Effects of Complex DNA and MVs with GTF Extracted from Streptococcus mutans on the Oral Biofilm
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
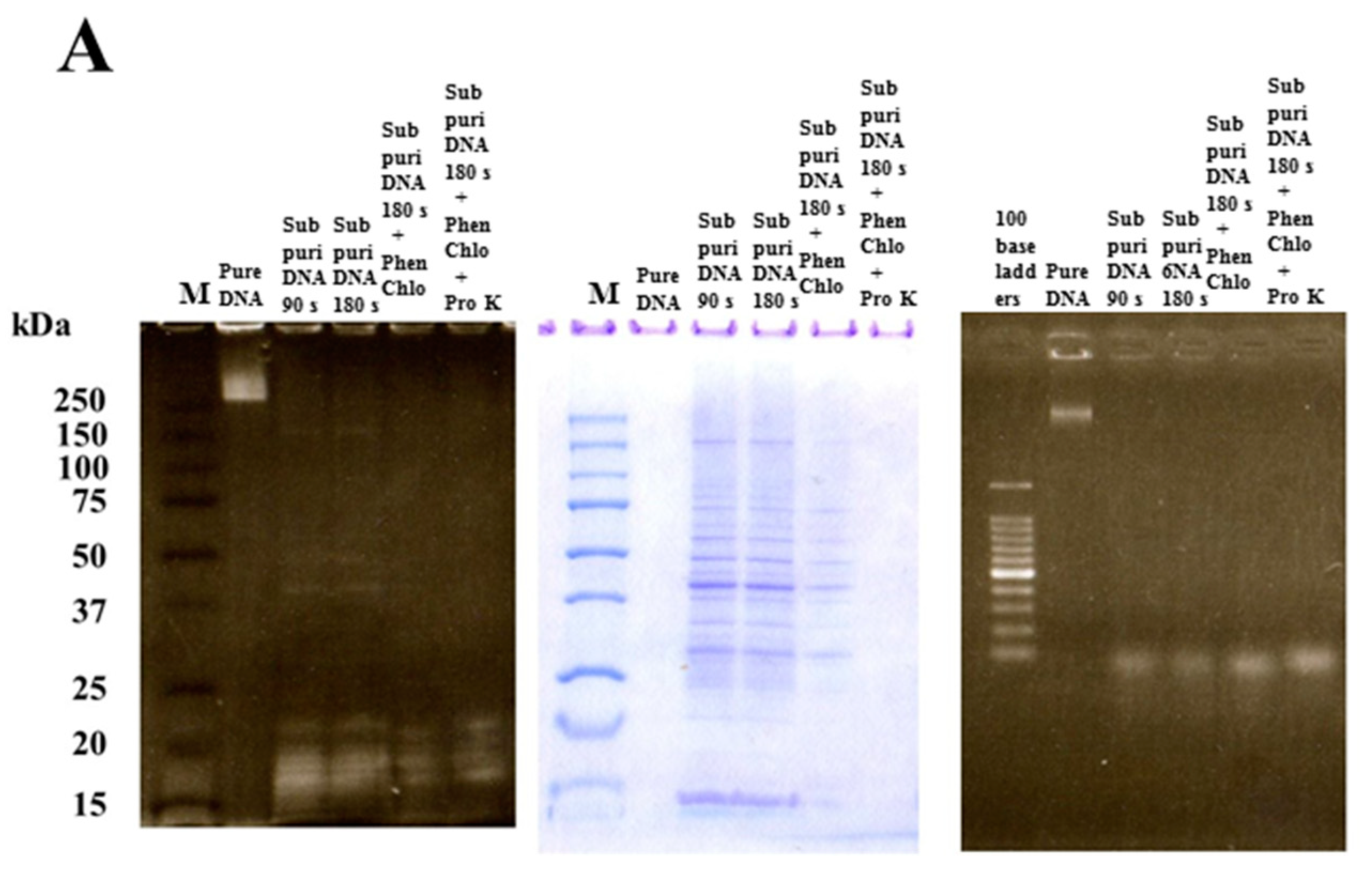
2.2. Extraction of Four Types of DNA
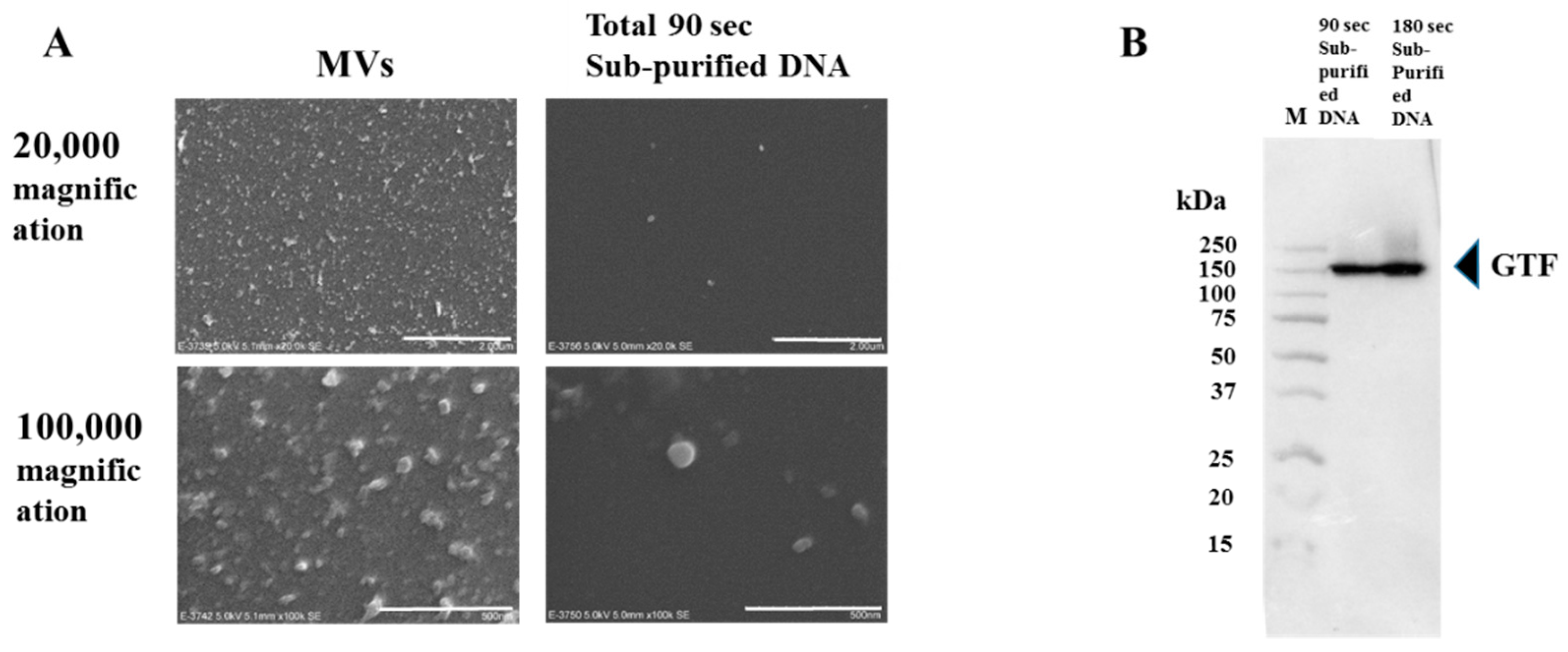
2.3. Extraction of MVs
2.4. Human Saliva Collection
2.5. Biofilm Formation
2.6. SDS-PAGE and Native-PAGE
2.7. Western Blotting
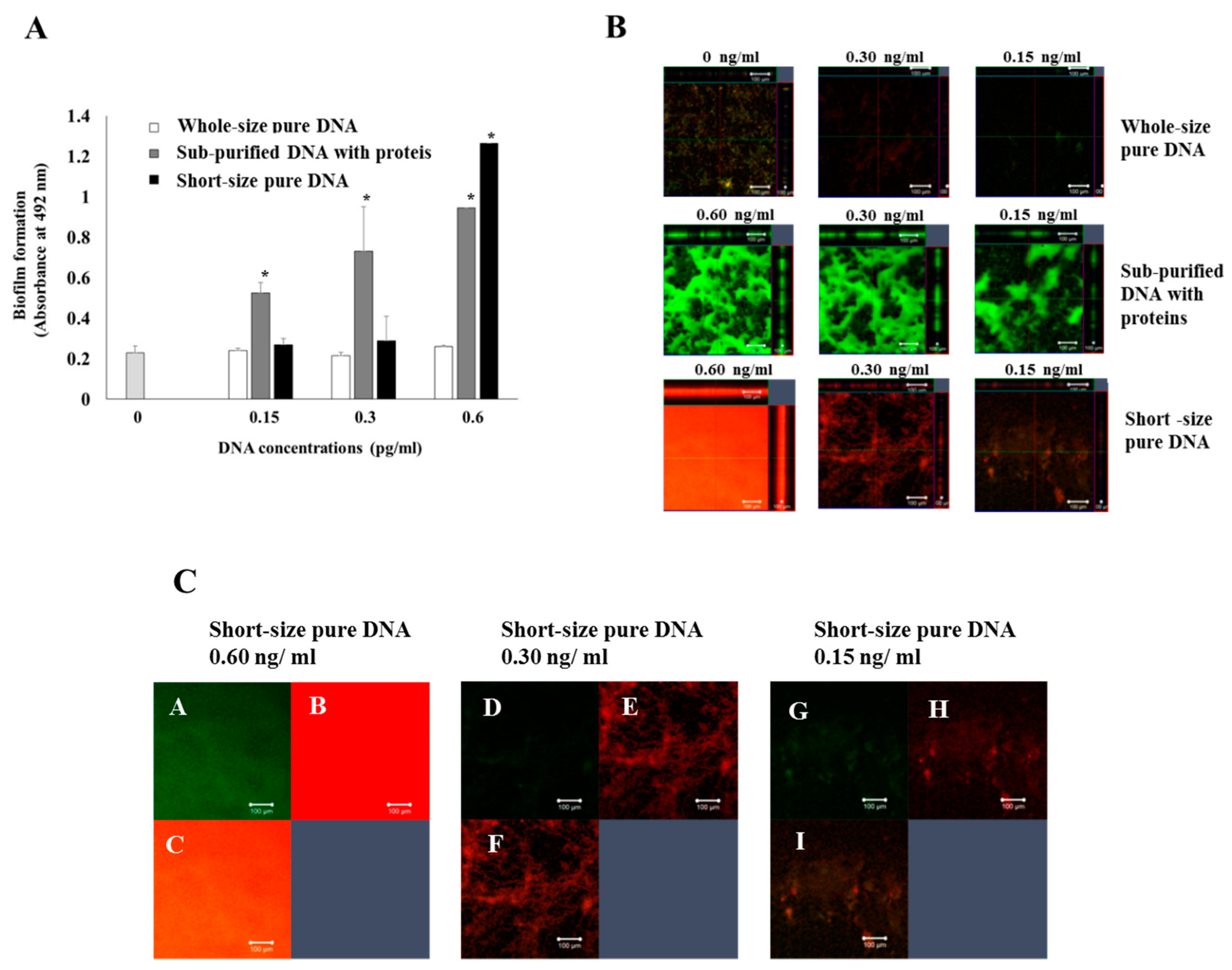
2.8. Observation of Live/Dead Cells and Glucan in Biofilm Formation
2.9. Statistical Analyses
3. Results
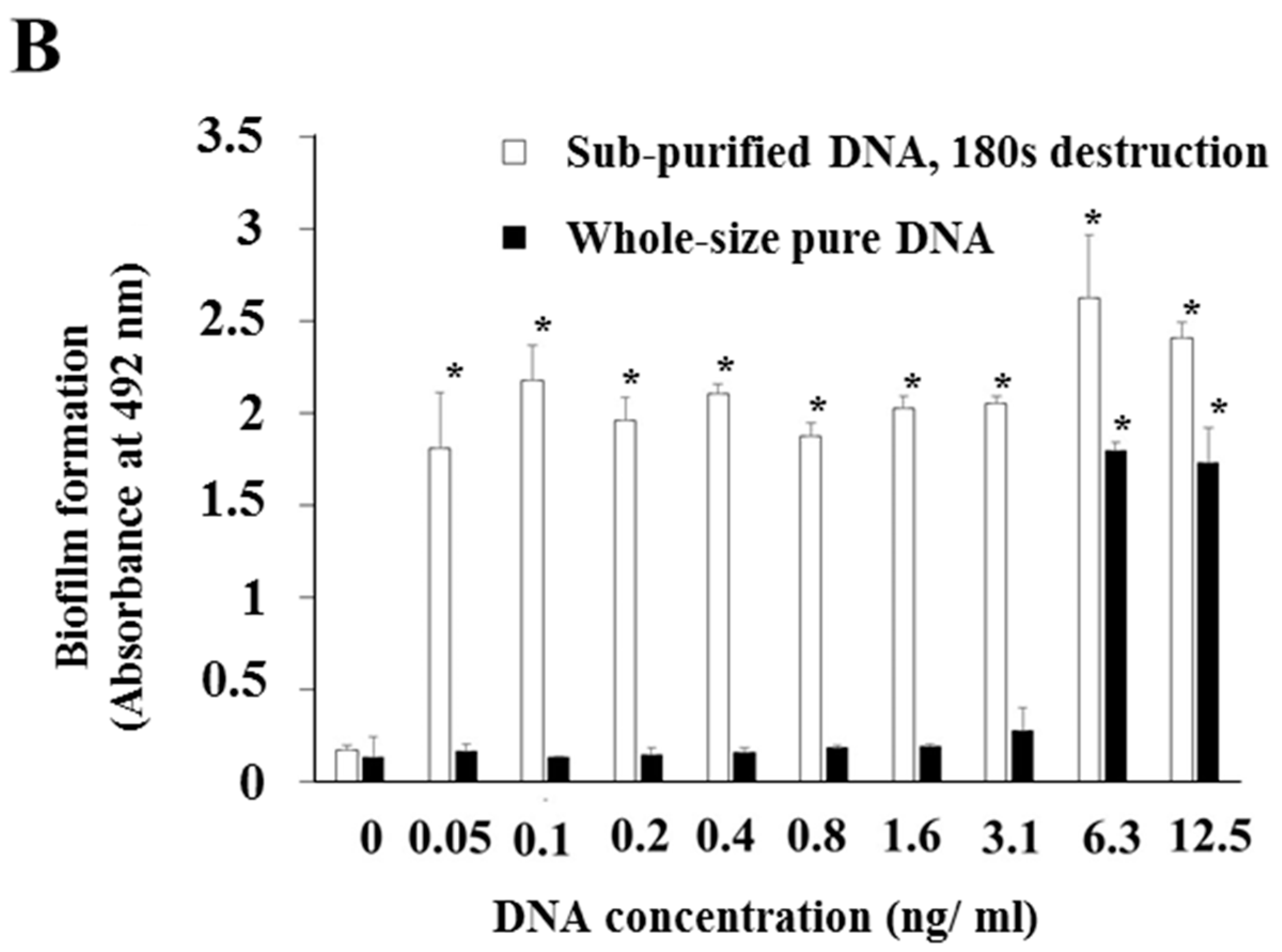
3.1. Effects of Various Types of DNA on Biofilm Formation
3.2. Effects of DNA with Proteins and Whole-Size Pure DNA on Biofilm Formation
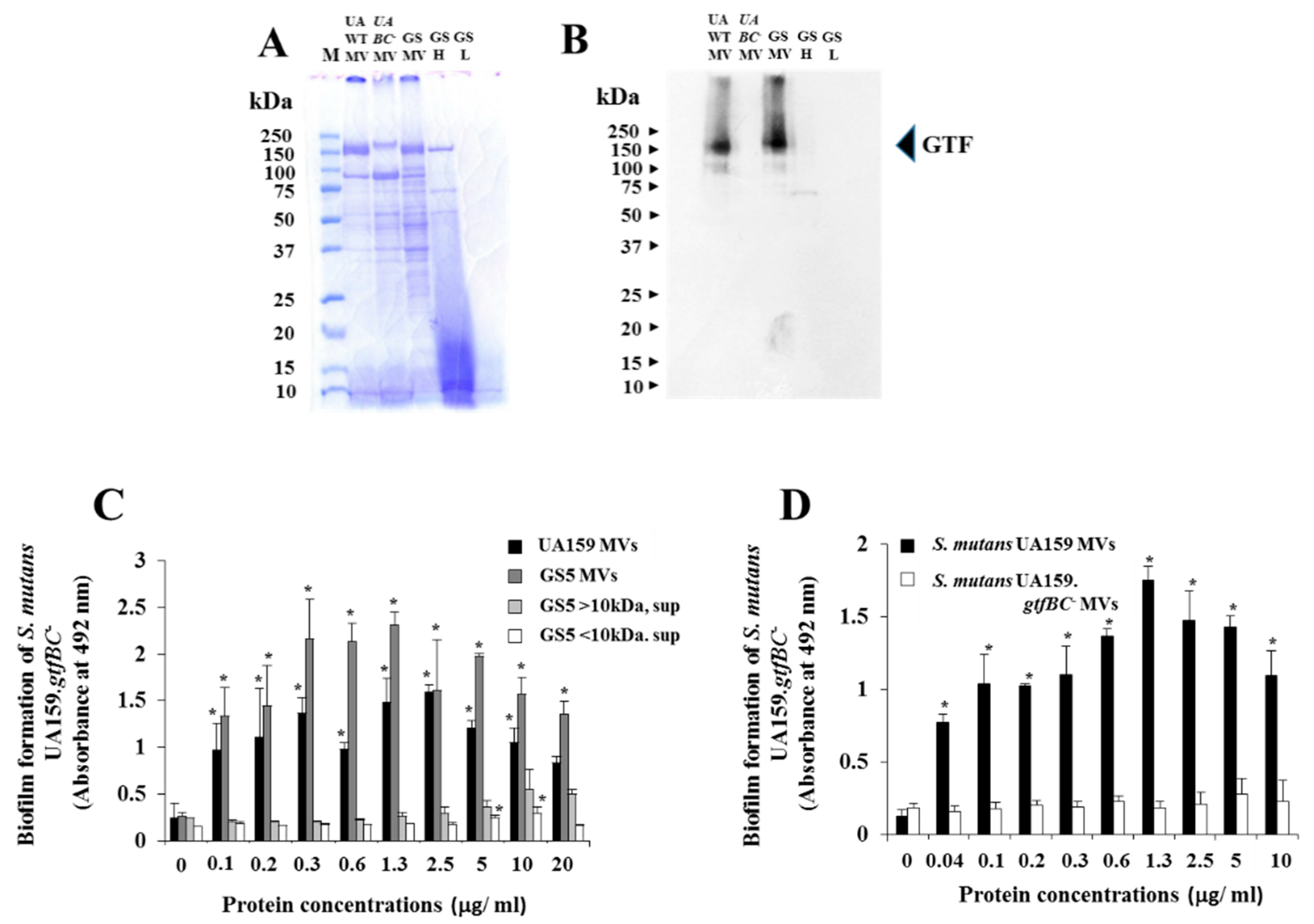
3.3. Effects of GTF-I and GTF-SI on MVs on the Biofilm Formation of S. Mutans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Burne, R. Oral Streptococci. Products of their environment. J. Dent. Res. 1998, 77, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 1986, 50, 353–380. [Google Scholar] [PubMed]
- Nyvad, B.; Kilian, M. Comparison of the initial Streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 1990, 24, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef] [PubMed]
- Kroes, I.; Lepp, P.W.; Relman, D.A. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 1999, 96, 14547–14552. [Google Scholar] [CrossRef] [PubMed]
- Paster, B.J.; Boches, S.K.; Galvin, J.L.; Ericson, R.E.; Lau, C.N.; Levanos, V.A.; Sahasrabudhe, A.; Dewhirst, F.E. Bacterial diversity in human subgingival plaque. J. Bacteriol. 2001, 183, 3770–3783. [Google Scholar] [CrossRef]
- Hamada, S.; Slade, H.D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 1980, 44, 331–384. [Google Scholar]
