Antibacterial Evaluation, In Silico Characters and Molecular Docking of Schiff Bases Derived from 5-aminopyrazoles
Abstract
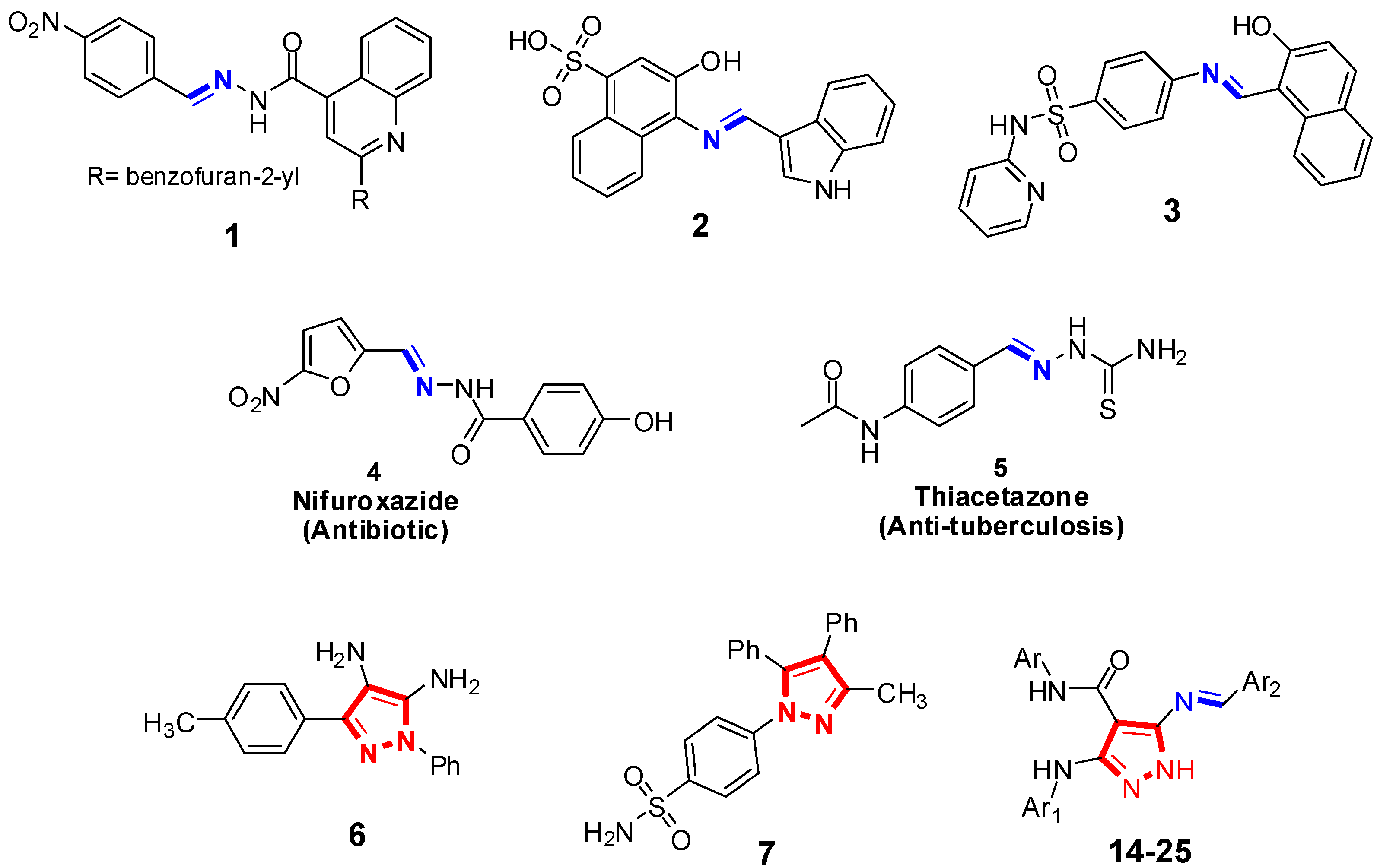
1. Introduction
2. Results and Discussion
2.1. Chemistry
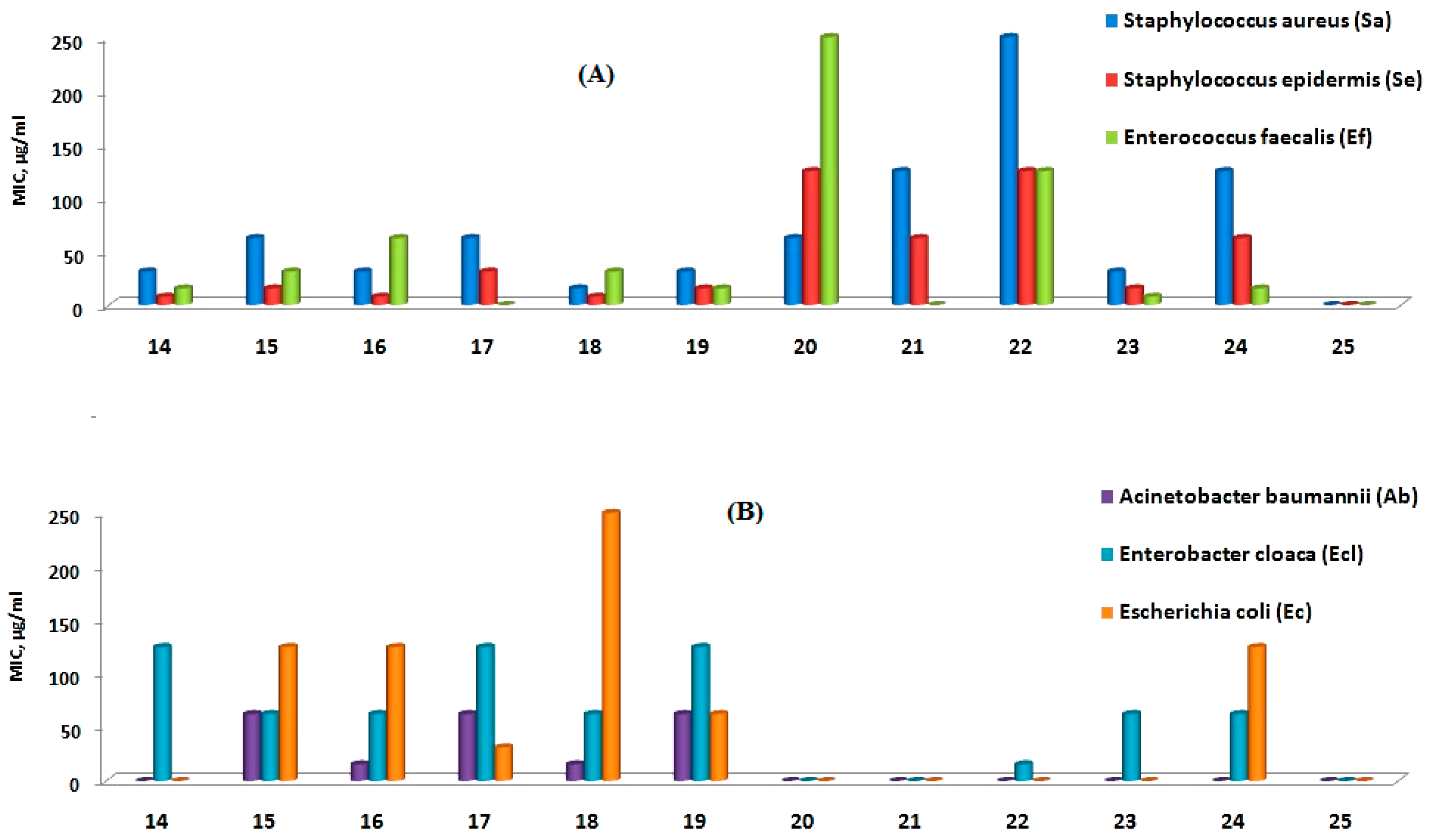
2.2. Antibacterial Evaluation
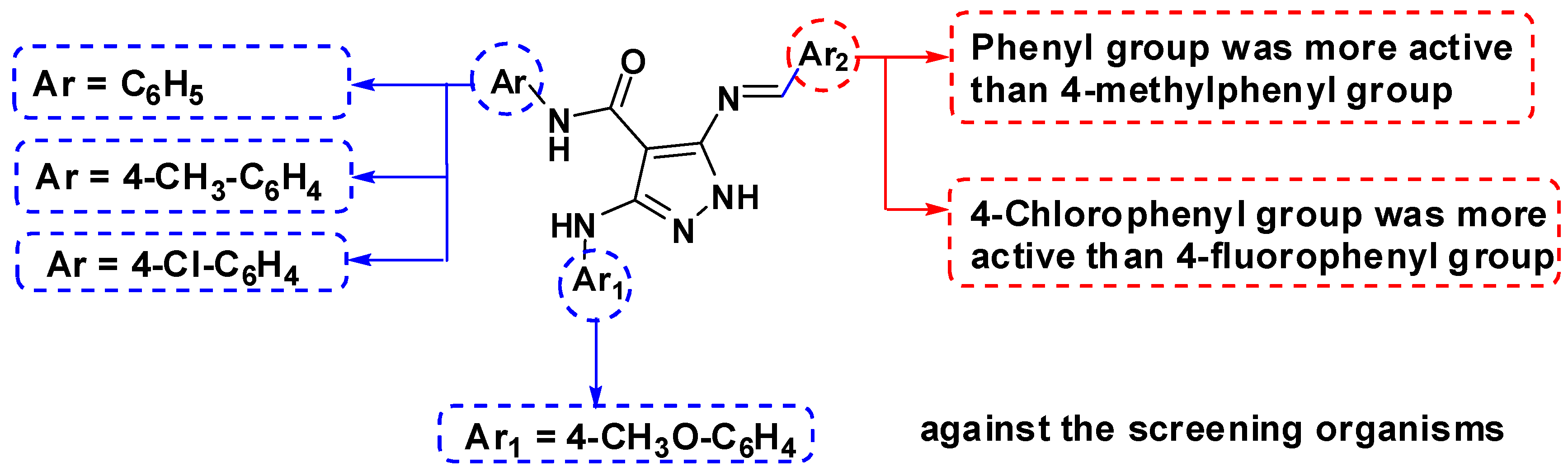
2.3. Structure-Activity Relationship (SAR)
2.4. In Silico ADMET Properties of Schiff Bases 14-25
2.5. In Vitro Kinase Assessment
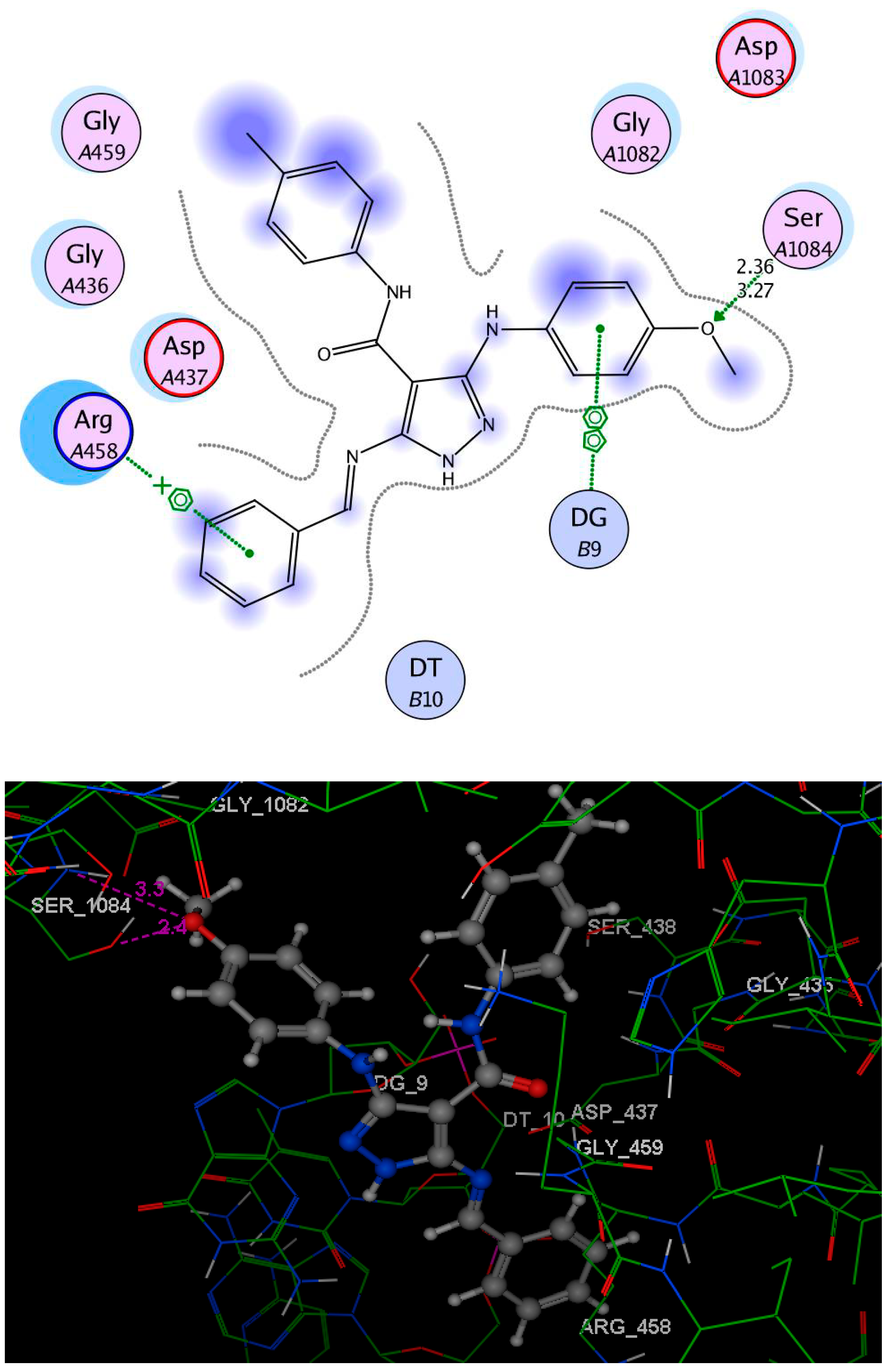
2.6. Molecular Docking Study
3. Materials and Methods
3.1. Chemicals
3.2. In Vitro Antibacterial Evaluation
3.2.1. Test Microorganisms
3.2.2. Antibacterial Activity
3.2.3. Minimum Inhibitory Concentration (MIC) of the Active Compounds
3.3. In Vitro Kinase Assessment
3.4. Molecular Docking Study
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Asadi, Z.; Nasrollahi, N. The effect of metal and substituent on DNA binding, cleavage activity, and cytotoxicity of new synthesized Schiff base ligands and Zn(II) complex. J. Mol. Struct. 2017, 1147, 582–593. [Google Scholar] [CrossRef]
- Jamshidvand, A.; Sahihi, M.; Mirkhani, V.; Moghadam, M.; Mohammadpoor-Baltork, I.; Tangestaninejad, S.; Rudbari, H.A.; Kargar, H.; Keshavarzi, R.; Gharaghani, S. Studies on DNA binding properties of new Schiff base ligands using spectroscopic, electrochemical and computationalmethods: Influence of substitutions on DNA-binding. J. Mol. Liq. 2018, 253, 61–71. [Google Scholar] [CrossRef]
- Zhao, F.; Wang, W.; Lu, W.; Xu, L.; Yang, S.; Cai, X.-M.; Zhou, M.; Lei, M.; Ma, M.; Xu, H.-J.; et al. High anticancer potency on tumor cells of dehydroabietylamine Schiff-base derivatives and a copper(II) complex. Eur. J. Med. Chem. 2018, 146, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Sangeeta, S.; Ahmad, K.; Noorussabah, N.; Bharti, S.; Mishra, M.K.; Sharma, S.R.; Choudhary, M. Synthesis, crystal structures, molecular docking and urease inhibition studies of Ni(II) and Cu(II) Schiff base complexes. J. Mol. Struct. 2018, 1156, 1–11. [Google Scholar] [CrossRef]
