Interspecies Variation in NCMN-O-Demethylation in Liver Microsomes from Various Species
Abstract
1. Introduction
2. Results
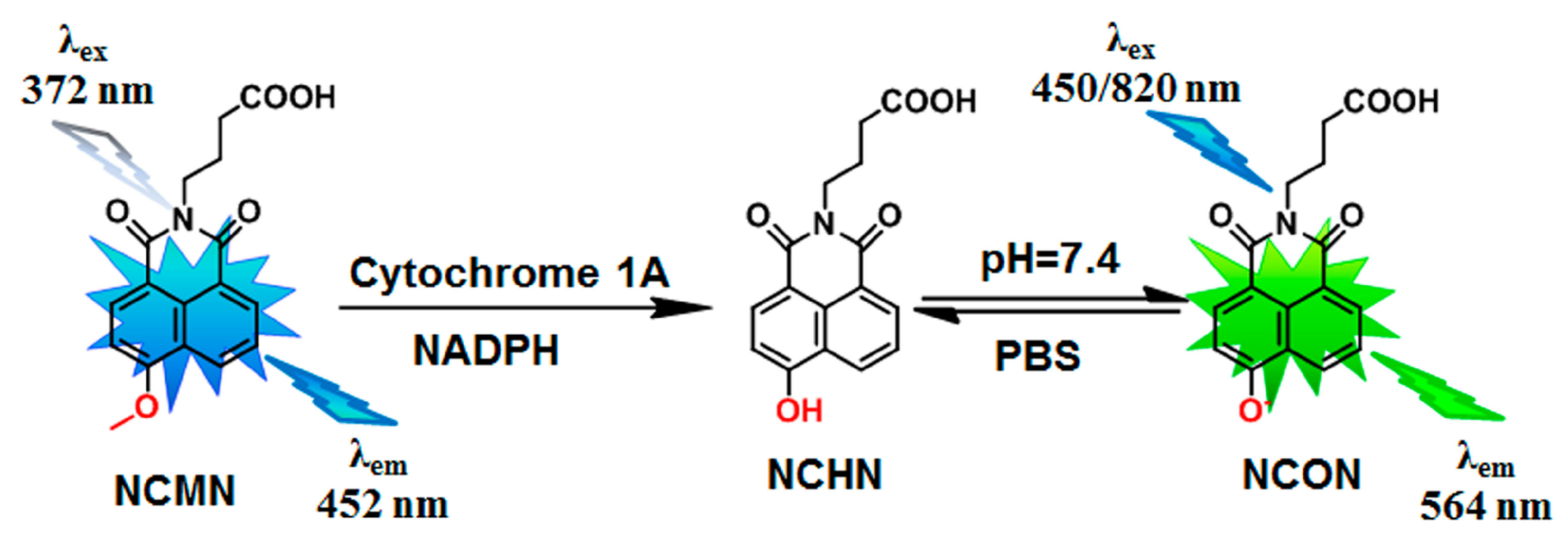
2.1. NCMN-O-Dealkylation in Liver Microsomes from Various Species
2.2. Inhibitory Effects of CYP450 Inhibitors on NCMN-O-Dealkylation
2.3. Inhibition of Furafylline on NCMN-O-Dealkylation in Liver Microsomes from Various Species
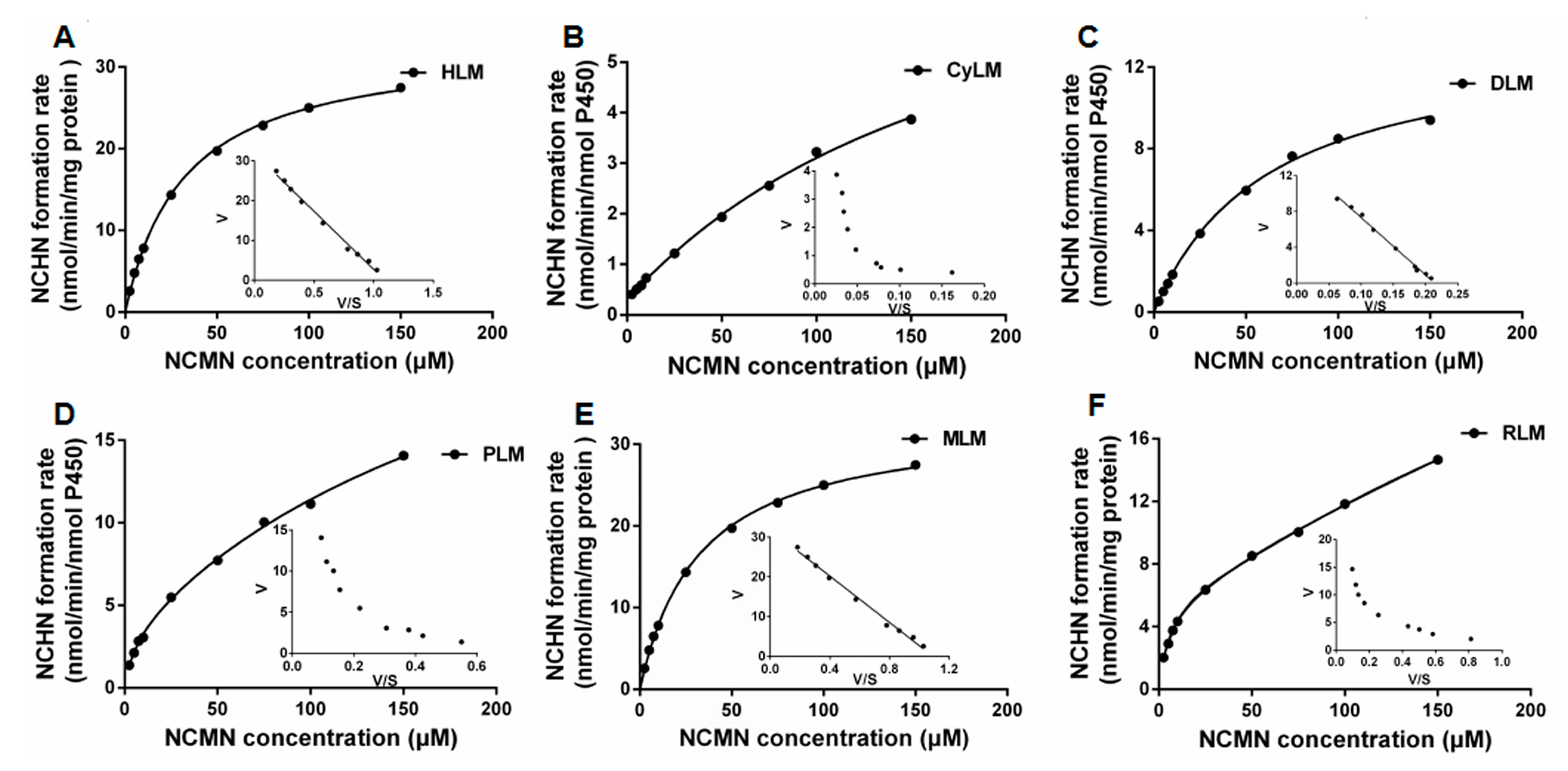
2.4. Kinetic Analyses of NCMN-O-Demethylation in Liver Microsomes from Various Species
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Incubation Conditions
4.3. Kinetic Analyses
4.4. Chemical Inhibition Assays
4.5. Fluorescence Detection and LC-ESI-MS Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Cytochrome P450: What have we learned and what are the future issues? Drug Metab. Rev. 2004, 36, 159–197. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Guengerich, F.P. Selection of human cytochrome P450 1A2 mutants with enhanced catalytic activity for heterocyclic amine N-hydroxylation. Biochemistry 2004, 43, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Guengerich, F.P. Cytochrome P450 activation of arylamines and heterocyclic amines. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 27–49. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Lu, A.Y. CYP1A induction and human risk assessment: An evolving tale of in vitro and in vivo studies. Drug Metab. Dispos. 2007, 35, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Michaud, V.; Frappier, M.; Dumas, M.C.; Turgeon, J. Metabolic activity and mRNA levels of human cardiac CYP450s involved in drug metabolism. PLoS ONE 2010, 5, e15666. [Google Scholar] [CrossRef]
- Ohtsuki, S.; Schaefer, O.; Kawakami, H.; Inoue, T.; Liehner, S.; Saito, A.; Ishiguro, N.; Kishimoto, W.; Ludwig-Schwellinger, E.; Ebner, T.; et al. Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: Comparison with mRNA levels and activities. Drug Metab. Dispos. 2012, 40, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, H.; Ohtsuki, S.; Kamiie, J.; Suzuki, T.; Abe, T.; Terasaki, T. Simultaneous absolute quantification of 11 cytochrome P450 isoforms in human liver microsomes by liquid chromatography tandem mass spectrometry with in silico target peptide selection. J. Pharm. Sci. 2011, 100, 341–352. [Google Scholar] [CrossRef]