- Bowen, W.; Koo, H. Biology of Streptococcus mutans-derived glucosyltransferases: Role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011, 45, 69–86. [Google Scholar] [CrossRef]
- Banas, J.A. Virulence properties of Streptococcus mutans. Front. Biosci. 2004, 9, 1267–1277. [Google Scholar] [CrossRef]
- Aoki, H.; Shiroza, T.; Hayakawa, M.; Sato, S.; Kuramitsu, H.K. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect. Immun. 1986, 53, 587–594. [Google Scholar]
- Lynch, D.J.; Fountai, T.L.; Mazurkeiwicz, J.E.; Banas, J.A. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol. Lett. 2007, 268, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Duque, C.; Stipp, R.N.; Wang, B.; Smith, D.J.; Höfling, J.F.; Kuramitsu, H.K.; Duncan, M.J.; Mattos-Graner, R.O. Downregulation of GbpB, a component of the VicRK regulon, affects biofilm formation and cell surface characteristics of Streptococcus mutans. Infect. Immun. 2011, 79, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.-J.; Ahn, S.-J.; Wen, Z.T.; Brady, L.J.; Burne, R.A. Characteristics of Biofilm Formation by Streptococcus mutans in the Presence of Saliva. Infect. Immun. 2008, 76, 4259–4268. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.A.; Cvitkovitch, D.G.; Lévesque, C.M. Cell death in Streptococcus mutans biofilms: A link between CSP and extracellular DNA. FEMS Microbiol. Lett. 2009, 299, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Sharma, P.K.; Busscher, H.J.; Van Der Mei, H.C.; Krom, B.P. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 2010, 76, 3405–3408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Ou, M.; Wang, W.; Ling, J. Effects of quorum sensing on cell viability in Streptococcus mutans biofilm formation. Biochem. Biophys. Res. Commun. 