- Demirci, S.; Doğan, A.; Türkmen, N.B.; Telci, D.; Rizvanov, A.A.; Şahin, F. Schiff base-Poloxamer P85 combination demonstrates chemotherapeutic effect on prostate cancer cells in vitro. Biomed. Pharmacother. 2017, 86, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Rauf, A.; Shah, A.; Khan, A.A.; Shah, A.H.; Abbasi, R.; Qureshi, I.Z.; Ali, S. Synthesis, pH dependent photometric and electrochemical investigation, redox mechanism and biological applications of novel Schiff base and its metallic derivatives. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017, 176, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-T.; Sheng, J.; Yin, D.-W.; Xin, H.; Yang, X.-M.; Qiao, Q.-Y.; Yang, Z.-J. Ferrocenyl chalcone-based Schiff bases and their metal complexes: Highly efficient, solvent-free synthesis, characterization, biological research. J. Organomet. Chem. 2018, 856, 27–33. [Google Scholar] [CrossRef]
- Raman, N.; Raja, J.D.; Sakthivel, A. Synthesis, spectral characterization of Schiff base transition metal complexes: DNA cleavage and antimicrobial activity studies. J. Chem. Sci. 2007, 119, 303–310. [Google Scholar] [CrossRef]
- Iftikhar, B.; Javed, K.; Khan, M.S.U.; Akhter, Z.; Mirza, B.; Mckee, V. Synthesis, characterization and biological assay of Salicylaldehyde Schiff base Cu(II) complexes and their precursors. J. Mol. Struct. 2018, 1155, 337–348. [Google Scholar] [CrossRef]
- Fekri, R.; Salehi, M.; Asadi, A.; Kubicki, M. Synthesis, characterization, anticancer and antibacterial evaluation of Schiff base ligands derived from hydrazone and their transition metal complexes. Inorg. Chim. Acta 2019, 484, 245–254. [Google Scholar] [CrossRef]
- Tamer, A.M.; Hassan, M.A.; Omer, M.A.; Baset, W.M.A.; Hassan, M.E.; El-Shafeey, M.E.A.; Mohy Elding, M.S. Synthesis, characterization and antimicrobial evaluation of two aromatic chitosan Schiff base derivatives. Process Biochem. 2016, 51, 1721–1730. [Google Scholar] [CrossRef]
- Bodke, Y.D.; Shankerrao, S.; Kenchappa, R.; Telkar, S. Synthesis, Antibacterial and Antitubercular Activity of Novel Schiff Bases of 2-(1-Benzofuran-2-yl)quinoline-4-carboxylic Acid Derivatives1. Russ. J. Gen. Chem. 2017, 87, 1843–1849. [Google Scholar] [CrossRef]