- Proctor, R.N. Tobacco and the global lung cancer epidemic. Nat. Rev. Cancer 2001, 1, 82–86. [Google Scholar] [CrossRef]
- Dobrinas, M.; Cornuz, J.; Oneda, B.; Kohler, S.M.; Puhl, M.; Eap, C.B. Impact of smoking, smoking cessation, and genetic polymorphisms on CYP1A2 activity and inducibility. Clin. Pharmacaol. Harmacol. Ther. 2011, 90, 117–125. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Z.; Han, S.; Lu, Y.; Feng, F.; Yuan, J. CYP1A2 rs762551 polymorphism contributes to cancer susceptibility: A meta-analysis from 19 case-control studies. BMC Cancer 2012, 12, 528. [Google Scholar] [CrossRef] [PubMed]
- Sergentanis, T.N.; Economopoulos, K.P. Four polymorphisms in cytochrome P450 1A1 (CYP1A1) gene and breast cancer risk: A meta-analysis. Breast Cancer Res. Treat. 2010, 122, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Berthou, F.; Guillois, B.; Riche, C.; Dreano, Y.; Jacqz-Aigrain, E.; Beaune, P.H. Interspecies variations in caffeine metabolism related to cytochrome P4501A enzymes. Xenobiotica 1992, 22, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Martignoni, M.; Groothuis, G.M.; de Kanter, R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug. Metab. Toxicol. 2006, 2, 875–894. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Feng, L.; Jin, Q.; Cheng, H.; Li, Y.; Ning, J.; Yu, Y.; Ge, G.; Cui, J.; Yang, L. A practical strategy to design and develop an isoform-specific fluorescent probe for a target enzyme: CYP1A1 as a case study. Chem. Sci. 2017, 8, 2795–2803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fan, J.; Wang, J.; Zhang, S.; Dou, B.; Peng, X. An off-on COX-2-specific fluorescent probe: Targeting the Golgi apparatus of cancer cells. J. Am. Chem. Soc. 2013, 135, 11663–11669. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Feng, L.; Wang, D.D.; Dai, Z.R.; Wang, P.; Zou, L.W.; Liu, Z.H.; Wang, J.Y.; Yu, Y.; Ge, G.B.; et al. A Two-Photon Ratiometric Fluorescent Probe for Imaging Carboxylesterase 2 in Living Cells and Tissues. ACS Appl. Mater. Interfaces 2015, 7, 28474–28481. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Yamazaki, H.; Mimura, M.; Wakamiya, N.; Ueng, Y.F.; Guengerich, F.P.; Inui, Y. Characterization of microsomal cytochrome P450 enzymes involved in the oxidation of xenobiotic chemicals in human fetal liver and adult lungs. Drug Metab. Dispos. 1996, 24, 515–522. [Google Scholar]