2009, 379, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Petersen, F.C.; Tao, L.; Scheie, A.A. DNA Binding-Uptake System: A link between Cell-to-Cell communication and biofilm formation. J. Bacteriol. 2005, 187, 4392–4400. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rutz, K.; Kunze, B.; Tomasch, J.; Surapaneni, S.K.; Schulz, S.; Wagner-Döbler, I. The biofilm inhibitor carolacton disturbs membrane integrity and cell division of Streptococcus mutans through the Serine/Threonine protein kinase PknB. J. Bacteriol. 2011, 193, 5692–5706. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liao, S.; Klein, M.I.; Heim, K.P.; Fan, Y.; Bitoun, J.P.; Ahn, S.-J.; Burne, R.A.; Koo, H.; Brady, L.J.; Wen, Z.T. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J. Bacteriol. 2014, 196, 2355–2366. [Google Scholar] [CrossRef]
- Rivera, J.; Cordero, R.J.B.; Nakouzi, A.S.; Frases, S.; Nicola, A.; Casadevall, A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. USA 2010, 107, 19002–19007. [Google Scholar] [CrossRef]
- Prados-Rosales, R.; Carreño, L.J.; Batista-Gonzalez, A.; Baena, A.; Venkataswamy, M.M.; Xu, J.; Yu, X.; Wallstrom, G.; Magee, D.M.; LaBaer, J.; et al. Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. mBio 2014, 5, e01921-14. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Cárcamo-Oyarce, G.; Yamamoto, T.; Eisenstein, F.; Hsiao, C.-C.; Kurosawa, M.; Gademann, K.; Pilhofer, M.; Nomura, N.; Eberl, L. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat. Commun. 2017, 8, 481. [Google Scholar] [CrossRef] [PubMed]