- Sinha, D.; Tiwari, A.K.; Singh, S.; Shukla, G.; Mishra, P.; Chandra, H.; Mishra, A.K. Synthesis, characterization and biological activity of Schiff base analogues of indole-3-carboxaldehyde. Eur. J. Med. Chem. 2008, 43, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Mandal, S.M.; Mondal, T.K.; Sinha, C. Spectroscopic characterization, antimicrobial activity, DFT computation and docking studies of sulfonamide Schiff bases. J. Mol. Struct. 2017, 1127, 557–567. [Google Scholar] [CrossRef]
- Hassan, A.S.; Hafez, T.S.; Ali, M.M.; Khatab, T.K. Design, synthesis and cytotoxic activity of some new pyrazolines bearing benzofuran and pyrazole moieties. Res. J. Pharm. Biol. Chem. Sci. 2016, 7, 417–429. [Google Scholar]
- Hassan, A.S.; Mady, M.F.; Awad, H.M.; Hafez, T.S. Synthesis and antitumor activity of some new pyrazolo [1,5-a]pyrimidines. Chin. Chem. Lett. 2017, 28, 388–393. [Google Scholar] [CrossRef]
- Abd El Razik, H.A.; Badr, M.H.; Attia, H.A.; Mouneir, S.M.; Abu-Serie, M.M. Benzodioxole-pyrazole hybrids as anti-inflammatory and analgesic agents with COX-1,2/5-LOX inhibition and antioxidant potential. Arch. Pharm. Chem. Life Sci. 2017, 350, e1700026. [Google Scholar] [CrossRef]
- Hassan, A.S.; Hafez, T.S.; Osman, S.A.M.; Ali, M.M. Synthesis and in vitro cytotoxic activity of novel pyrazolo[1,5-a]pyrimidines and related Schiff bases. Turk. J. Chem. 2015, 39, 1102–1113. [Google Scholar] [CrossRef]
- Rizk, H.F.; EI-Badawi, M.A.; Ibrahim, S.A.; EI-Borai, M.A. Cyclization of 4,5-diamino pyrazole derivatives and their antibacterial activities. Chin. J. Chem. 2011, 29, 1451–1459. [Google Scholar] [CrossRef]
- Sharshira, E.M.; Hamada, N.M.M. Synthesis and Antimicrobial Evaluation of Some Pyrazole Derivatives. Molecules 2012, 17, 4962–4971. [Google Scholar] [CrossRef]
- El-Naggar, M.; Hassan, A.S.; Awad, H.M.; Mady, M.F. Design, Synthesis and Antitumor Evaluation of Novel Pyrazolopyrimidines and Pyrazoloquinazolines. Molecules 2018, 23, 1249. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.S.; Hafez, T.S. Antimicrobial Activities of Ferrocenyl Complexes: A Review. J. App. Pharm. Sci. 2018, 8, 156–165. [Google Scholar]
