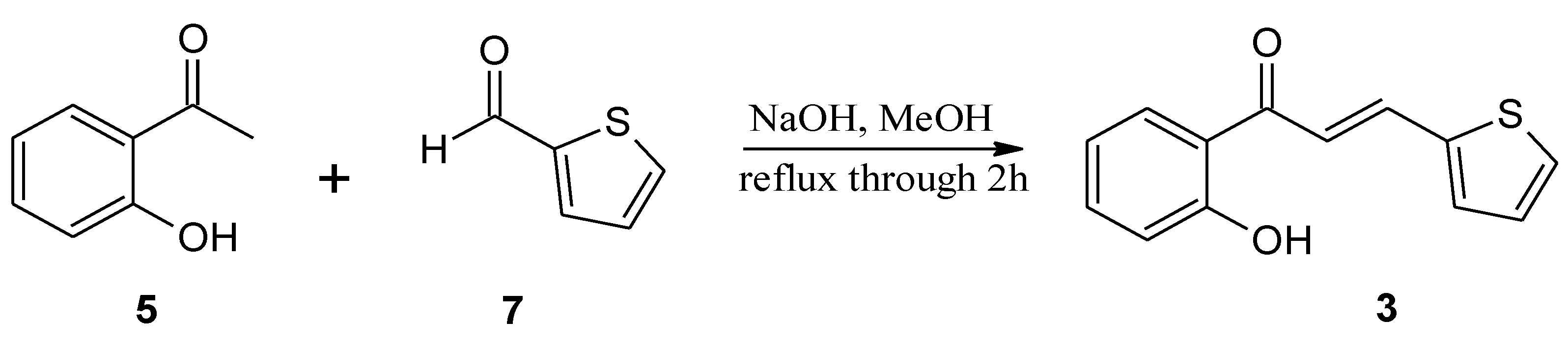
3.1. Substrate
3.2. Microorganisms
3.3. Analysis
3.4. Gas Chromatography (GC)
3.5. NMR Analysis
3.6. Screening
3.7. Semi-Preparative Scale
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Winnig, M.; Bufe, B.; A Kratochwil, N.; Slack, J.P.; Meyerhof, W. The binding site for neohesperidin dihydrochalcone at the human sweet taste receptor. BMC Struct. Boil. 2007, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Krammer, G.; Ley, J.; Riess, T.; Haug, M.; Paetz, S.; Kindel, G.; Schmidtmann, R. Use of 4-hydroxydihydrochalcons and their salts for enhancing an impression of sweetness. U.S. Patent 9,445,606, 20 September 2016. [Google Scholar]
- Orlíková, B.; Tasdemir, D.; Golais, F.; Dicato, M.; Diederich, M. Dietary chalcones with chemopreventive and chemotherapeutic potential. Genes Nutr. 2011, 6, 125–147. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Zhang, Y.; Chen, X.; Wang, Y.; Chen, W.; Xu, Q.; Li, P.; Ma, F. Extraction, identification, and antioxidant and anticancer tests of seven dihydrochalcones from Malus ‘Red Splendor’ fruit. Food Chem. 2017, 231, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Sofic, E.; Prior, R.L. Antioxidant and prooxidant behavior of flavonoids: structure-activity relationships. Free Radical Biol. Med. 1997, 22, 749–760. [Google Scholar] [CrossRef]
- Corrêa, M.J.C.; Nunes, F.M.; Bitencourt, H.R.; Borges, F.C.; Guilhon, G.M.S.P.; Arruda, M.S.P.; Marinho, A.M.R.; Santos, A.S.; Alves, C.N.; Brasil, D.S.B.; et al. Biotransformation of chalcones by the endophytic fungus Aspergillus flavus isolated from Paspalum maritimum trin. J. Braz. Chem. Soc. 2011, 22, 1333–1338. [Google Scholar] [CrossRef]
- Gall, M.; Thomsen, M.; Peters, C.; Pavlidis, I.V.; Jonczyk, P.; Grünert, P.P.; Beutel, S.; Scheper, T.; Gross, E.; Backes, M.; et al. Enzymatic conversion of flavonoids using bacterial chalcone isomerase and enoate reductase. Angew. Chem. Int. Ed. 2014, 53, 1439–1442. [Google Scholar] [CrossRef] [PubMed]
- Awouafack, M.D.; Kusari, S.; Lamshöft, M.; Ngamga, D.; Tane, P.; Spiteller, M. Semi-synthesis of dihydrochalcone derivatives and their in vitro antimicrobial activities. Planta Med. 2010, 76, 640–643. [Google Scholar] [CrossRef][Green Version]
- Silva, V.D.; Stambuk, B.U.; Nascimento, M.D.G. Efficient chemoselective biohydrogenation of 1,3-diaryl-2-propen-1-ones catalyzed by Saccharomyces cerevisiae yeasts in biphasic system. J. Mol. Catal. B: Enzym. 2010, 63, 157–163. [Google Scholar] [CrossRef]
- Zhang, L.-Q.; Yang, X.-W.; Zhang, Y.-B.; Zhai, Y.-Y.; Xu, W.; Zhao, B.; Liu, D.-L.; Yu, H.-J. Biotransformation of phlorizin by human intestinal flora and inhibition of biotransformation products on tyrosinase activity. Food Chem. 2012, 132, 936–942. [Google Scholar] [CrossRef]
- Noe, C.R.; Knollmiiller, M.; Oberhauser, B.; Steinbauer, G. Eine Methode zur Bestimmung der Absolutkonfiguration chiraler a-hydroxysubstituierter Nitrile, Alkine und Aldehyde. Chem. Ber. 1986, 119, 729–743. [Google Scholar] [CrossRef]