- Olaya-Abril, A.; Prados-Rosales, R.; McConnell, M.J.; Martín-Peña, R.; González-Reyes, J.A.; Jiménez-Munguía, I.; Gómez-Gascón, L.; Fernandez, J.; Luque-Garcia, J.L.; García-Lidón, C.; et al. Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J. Proteom. 2014, 106, 46–60. [Google Scholar] [CrossRef]
- Kawarai, T.; Narisawa, N.; Suzuki, Y.; Nagasawa, R.; Senpuku, H. Streptococcus mutans biofilm formation is dependent on extracellular DNA in primary low pH conditions. J. Oral Biosci. 2016, 58, 55–61. [Google Scholar] [CrossRef]
- Das, T.; Sehar, S.; Manefield, M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ. Microbiol. Rep. 2013, 5, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, R.; Sato, T.; Senpuku, H. Raffinose induces biofilm formation by Streptococcus mutans in low concentrations of sucrose by increasing production of extracellular DNA and fructan. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Nagasawa, R.; Senpuku, H. Inhibiting effects of fructanase on competence-stimulating peptide-dependent quorum sensing system in Streptococcus mutans. J. Infect. Chemother. 2017, 23, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Nakao, R.; Hasegawa, H.; Ochiai, K.; Takashiba, S.; Ainai, A.; Ohnishi, M.; Watanabe, H.; Senpuku, H. Outer membrane vesicles of Porphyromonas gingivalis elicit a mucosal immune response. PLoS ONE 2014, 6, e26163. [Google Scholar] [CrossRef] [PubMed]
- Motegi, M.; Takagi, Y.; Yonezawa, H.; Hanada, N.; Terajima, J.; Watanabe, H.; Senpuku, H. Assessment of genes associated with Streptococcus mutans Biofilm Morphology. Appl. Environ. Microbiol. 2006, 72, 6277–6287. [Google Scholar] [CrossRef] [PubMed]