- Hassan, A.S.; Masoud, D.M.; Sroor, F.M.; Askar, A.A. Synthesis and biological evaluation of pyrazolo[1,5-a]pyrimidine-3-carboxamide as antimicrobial agents. Med. Chem. Res. 2017, 26, 2909–2919. [Google Scholar] [CrossRef]
- Hassan, A.S.; Moustafa, G.O.; Awad, H.M. Synthesis and in vitro anticancer activity of pyrazolo[1,5-a]pyrimidines and pyrazolo[3,4-d][1,2,3]triazines. Synth. Commun. 2017, 47, 1963–1972. [Google Scholar] [CrossRef]
- Khatab, T.K.; Hassan, A.S.; Hafez, T.S. V2O5/SiO2 as an efficient catalyst in the synthesis of 5-aminopyrazole derivatives under solvent free condition. Bull. Chem. Soc. Ethiop. 2019, 33, 135–142. [Google Scholar] [CrossRef]
- Magd-El-Din, A.A.; Mousa, H.A.; Labib, A.A.; Hassan, A.S.; Abd El-All, A.S.; Ali, M.M.; El-Rashedy, A.A.; El-Desoky, A.H. Benzimidazole-Schiff bases and their complexes: Synthesis, anticancer activity and molecular modeling as Aurora kinase inhibitor. Z. Naturforsch. C 2018, 73, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.S.; Moustafa, G.O.; Askar, A.A.; Naglah, A.M.; Al-Omar, M.A. Synthesis and antibacterial evaluation of fused pyrazoles and Schiff bases. Synth. Commun. 2018, 48, 2761–2772. [Google Scholar] [CrossRef]
- Abd El-All, A.S.; Hassan, A.S.; Osman, S.A.; Yosef, H.A.A.; Abdel-Hady, W.H.; El-Hashash, M.A.; Atta-Allah, S.R.; Ali, M.M.; El Rashedy, A.A. Synthesis, characterization and biological evaluation of new fused triazine derivatives based on 6-methyl-3-thioxo-1,2,4-triazin-5-one. Acta Pol. Pharm. 2016, 73, 79–92. [Google Scholar] [PubMed]
- Hassan, A.S.; Osman, S.A.; Hafez, T.S. 5-Phenyl-2-furaldehyde: Synthesis, Reactions and Biological Activities. Egypt. J. Chem. 2015, 58, 113–139. [Google Scholar]
- Osman, S.A.; Mousa, H.A.; Yosef, H.A.A.; Hafez, T.S.; El-Sawy, A.A.; Abdallah, M.M.; Hassan, A.S. Synthesis, characterization and cytotoxicity of mixed ligand Mn(II), Co(II) and Ni(II) complexes. J. Serb. Chem. Soc. 2014, 79, 953–964. [Google Scholar] [CrossRef]