- Smith, G.B.; Harper, P.A.; Wong, J.M.; Lam, M.S.; Reid, K.R.; Petsikas, D.; Massey, T.E. Human lung microsomal cytochrome P4501A1 (CYP1A1) activities: Impact of smoking status and CYP1A1, aryl hydrocarbon receptor, and glutathione S-transferase M1 genetic polymorphisms. Cancer Epidemiol. Prev. Biomark. 2001, 10, 839–853. [Google Scholar]
- Sípal, Z.; Ahlenius, T.; Bergstrand, A.; Rodriquez, L.; Jakobsson, S.W. Oxidative biotransformation of benzo(a)pyrene by human lung microsomal fractions prepared from surgical specimens. Xenobiotica 1979, 9, 633–645. [Google Scholar]
- Devereux, T.R.; Massey, T.E.; Van Scott, M.R.; Yankaskas, J.; Fouts, J.R. Xenobiotic metabolism in human alveolar type II cells isolated by centrifugal elutriation and density gradient centrifugation. Cancer Res. 1986, 46, 5438–5443. [Google Scholar] [PubMed]
- Dai, Z.; Ge, G.; Feng, L.; Ning, J.; Hu, L.; Jin, Q.; Wang, D.; Lv, X.; Dou, T.; Cui, J.; et al. A Highly Selective Ratiometric Two-Photon Fluorescent Probe for Human Cytochrome P450 1A. J. Am. Chem. Soc. 2015, 137, 14488–14495. [Google Scholar] [CrossRef] [PubMed]
- Fuhr, U.; Jetter, A.; Kirchheiner, J. Appropriate phenotyping procedures for drug metabolizing enzymes and transporters in humans and their simultaneous use in the "cocktail" approach. Clin. Pharmacol. Ther. 2007, 81, 270–283. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F.; Wang, B.; Yang, L.P.; Liu, J.P. Structure, function, regulation and polymorphism and the clinical significance of human cytochrome P450 1A2. Drug Metab. Rev. 2010, 42, 268–354. [Google Scholar] [CrossRef] [PubMed]
- Wogan, G.N.; Hecht, S.S.; Felton, J.S.; Conney, A.H.; Loeb, L.A. Environmental and chemical carcinogenesis. Semin. Cancer Biol. 2004, 14, 473–486. [Google Scholar] [CrossRef]
- Shimada, T.; Fujii-Kuriyama, Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004, 95, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, M.C.; El-Sohemy, A.; Kabagambe, E.K.; Campos, H. Coffee, CYP1A2 genotype, and risk of myocardial infarction. JAMA 2006, 295, 1135–1141. [Google Scholar] [CrossRef]