- Hamada, S.; Horikoshi, T.; Minami, T.; Okahashi, N.; Koga, T. Purification and characterization of cell-associated glucosyltransferase synthesizing water-insoluble glucan from serotype c Streptococcus mutans. J. Gen. Microbiol. 1989, 135, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Xiao, J.; Klein, M.I.; Jeon, J.G. Exopolysaccharides Produced by Streptococcus mutans Glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 2010, 192, 3024–3032. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E.; Andersen, R.N.; Blehert, D.S.; Egland, P.G.; Foster, J.S.; Palmer, R.J. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 486–505. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.I.; Hwang, G.; Santos, P.H.S.; Campanella, O.H.; Koo, H. Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Front. Microbiol. 2015, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.V.; Scott-Anne, K.; Whelehan, M.; Berkowitz, R.; Feng, C.; Bowen, W. Salivary glucosyltransferase B as a possible marker for caries activity. Caries Res. 2007, 41, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.I.; DeBaz, L.; Agidi, S.; Lee, H.; Xie, G.; Lin, A.H.-M.; Hamaker, B.R.; Lemos, J.A.; Koo, H. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoS ONE 2010, 5, e13478. [Google Scholar] [CrossRef] [PubMed]
- Kesty, N.C.; Mason, K.M.; Reedy, M.; Miller, S.E.; Kuehn, M.J. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 2004, 23, 4538–4549. [Google Scholar] [CrossRef] [PubMed]
- Steinberger, R.E.; Holden, P.A. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 2015, 71, 5404–5410. [Google Scholar] [CrossRef]
- Rainey, K.; Michalek, S.M.; Wen, Z.T.; Wu, H. Glycosyltransferase mediated matrix dynamics and virulence of Streptooccus mutans. Appl. Environ. Microbiol. 2019, 85, e02247-18. [Google Scholar] [CrossRef]
- Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Allesen-Holm, M.; Tolker-Nielsen, T.; et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006, 59, 1114–1128. [Google Scholar]
- Jung, C.-J.; Hsu, R.-B.; Shun, C.-T.; Hsu, C.-C.; Chia, J.-S. AtlA mediates extracellular DNA release, which contributes to Streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect. Immun. 2017, 85, e00252-17. [Google Scholar] [CrossRef]
- Rainey, M.M.; Korostyshevsky, D.; Lee, S.; Perlstein, E.O. The antidepressant sertraline targets intracellular vesiculogenic membranes in Yeast. Genetics 2010, 185, 1221–1233. [Google Scholar] [CrossRef] [PubMed]
- Foulston, L.; Elsholz, A.K.W.; DeFrancesco, A.S.; Losick, R. The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. mBio 2014, 5, e01667-14. [Google Scholar] [CrossRef] [PubMed]
- Dengler, V.; Foulston, L.; DeFrancesco, A.S.; Losick, R. An Electrostatic Net Model for the Role of extracellular DNA in biofilm formation by Staphylococcus aureus. J. Bacteriol. 2015, 197, 3779–3787. [Google Scholar] [CrossRef] [PubMed]
- Marcato-Romain, C.-E.; Randrianjatovo-Gbalou, I.; Rouquette, P.; Lefebvre, D.; Girbal-Neuhauser, E. In situ analysis of Bacillus licheniformis biofilms: Amyloid-like polymers and eDNA are involved in the adherence and aggregation of the extracellular matrix. J. Appl. Microbiol. 2017, 122, 1262–1274. [Google Scholar]
- Carot, M.L.; Torresi, R.M.; Garcia, C.D.; Esplandiu, M.J.; Giacomelli, C.E. Electrostatic and hydrophobic interactions involved in CNT biofunctionalization with short ss-DNA. J. Phys. Chem. C 2010, 114, 4459–4465. [Google Scholar] [CrossRef] [PubMed]