- Hafez, T.S.; Osman, S.A.; Yosef, H.A.A.; Abd El-All, A.S.; Hassan, A.S.; El-Sawy, A.A.; Abdallah, M.M.; Youns, M. Synthesis, structural elucidation and in vitro antitumor activities of some pyrazolopyrimidines and Schiff bases derived from 5-amino-3-(arylamino)-1H-pyrazole-4-carboxamides. Sci. Pharm. 2013, 81, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Osman, S.A.; Yosef, H.A.A.; Hafez, T.S.; El-Sawy, A.A.; Mousa, H.A.; Hassan, A.S. Synthesis and antibacterial activity of some novel chalcones, pyrazoline and 3-cyanopyridine derivatives based on khellinone as well as Ni(II), Co(II) and Zn(II) complexes. Aust. J. Basic Appl. Sci. 2012, 6, 852–863. [Google Scholar]
- Elgemeie, G.H.; Elsayed, S.H.; Hassan, A.S. Design and synthesis of the first thiophene thioglycosides. Synth. Commun. 2009, 39, 1781–1792. [Google Scholar] [CrossRef]
- Elgemeie, G.H.; Elsayed, S.H.; Hassan, A.S. Direct route to a new class of acrylamide thioglycosides and their conversions to pyrazole derivatives. Synth. Commun. 2008, 38, 2700–2706. [Google Scholar] [CrossRef]
- Moustafa, G.O.; Khalaf, H.; Naglah, A.; Al-Wasidi, A.; Al-Jafshar, N.; Awad, H. The Synthesis of Molecular Docking Studies, In Vitro Antimicrobial and Antifungal Activities of Novel Dipeptide Derivatives Based on N-(2-(2-Hydrazinyl-2-oxoethylamino)-2-oxoethyl)-nicotinamide. Molecules 2018, 23, 761. [Google Scholar] [CrossRef]
- Al-Salem, H.S.A.; Naglah, A.M.; Moustafa, G.O.; Mahmoud, A.Z.; Al-Omar, M.A. Synthesis of Novel Tripeptides Based on Dibenzofuran-2-Sulfonyl-[Aromatic and Hydroxy Aromatic Residues]: Towards Antimicrobial and Antifungal Agents. J. Comput. Theor. Nanosci. 2017, 14, 3958–3966. [Google Scholar] [CrossRef]
- Naglah, A.M.; Moustafa, G.O.; Al-Omar, M.A.; Al-Salem, H.S.A.; Hozzein, W.N. Synthesis, Characterization andInVitroAntimicrobial Investigation of Novel Amino Acids and Dipeptides Based on Dibenzofuran-2-Sulfonyl-Chloride. J. Comput. Theor. Nanosci. 2017, 14, 3183–3190. [Google Scholar] [CrossRef]