- Ecker, G.; Chiba, P.; Hitzler, M.; Schmid, D.; Visser, K.; Cordes, H.P.; Csöllei, J.; Seydel, J.K.; Schaper, K.-J. Structure−Activity Relationship Studies on Benzofuran Analogs of Propafenone-Type Modulators of Tumor Cell Multidrug Resistance. J. Med. Chem. 1996, 39, 4767–4774. [Google Scholar] [CrossRef] [PubMed]
- Ecker, G.; Noe, C.R.; Fleischhacker, W. Improved synthesis of the enantiomers of propafenone using chiral building blocks. Chem. Monthly 1997, 128, 53–59. [Google Scholar] [CrossRef]
- Shallenberger, R.S. Hydrogen Bonding and the Varying Sweetness of the Sugars. J. Food Sci. 1963, 28, 584–589. [Google Scholar] [CrossRef]
- Belitz, H.-D.; Grosch, W.; Schieberle, P. Food Chemistry, 4th ed.; Springer-Verlag: Berlin, Germany, 2009. [Google Scholar]
- Whitelaw, M.L.; Chung, H.-J.; Daniel, J.R. Synthesis and sensory evaluation of ring-substituted dihydrochalcone sweetener. J. Agric. Food Chem. 1991, 39, 663–667. [Google Scholar] [CrossRef]
- Portmann, M.-O.; Kilcast, D. Psychophysical characterization of new sweeteners of commercial importance for the EC food industry. Food Chem. 1996, 56, 291–302. [Google Scholar] [CrossRef]
- Hunter, J.; Rice, S.; Lowe, R.; Pask, C.M.; Warriner, S.; Sridharan, V. Iridium catalyzed alkylation of 2′-hydroxyacetophenone with alcohols under thermal or microwave conditions. Tetrahedron Lett. 2017, 58, 4400–4402. [Google Scholar] [CrossRef]
- Chen, P.; Li, W.; Wang, Y. Atmospheric hydrogenation of α, β-unsaturated ketones catalyzed by highly efficient and recyclable Pd nanocatalyst. Catal. Commun. 2019, 125, 10–14. [Google Scholar] [CrossRef]
- Stompor, M.; Kałużny, M.; Żarowska, B. Biotechnological methods for chalcone reduction using whole cells of Lactobacillus, Rhodococcus and Rhodotorula strains as a way to produce new derivatives. Appl. Microbiol. Biotechnol. 2016, 100, 8371–8384. [Google Scholar] [CrossRef]
- Li, W.; Wu, X.-F. ChemInform Abstract: Ruthenium-Catalyzed Conjugate Hydrogenation of α,β-Enones by in situ Generated Dihydrogen from Paraformaldehyde and Water. Eur. J. Org. Chem. 2015, 2015, 331–335. [Google Scholar] [CrossRef]
- Chen, S.; Lu, G.; Cai, C. A base-controlled chemoselective transfer hydrogenation of α,β-unsaturated ketones catalyzed by [IrCp*Cl2]2 with 2-propanol. RSC Adv. 2015, 5, 13208–13211. [Google Scholar] [CrossRef]
- Rosa, G.P.; Seca, A.M.L.; Barreto, M.D.C.; Pinto, D.C.G.A.; Pinto, D.C.G.A. Chalcone: A Valuable Scaffold Upgrading by Green Methods. ACS Sustain. Chem. Eng. 2017, 5, 7467–7480. [Google Scholar] [CrossRef]
- Winkler, C.K.; Faber, K.; Hall, M. Biocatalytic reduction of activated C C-bonds and beyond: emerging trends. Curr. Opin. Chem. Boil. 2018, 43, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, S.; Romano, D.; Amaretti, A.; Molinari, F.; Rossi, M. Enoate reductases from non conventional yeasts: Bioconversion, cloning, and functional expression in Saccharomyces cerevisiae. J. Biotechnol. 2011, 156, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Janeczko, T.; Dymarska, M.; Siepka, M.; Gniłka, R.; Leśniak, A.; Popłoński, J.; Kostrzewa-Susłow, E. Enantioselective reduction of flavanone and oxidation of cis- and trans-flavan-4-ol by selected yeast cultures. J. Mol. Catal. B: Enzym. 2014, 109, 47–52. [Google Scholar] [CrossRef]