- Balladur, V.; Theretz, A.; Mandrand, B. Determination of the main forces driving DNA oligonucleotide adsorption onto aminated silica wafers. J. Colloid Interface Sci. 1997, 194, 408–418. [Google Scholar] [CrossRef]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Hall-Stoodley, L.; Stoodley, P. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Genet. 2017, 15, 740–755. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds, MVs are available from the authors. |


| Strains | None | UA159 MVs | UA159.gtfBC− MVs | Activity | |
|---|---|---|---|---|---|
| S. mitis | ATCC 903 | 0.241 ± 0.027 | 1.370 ± 0.101 * | 0.242 ± 0.048 | UP |
| ATCC 6249 | 0.411 ± 0.045 | 0.432 ± 0.082 | 0.473 ± 0.082 | ||
| S. oralis | ATCC 35037 | 0.149 ± 0.012 | 0.685 ± 0.079 * | 0.127 ± 0.002 | UP |
| S. sanguinis | ATCC 10556 | 1.147 ± 0.256 | 2.029 ± 0.082 * | 1.430 ± 0.240 | UP |
| ST 205 | 0.270 ± 0.035 | 1.164 ± 0.221 * | 0.356 ± 0.044 | UP | |
| ST 134 | 0.175 ± 0.006 | 0.255 ± 0.044 | 0.160 ± 0.030 | ||
| S. gordonii | ATCC 10558 | 0.406 ± 0.018 | 1.727 ± 0.341 * | 0.398 ± 0.005 | UP |
| S. anginosus | ATCC 33397 | 1.256 ± 0.254 | 1.175 ± 0.026 | 1.115 ± 0.581 | - |
| S. intermedius | ATCC 27335 | 0.153 ± 0.010 | 0.330 ± 0.070 | 0.150 ± 0.036 | - |
| S. pyogenes | K33 | 0.289 ± 0.011 | 0.377 ± 0.182 | 0.358 ± 0.163 | - |
| K32 | 0.574 ± 0.107 | 0.484 ± 0.091 | 0.529 ± 0.330 | - | |
| S. pneumoniae | GTC 261 | 0.178 ± 0.015 | 0.127 ± 0.010 | 0.212 ± 0.133 | - |
| S. salivarius | JCM 5907 | 0.492 ± 0.018 | 0.584 ± 0.020 | 0.566 ± 0.012 | - |
| ATCC 9759 | 0.124 ± 0.021 | 0.299 ± 0.070 | 0.163 ± 0.020 | - | |
| HT9R | 0.222 ± 0.029 | 0.277 ± 0.005 | 0.244 ± 0.007 | - | |
| A. naeslundii | x600 | 0.415 ± 0.092 | 1.916 ± 0.582 * | 1.059 ± 0.423 | UP |
| A. oris | MG1 | 1.087 ± 0.222 | 2.264 ± 0.207 * | 0.998 ± 0.157 | UP |