- Androutsopoulos, V.P.; Tsatsakis, A.M.; Spandidos, D.A. Cytochrome P450 CYP1A1: Wider roles in cancer progression and prevention. BMC Cancer 2009, 9, 187. [Google Scholar] [CrossRef]
- Sharer, J.E.; Shipley, L.A.; Vandenbranden, M.R.; Binkley, S.N.; Wrighton, S.A. Comparisons of Phase I and Phase II in vitro hepatic enzyme activities of human, dog, rhesus monkey, and cynomolgus monkey. Drug Metab. Dispos. 1995, 23, 1231–1241. [Google Scholar]
- Santostefano, M.J.; Ross, D.G.; Savas, U.; Jefcoate, C.R.; Birnbaum, L.S. Differential time-course and dose-response relationships of TCDD-induced CYP1B1, CYP1A1, and CYP1A2 proteins in rats. Biochem. Biophys. Res. Commun. 1997, 233, 20–24. [Google Scholar] [CrossRef]
- Choudhary, D.; Jansson, I.; Schenkman, J.B.; Sarfarazi, M.; Stoilov, I. Comparative expression profiling of 40 mouse cytochrome P450 genes in embryonic and adult tissues. Arch. Biochem. Biophys. 2003, 414, 91–100. [Google Scholar] [CrossRef]
- Graham, R.A.; Downey, A.; Mudra, D.; Krueger, L.; Carroll, K.; Chengelis, C.; Madan, A.; Parkinson, A. In Vivo and in vitro induction of cytochrome P450 enzymes in beagle dogs. Drug Metab. Dispos. 2002, 30, 1206–1213. [Google Scholar] [CrossRef] [PubMed]
- Zuber, R.; Anzenbacherová, E.; Anzenbacher, P. Cytochromes P450 and experimental models of drug metabolism. J. Cell. Mol. Med. 2002, 6, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, B.S.; Suvaithenamudhan, S.; Akbarsha, M.A.; Parthasarathy, S. Analysis of species-selectivity of human, mouse and rat Cytochrome P450 1A and 2B subfamily enzymes using molecular modeling, docking and dynamics simulations. Cell. Biochem. Biophys. 2018, 76, 91–110. [Google Scholar] [CrossRef] [PubMed]
- Rendic, S.; Guengerich, F.P. Contributions of Human Enzymes in Carcinogen Metabolism. Chem. Res. Toxicol. 2012, 25, 1316–1383. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, J.; Liu, J.; Foroozesh, M.; Klein Stevens, C.L. Inhibition of Cytochrome P450 Enzymes by Quinones and Anthraquinones. Chem. Res. Toxicol. 2012, 25, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Taylor, S.F.; Dupart, P.S.; Arnold, C.L.; Sridhar, J.; Jiang, Q.; Wang, Y.; Skripnikova, E.V.; Zhao, M.; Foroozesh, M. Pyranoflavones: A group of small-molecule probes for exploring the active site cavities of cytochrome P450 enzymes 1A1, 1A2, and 1B1. J. Med. Chem. 2013, 56, 4082–4092. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sridhar, J.; Foroozesh, M. Cytochrome P450 family 1 inhibitors and structure-activity relationships. Molecules 2013, 18, 14470–14495. [Google Scholar] [CrossRef]