- Abo-Ghalia, M.H.; Moustafa, G.O.; Alwasidi, A.S.; Naglah, A.M. Cytotoxic Investigation of Isophthaloyl Cyclopentapeptides. Lat. Am. J. Pharm. 2017, 36, 1957–1962. [Google Scholar]
- Moustafa, G.O.; El-Sawy, A.A.; Abo-Ghalia, M.H. Synthesis of novel cyclopeptide candidates: I-cyclo-[Nα_-isophthaloyl-bis-(Glycine-amino acid)-l-lysine] derivatives with expected anticancer activity. Egypt. J. Chem. 2013, 56, 473–494. [Google Scholar]
- Cappuccino, J.G.; Sherman, N. Microbiology, Laboratory Manual; Pearson Education, Inc.: New Delhi city, India, 2004; pp. 282–283. [Google Scholar]
- Cooper, K. The Theory of Antibiotic Inhibition Zones. In Analytical Microbiology; Academic press: New York, NY, USA; London, UK, 1963; pp. 1–85. [Google Scholar]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Amr, A.E.; Abo-Ghalia, M.H.; Moustafa, G.O.; Al-Omar, M.A.; Nossier, E.S.; Elsayed, E.A. Design, synthesis and docking studies of novel macrocyclic pentapeptides as anticancer multi-targeted kinase inhibitors. Molecules 2018, 23, 2416. [Google Scholar] [CrossRef] [PubMed]
- Bax, B.D.; Chan, P.F.; Eggleston, D.S.; Fosberry, A.; Gentry, D.R.; Gorrec, F.; Giordano, I.; Hann, M.M.; Hennessy, A.; Hibbs, M.; et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 2010, 466, 935–940. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, M.; Sallam, H.A.; Shaban, S.S.; Abdel-Wahab, S.S.; Amr, A.E.; Azab, M.E.; Nossier, E.S.; Al-Omar, M.A. Design, Synthesis, and Molecular Docking Study of Novel Heterocycles Incorporating 1,3,4-Thiadiazole Moiety as Potential Antimicrobial and Anticancer Agents. Molecules 2019, 24, 1066. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.S.; Hafez, T.S.; Osman, S.A. Synthesis, characterization, and cytotoxicity of some new 5-aminopyrazole and pyrazolo[1,5-a]pyrimidine derivatives. Sci. Pharm. 2015, 83, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.S.; Awad, H.M.; Magd-El-Din, A.A.; Hafez, T.S. Synthesis and in vitro antitumor evaluation of novel Schiff bases. Med. Chem. Res. 2018, 27, 915–927. [Google Scholar] [CrossRef]