| N. cinerea | 23-1 | 0.344 ± 0.018 | 0.261 ± 0.011 | 0.339 ± 0.011 | - |
| N. flavescens | ATCC 13120 | 0.353 ± 0.012 | 0.231 ± 0.044 | 0.376 ± 0.010 | - |
| N. mucosa | 16-2 | 1.889 ± 0.260 | 1.333 ± 0.133 | 1.590 ± 0.172 | DOWN |
| N. subflava | #2 | 1.071 ± 0.101 | 1.318 ± 0.237 | 1.301 ± 0.208 | - |
| C. albicans | SC5314 | 0.393 ± 0.046 | 0.292 ± 0.063 | 0.433 ± 0.181 | - |
| SC5312 | 0.178 ± 0.029 | 0.288 ± 0.049 | 0.171 ± 0.015 | - | |
| S. aureus | Cowan I | 0.283 ± 0.004 | 0.328 ± 0.018 | 0.262 ± 0.043 | - |
| ATCC 6538P | 0.211 ± 0.036 | 0.269 ± 0.036 | 0.208 ± 0.010 | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Senpuku, H.; Nakamura, T.; Iwabuchi, Y.; Hirayama, S.; Nakao, R.; Ohnishi, M. Effects of Complex DNA and MVs with GTF Extracted from Streptococcus mutans on the Oral Biofilm. Molecules 2019, 24, 3131. https://doi.org/10.3390/molecules24173131
Senpuku H, Nakamura T, Iwabuchi Y, Hirayama S, Nakao R, Ohnishi M. Effects of Complex DNA and MVs with GTF Extracted from Streptococcus mutans on the Oral Biofilm. Molecules. 2019; 24(17):3131. https://doi.org/10.3390/molecules24173131
Chicago/Turabian StyleSenpuku, Hidenobu, Tomoyo Nakamura, Yusuke Iwabuchi, Satoru Hirayama, Ryoma Nakao, and Makoto Ohnishi. 2019. "Effects of Complex DNA and MVs with GTF Extracted from Streptococcus mutans on the Oral Biofilm" Molecules 24, no. 17: 3131. https://doi.org/10.3390/molecules24173131
APA StyleSenpuku, H., Nakamura, T., Iwabuchi, Y., Hirayama, S., Nakao, R., & Ohnishi, M. (2019). Effects of Complex DNA and MVs with GTF Extracted from Streptococcus mutans on the Oral Biofilm. Molecules, 24(17), 3131. https://doi.org/10.3390/molecules24173131