- Zhou, Q.; Yan, X.F.; Zhang, Z.M.; Pan, W.S.; Zeng, S. Rational prescription of drugs within similar therapeutic or structural class for gastrointestinal disease treatment: Drug metabolism and its related interactions. World J. Gastroenterol. 2007, 13, 5618–5628. [Google Scholar] [CrossRef]
- Toon, S.; Hopkins, K.J.; Garstang, F.M.; Aarons, L.; Sedman, A.; Rowland, M. Enoxacin-warfarin interaction: Pharmacokinetic and stereochemical aspects. Clin. Pharmacol. Ther. 1987, 42, 33–41. [Google Scholar] [CrossRef]
- Taura, K.; Naito, E.; Ishii, Y.; Mori, M.; Oguri, K.; Yamada, H.; Medicine, S.O.; Sciences, A.S.O.P.; Sciences, C.O.P.; University, K.; et al. Cytochrome P450 1A1 (CYP1A1) inhibitor alpha-naphthoflavone interferes with UDP-glucuronosyltransferase (UGT) activity in intact but not in permeabilized hepatic microsomes from 3-methylcholanthrene-treated rats: Possible involvement of UGT-P450 interactions. Biol. Pharm. Bull. 2004, 27, 56–60. [Google Scholar] [PubMed]
- Bogaards, J.J.P.; Bertrand, M.; Jackson, P.; Oudshoorn, M.J.; Weaver, R.J.; Van Bladeren, P.J.; Walther, B. Determining the best animal model for human cytochrome P450 activities: A comparison of mouse, rat, rabbit, dog, micropig, monkey and man. Xenobiotica 2000, 30, 1131–1152. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.A.; Szklarz., G.D.; Scott, E.E. Human Cytochrome P450 1A1 Structure and Utility in Understanding Drug. J. Biol. Chem. 2013, 288, 12932–12943. [Google Scholar] [CrossRef] [PubMed]
- Balani, S.K.; Zhu, T.; Yang, T.J.; Liu, Z.; He, B.; Lee, F.W. Effective dosing regimen of 1-aminobenzotriazole for inhibition of antipyrine clearance in rats, dogs, and monkeys. Drug Metab. Dispos. 2002, 30, 1059–1062. [Google Scholar] [CrossRef] [PubMed]
- Bjornsson, T.D.; Callaghan, J.T.; Einolf, H.J.; Fischer, V.; Gan, L.; Grimm, S.; Kao, J.; King, S.P.; Miwa, G.; Ni, L.; et al. The conduct of in vitro and in vivo drug-drug interaction studies: A PhRMA perspective. J. Clin. Pharmacol. 2003, 43, 443–469. [Google Scholar] [CrossRef] [PubMed]