- Kozłowska, J.; Potaniec, B.; Żarowska, B.; Anioł, M. Microbial transformations of 4′-methylchalcones as an efficient method of obtaining novel alcohol and dihydrochalcone derivatives with antimicrobial activity. RSC Adv. 2018, 8, 30379–30386. [Google Scholar] [CrossRef]
- Żyszka-Haberecht, B.; Poliwoda, A.; Lipok, J. Biocatalytic hydrogenation of the C=C bond in the enone unit of hydroxylated chalcones—process arising from cyanobacterial adaptations. Appl. Microbiol. Biotechnol. 2018, 102, 7097–7111. [Google Scholar] [CrossRef]
- Hall, M.; Stueckler, C.; Hauer, B.; Stuermer, R.; Friedrich, T.; Breuer, M.; Kroutil, W.; Faber, K. Asymmetric Bioreduction of Activated C=C Bonds UsingZymomonas mobilis NCR Enoate Reductase and Old Yellow Enzymes OYE 1–3 from Yeasts. Eur. J. Org. Chem. 2008, 2008, 1511–1516. [Google Scholar] [CrossRef]
- Kohli, R.M.; Massey, V. The oxidative half-reaction of Old Yellow Enzyme. J. Biol. Chem. 1998, 273, 32763–32770. [Google Scholar] [CrossRef]
- Karplus, P.A.; Fox, K.M.; Massey, V. Flavoprotein structure and mechanism. 8. Structure-function relations for old yellow enzyme. FASEB J. 1995, 9, 1518–1526. [Google Scholar] [CrossRef]
- Janeczko, T.; Gładkowski, W.; Kostrzewa-Susłow, E. Microbial transformations of chalcones to produce food sweetener derivatives. J. Mol. Catal. B: Enzym. 2013, 98, 55–61. [Google Scholar] [CrossRef]
- Janeczko, T.; Kostrzewa-Susłow, E. Enantioselective reduction of propiophenone formed from 3-chloropropiophenone and stereoinversion of the resulting alcohols in selected yeast cultures. Tetrahedron Asymmetry 2014, 25, 1264–1269. [Google Scholar] [CrossRef]
- Łużny, M.; Kozłowska, E.; Dymarska, M.; Kostrzewa-Susłow, E.; Janeczko, T. Sposób wytwarzania 1-(2’-hydrokyfenylo)-3-(3”-metoksyfenylo)-propan-1-onu. P.426760. 23 August 2018. [Google Scholar]
- Łużny, M.; Kozłowska, E.; Popłoński, J.; Dymarska, M.; Kostrzewa-Susłow, E.; Janeczko, T. Sposób wytwarzania 1-(2’-hydrokyfenylo)-3-(4”-metoksyfenylo)-propan-1-onu. P.426763. 23 August 2018. [Google Scholar]
- Łużny, M.; Kozłowska, E.; Dymarska, M.; Kostrzewa-Susłow, E.; Janeczko, T. Sposób wytwarzania 1-(2’-hydrokyfenylo)-3-(4”-metoksyfenylo)-propan-1-onu. P.426770. 23 August 2018. [Google Scholar]
- Kostrzewa-Susłow, E.; Dymarska, M.; Guzik, U.; Wojcieszyńska, D.; Janeczko, T. Stenotrophomonas maltophilia: A Gram-Negative Bacterium Useful for Transformations of Flavanone and Chalcone. Molecules 2017, 22, 1830. [Google Scholar] [CrossRef] [PubMed]