- Othman, I.M.M.; Gad-Elkareem, M.A.M.; El-Naggar, M.; Nossier, E.S.; Amr, A.E. Novel phthalimide based analogues: Design, synthesis, biological evaluation, and molecular docking studies. J. Enzyme Inhib. Med. Chem. 2019, 34, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
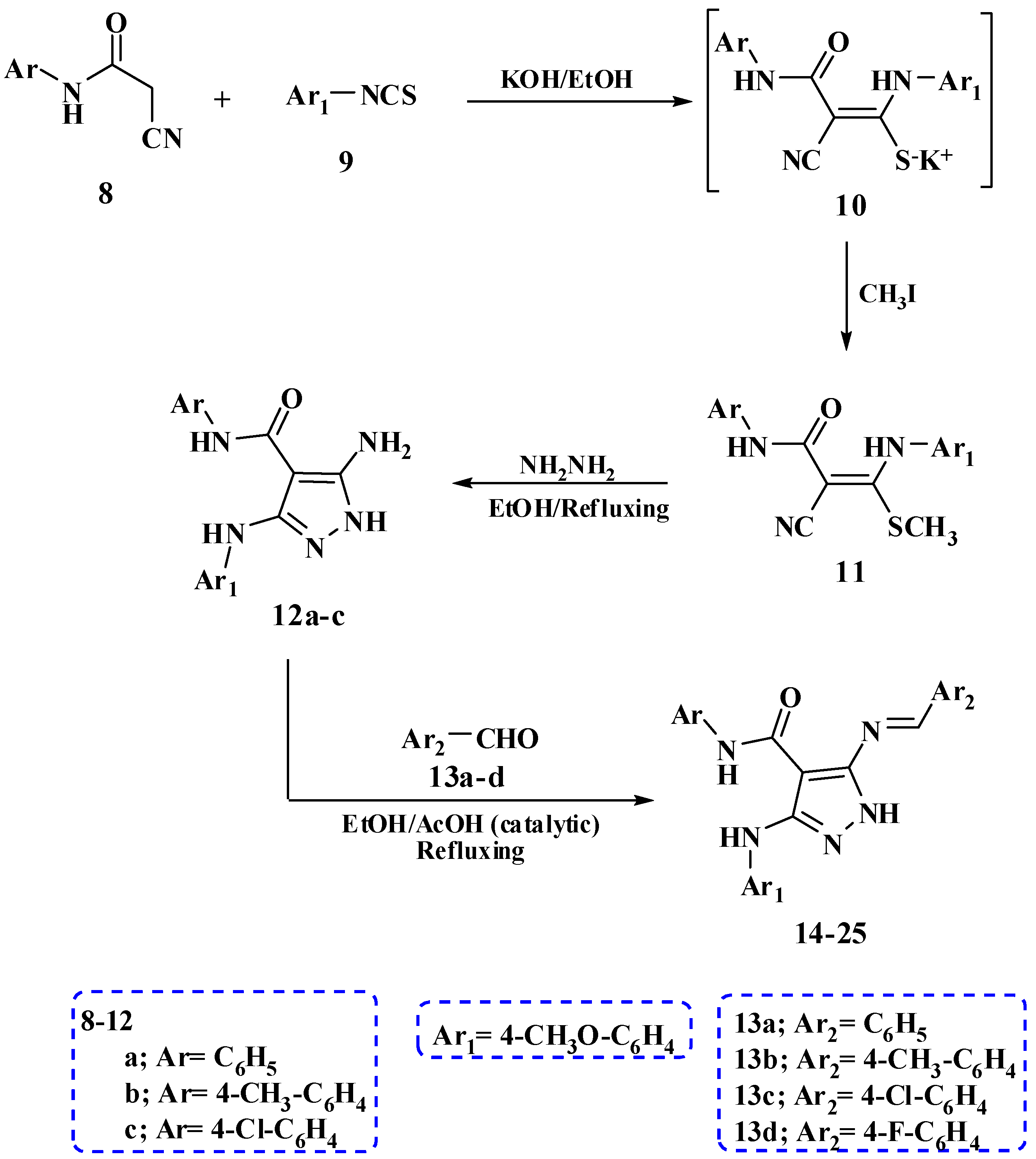

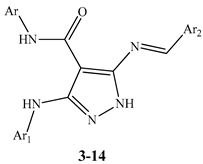
| Compounds | Ar | Ar1 | Ar2 |
|---|---|---|---|
| 14 | Ph | 4-MeOC6H4- | Ph |
| 15 | Ph | 4-MeOC6H4- | 4-MeC6H4- |
| 16 | Ph | 4-MeOC6H4- | 4-ClC6H4- |
| 17 | Ph | 4-MeOC6H4- | 4-FC6H4- |
| 18 | 4-MeC6H4- | 4-MeOC6H4- | Ph |
| 19 | 4-MeC6H4- | 4-MeOC6H4- | 4-MeC6H4- |
| 20 | 4-MeC6H4- | 4-MeOC6H4- | 4-ClC6H4- |
| 21 | 4-MeC6H4- | 4-MeOC6H4- | 4-FC6H4- |
| 22 | 4-ClC6H4- | 4-MeOC6H4- | Ph |
| 23 | 4-ClC6H4- | 4-MeOC6H4- | 4-MeC6H4- |
| 24 | 4-ClC6H4- | 4-MeOC6H4- | 4-ClC6H4- |
| 25 | 4-ClC6H4- | 4-MeOC6H4- | 4-FC6H4- |
| Comp. | Ar | Ar1 | Ar2 | Gram-Positive Bacteria | Gram-Negative Bacteria | ||||
|---|---|---|---|---|---|---|---|---|---|
| Sa | Se | Ef | Ab | Ecl | Ec | ||||
| 14 | Ph | 4-MeOC6H4- | Ph | 31.25 | 7.81 * | 15.62 | NA | 125 | NA |
| 15 | Ph | 4-MeOC6H4- | 4-MeC6H4- | 62.50 | 15.62 | 31.25 | 62.5 | 62.5 | 125 |
| 16 | Ph | 4-MeOC6H4- | 4-ClC6H4- | 31.25 | 7.81 * | 62.5 | 15.62 * | 62.5 | 125 |
| 17 | Ph | 4-MeOC6H4- | 4-FC6H4- | 62.5 | 31.25 | NA | 62.5 | 125 | 31.25 * |
| 18 | 4-MeC6H4- | 4-MeOC6H4- | Ph | 15.62 * | 7.81 * | 31.25 | 15.62 * | 62.5 | 250 |
| 19 | 4-MeC6H4- | 4-MeOC6H4- | 4-MeC6H4- | 31.25 | 15.62 | 15.62 | 62.5 | 125 | 62.5 |
| 20 | 4-MeC6H4- | 4-MeOC6H4- | 4-ClC6H4- | 62.5 | 125 | 250 | NA | NA | NA |
| 21 | 4-MeC6H4- | 4-MeOC6H4- | 4-FC6H4- | 125 | 62.5 | NA | NA | NA | NA |
| 22 | 4-ClC6H4- | 4-MeOC6H4- | Ph | 250 | 125 | 125 | NA | 15.62 * | NA |
| 23 | 4-ClC6H4- | 4-MeOC6H4- | 4-MeC6H4- | 31.25 | 15.62 | 7.81 * | NA | 62.5 | NA |
| 24 | 4-ClC6H4- | 4-MeOC6H4- | 4-ClC6H4- | 125 | 62.5 | 15.62 | NA | 62.5 | 125 |
| 25 | 4-ClC6H4- | 4-MeOC6H4- | 4-FC6H4- | NA | NA | NA | NA | NA | NA |
| Ciprofloxacin | 7.81 * | 15.62 * | 7.81 * | 15.62 * | 15.62 * | 7.81 * | |||
| Comp. | MW a | MLogP b | nHBA c | nHBD d | nRB e | TPSA f | nviolations g |