- Tassaneeyakul, W.; Birkett, D.J.; Veronese, M.E.; McManus, M.E.; Tukey, R.H.; Miners, J.O. Direct characterization of the selectivity of furafylline as an inhibitor of human cytochromes P450 1A1 and 1A2. Pharmacogenetics 1994, 4, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, S.J.; Bloomer, J.C.; Smith, G.J.; Ayrton, A.D.; Clarke, S.E.; Chenery, R.J. Ketoconazole and sulphaphenazole as the respective selective inhibitors of P4503A and 2C9. Xenobiotica 1995, 25, 261–270. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds (NCMN and NCHN) are available from the authors. |



| Enzyme Source | IC50 (µM) |
|---|---|
| HLM | 0.31 |
| CyLM | 56.36 |
| DLM | 26.77 |
| PLM | 14.61 |
| MLM | 5.73 |
| RLM | 1.98 |
| Enzyme source | Vm1 | Vm2 | Km1 | Km2 | Vm/Km |
|---|---|---|---|---|---|
| HLM | 25.61 ± 0.15 | 9.54 ± 0.24 | 2684.48 | ||
| CyLM | 8.43 ± 0.91 | 0.28 ± 0.08 | 198.70 ± 39.66 | 0.18 ± 0.09 | 1559.45 1 |
| DLM | 13.53 ± 0.34 | 61.79 ± 3.52 | 218.96 | ||
| PLM | 28.22 ± 5.82 | 3.35 ± 1.20 | 242.80 ± 111.80 | 5.70 ± 3.27 | 587.44 1 |
| MLM | 33.17 ± 0.42 | 22.80 ± 1.22 | 1454.82 | ||
| RLM | 78.42 ± 31.17 | 5.71 ± 0.45 | 1134.00 ± 482.10 | 5.66 ± 0.80 | 1007.94 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Z.; Sun, G.; Yang, J.; Hou, J.; Zhou, P.; Xie, W.; Ge, G.; Sun, X.; Yang, L. Interspecies Variation in NCMN-O-Demethylation in Liver Microsomes from Various Species. Molecules 2019, 24, 2765. https://doi.org/10.3390/molecules24152765
Dai Z, Sun G, Yang J, Hou J, Zhou P, Xie W, Ge G, Sun X, Yang L. Interspecies Variation in NCMN-O-Demethylation in Liver Microsomes from Various Species. Molecules. 2019; 24(15):2765. https://doi.org/10.3390/molecules24152765
Chicago/Turabian StyleDai, Ziru, Guibo Sun, Jiada Yang, Jie Hou, Ping Zhou, Weijie Xie, Guangbo Ge, Xiaobo Sun, and Ling Yang. 2019. "Interspecies Variation in NCMN-O-Demethylation in Liver Microsomes from Various Species" Molecules 24, no. 15: 2765. https://doi.org/10.3390/molecules24152765
APA StyleDai, Z., Sun, G., Yang, J., Hou, J., Zhou, P., Xie, W., Ge, G., Sun, X., & Yang, L. (2019). Interspecies Variation in NCMN-O-Demethylation in Liver Microsomes from Various Species. Molecules, 24(15), 2765. https://doi.org/10.3390/molecules24152765