- De Matos, I.L.; Nitschke, M.; Porto, A.L.M. Hydrogenation of Halogenated 2′-Hydroxychalcones by Mycelia of Marine-Derived Fungus Penicillium raistrickii. Mar. Biotechnol. 2019, 21, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Żyszka, B.; Anioł, M.; Lipok, J. Highly effective, regiospecific reduction of chalcone by cyanobacteria leads to the formation of dihydrochalcone: two steps towards natural sweetness. Microb. Cell Factories 2017, 16, 136. [Google Scholar] [CrossRef] [PubMed]
- Janeczko, T.; Popłoński, J.; Kozłowska, E.; Dymarska, M.; Huszcza, E.; Kostrzewa-Susłow, E. Application of α- and β-naphthoflavones as monooxygenase inhibitors of Absidia coerulea KCh 93, Syncephalastrum racemosum KCh 105 and Chaetomium sp. KCh 6651 in transformation of 17α-methyltestosterone. Bioorganic Chem. 2018, 78, 178–184. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |

| Time [Days] | Strain Number | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KCh 4 | KCh 71 | KCh 77 | KCh 82 | KCh 120 | KCh 242 | KCh 464 | KCh 909 | ||||||||||
| 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | ||
| 1 | 91 | 23 | 98 | >99 | 92 | 38 | 98 | 59 | 49 | 84 | 0 | 52 | 98 | 98 | 2 | 9 | |
| 3 | 99 | 33 | >99 | >99 | >99 | 60 | >99 | 84 | >99 | 99 | 0 | 67 | >99 | 99 | 14 | 12 | |
| 7 | >99 | 49 | >99 | >99 | >99 | 63 | >99 | 97 | >99 | 99 | 17 | 87 | >99 | 98 | 26 | 16 | |
| Time [h] | Strain Number | |||
|---|---|---|---|---|
| KCh4 | KCh71 | KCh120 | KCh464 | |
| 1 | 5 ± 1.1 | 52 ± 4.5 | 2 ± 0.5 | 40 ± 2.1 |
| 3 | 6 ± 0.1 | 88 ± 1.6 | 2 ± 0.4 | 84 ± 2.9 |
| 6 | 9 ± 0.5 | >99 ±0.0 | 4 ± 0.9 | 98 ± 0.5 |
| 12 | 10 ± 0.9 | >99 ± 0.0 | 9 ± 0.5 | 99 ± 0.6 |
| Time [h] | Strain Number | |||
|---|---|---|---|---|
| KCh71 | KCh120 | KCh242 | KCh464 | |
| 1 | 67 ± 3.9 | 0 ± 0.0 | 1 ± 0.6 | 35 ± 3.6 |
| 3 | >99 ± 0.0 | 10 ± 1.1 | 5 ± 0.5 | 64 ± 0.9 |
| 6 | >99 ± 0.0 | 14 ± 1.2 | 9 ± 0.6 | 83 ± 0.6 |
| 12 | >99 ± 0.0 | 25 ± 3.1 | 12 ± 0.9 | 98 ± 0.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łużny, M.; Krzywda, M.; Kozłowska, E.; Kostrzewa-Susłow, E.; Janeczko, T. Effective Hydrogenation of 3-(2”-furyl)- and 3-(2”-thienyl)-1-(2’-hydroxyphenyl)-prop-2-en-1-one in Selected Yeast Cultures. Molecules 2019, 24, 3185. https://doi.org/10.3390/molecules24173185
Łużny M, Krzywda M, Kozłowska E, Kostrzewa-Susłow E, Janeczko T. Effective Hydrogenation of 3-(2”-furyl)- and 3-(2”-thienyl)-1-(2’-hydroxyphenyl)-prop-2-en-1-one in Selected Yeast Cultures. Molecules. 2019; 24(17):3185. https://doi.org/10.3390/molecules24173185
Chicago/Turabian StyleŁużny, Mateusz, Martyna Krzywda, Ewa Kozłowska, Edyta Kostrzewa-Susłow, and Tomasz Janeczko. 2019. "Effective Hydrogenation of 3-(2”-furyl)- and 3-(2”-thienyl)-1-(2’-hydroxyphenyl)-prop-2-en-1-one in Selected Yeast Cultures" Molecules 24, no. 17: 3185. https://doi.org/10.3390/molecules24173185
APA StyleŁużny, M., Krzywda, M., Kozłowska, E., Kostrzewa-Susłow, E., & Janeczko, T. (2019). Effective Hydrogenation of 3-(2”-furyl)- and 3-(2”-thienyl)-1-(2’-hydroxyphenyl)-prop-2-en-1-one in Selected Yeast Cultures. Molecules, 24(17), 3185. https://doi.org/10.3390/molecules24173185