|---|---|---|---|---|---|---|---|
| Rule | <500 | ≤4.15 | ≤10 | ≤5 | ≤10 | <160 Å2 | 0 |
| 14 | 411.46 | 3.65 | 4 | 3 | 8 | 91.40 | 0 |
| 15 | 425.48 | 3.86 | 4 | 3 | 8 | 91.40 | 0 |
| 16 | 445.90 | 4.13 | 4 | 3 | 8 | 91.40 | 0 |
| 17 | 429.45 | 4.02 | 5 | 3 | 8 | 91.40 | 0 |
| 18 | 425.48 | 3.86 | 4 | 3 | 8 | 91.40 | 0 |
| 19 | 439.51 | 4.06 | 4 | 3 | 8 | 91.40 | 0 |
| 20 | 459.93 | 4.33 | 4 | 3 | 8 | 91.40 | 1 |
| 21 | 443.47 | 4.23 | 5 | 3 | 8 | 91.40 | 1 |
| 22 | 445.90 | 4.13 | 4 | 3 | 8 | 91.40 | 0 |
| 23 | 459.93 | 4.33 | 4 | 3 | 8 | 91.40 | 1 |
| 24 | 480.35 | 4.60 | 4 | 3 | 8 | 91.40 | 1 |
| 25 | 463.89 | 4.50 | 5 | 3 | 8 | 91.40 | 1 |
| Compound | IC50 (Mean ± SEM) (µM) | ||
|---|---|---|---|
| DNA Gyrase | Topoisomerase IV | DHFR | |
| 18 | 1.68 ± 0.10 | 74.55 ± 1.20 | 0.08 ± 1.15 |
| Ciprofloxacin | 1.51 ± 0.18 | 24.14 ± 1.01 | ----- |
| Methotrexate | ----- | ----- | 0.14 ± 1.07 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, A.S.; Askar, A.A.; Nossier, E.S.; Naglah, A.M.; Moustafa, G.O.; Al-Omar, M.A. Antibacterial Evaluation, In Silico Characters and Molecular Docking of Schiff Bases Derived from 5-aminopyrazoles. Molecules 2019, 24, 3130. https://doi.org/10.3390/molecules24173130
Hassan AS, Askar AA, Nossier ES, Naglah AM, Moustafa GO, Al-Omar MA. Antibacterial Evaluation, In Silico Characters and Molecular Docking of Schiff Bases Derived from 5-aminopyrazoles. Molecules. 2019; 24(17):3130. https://doi.org/10.3390/molecules24173130
Chicago/Turabian StyleHassan, Ashraf S., Ahmed A. Askar, Eman S. Nossier, Ahmed M. Naglah, Gaber O. Moustafa, and Mohamed A. Al-Omar. 2019. "Antibacterial Evaluation, In Silico Characters and Molecular Docking of Schiff Bases Derived from 5-aminopyrazoles" Molecules 24, no. 17: 3130. https://doi.org/10.3390/molecules24173130
APA StyleHassan, A. S., Askar, A. A., Nossier, E. S., Naglah, A. M., Moustafa, G. O., & Al-Omar, M. A. (2019). Antibacterial Evaluation, In Silico Characters and Molecular Docking of Schiff Bases Derived from 5-aminopyrazoles. Molecules, 24(17), 3130. https://doi.org/10.3390/molecules24173130





